Abstract
The dramatic changes in the epidemiology of Clostridium difficile infection (CDI) during recent years, with increases in incidence and severity of disease in several countries, have made CDI a global public health challenge. Increases in CDI incidence have been largely attributed to the emergence of a previously rare and more virulent strain, BI/NAP1/027. Increased toxin production and high-level resistance to fluoroquinolones have made this strain a very successful pathogen in healthcare settings. In addition, populations previously thought to be at low risk are now being identified as having severe CDI. Recent genetic analysis suggests that C. difficile has a highly fluid genome with multiple mechanisms to modify its content and functionality, which can make C. difficile adaptable to environmental changes and potentially lead to the emergence of more virulent strains. In the face of these changes in the epidemiology and microbiology of CDI, surveillance systems are necessary to monitor trends and inform public health actions.
Clostridium difficile, a spore-forming gram-positive anaerobic bacillus, was initially detected in the fecal flora of healthy newborns in 1935 [1]. Clostridium difficile was thought to be nonpathogenic until 1978, when Bartlett et al [2] identified C. difficile as the source of cytotoxin in the stools of patients with pseudomembranous colitis, a disorder frequently associated with antimicrobial use [3]. Understanding of the pathogenesis, epidemiology, diagnosis, and clinical management of C. difficile infection (CDI) has grown tremendously since 1978, particularly in the last several years. During the past decade, the epidemiology of C. difficile has changed rapidly and new methods for CDI diagnostic testing have emerged [4–6]. In this article, we review the recent changes in C. difficile epidemiology, discuss new diagnostic approaches and their potential impact on CDI rates, and describe the 2 existent surveillance systems for monitoring CDI in the United States.
PATHOGENESIS
Clostridium difficile is acquired through ingestion of spores usually transmitted from other patients through the hands of healthcare personnel or the environment [7, 8]. The spores resist the acidity of the stomach and germinate into the vegetative form in the small intestine. Disruption of normal gut flora, typically by exposure to antimicrobials, allows C. difficile to proliferate, causing a broad spectrum of clinical manifestations that can range from asymptomatic carriage to diarrhea of varying severity to fulminant colitis and even death [9].
The virulence of C. difficile is conferred primarily by 2 large exotoxins, toxins A and B, encoded by their genes, tcdA and tcdB, which are located, along with surrounding regulatory genes, on a 21-kilobase section of chromosomal DNA known as the pathogenicity locus (paloc). Toxin-negative C. difficile strains are considered nonpathogenic. In addition to toxins A and B, some strains also produce a third toxin known as binary toxin, encoded by ctdA and ctdB, located outside the paloc. The role of binary toxin in the pathogenesis of C. difficile remains unclear; however, the presence of this toxin among BI/NAP1/027 epidemic strains has raised concerns about its synergism with toxins A and B in causing severe colitis [10].
In most instances, exposure to both antimicrobials and toxin-producing C. difficile strains is necessary for the development of CDI. However, host factors also appear to play an important role in CDI development because some patients with both exposures do not become symptomatic [11, 12]. Although colonization of healthy nonhospitalized adults is uncommon (ie, rate <5%), colonization among hospitalized patients and especially nursing home residents may range from 25% to 55% [13, 14]. In contrast to the pathogenesis of many other healthcare-associated infections, asymptomatic carriage of C. difficile confers a decreased risk for subsequent CDI, which may be a result of natural immunity to CDI or colonization by harmless nontoxigenic strains [15].
EPIDEMIOLOGY OF CDI
Disease Burden
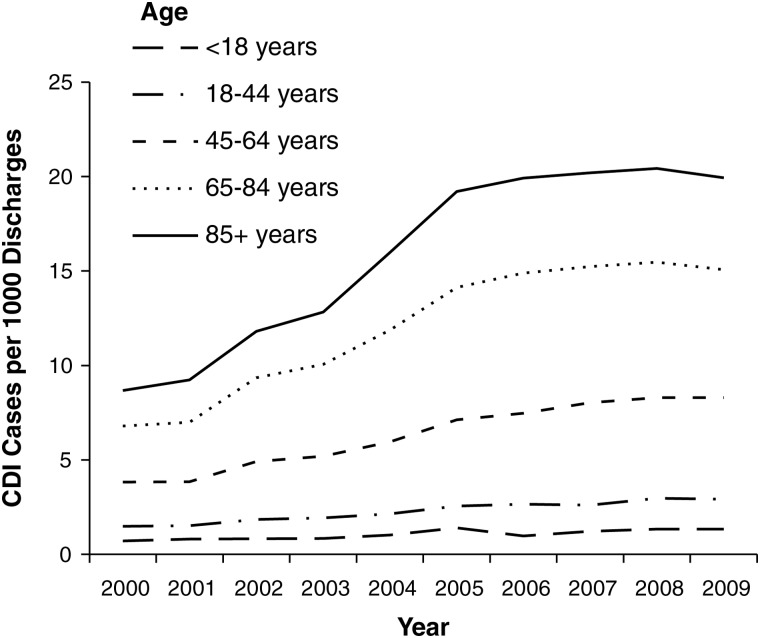
There has been a marked increase in CDI incidence and mortality across the United States, Canada, and Europe during the last decade [16–19]. Recent data from 28 community hospitals in the southern United States suggest that C. difficile has replaced methicillin-resistant Staphylococcus aureus as the most common cause of healthcare-associated infection [20]. US rates of hospital discharges with CDI listed as any diagnosis increased from 3.82 per 1000 discharges in 2000 to 8.75 per 1000 discharges in 2008; increases were especially prominent among those ≥65 years of age [21, 22] (Figure 1). However, in 2009 it appears that increases in the rate of CDI hospital discharges may be leveling off, with a 2.5% decrease in the point estimate from 8.75 per 1000 discharges in 2008 to 8.53 per 1000 discharges in 2009 [21, 22].
Figure 1.
Discharge rate for Clostridium difficile infection from US short-stay hospitals by age [22]. Abbreviation: CDI, Clostridium difficile infection.
Although rates of hospital discharges correlate with hospital laboratory–diagnosed CDI, they do not include CDI cases diagnosed and managed in the outpatient setting without the need for hospitalization. Based on mandatory reporting that occurred in Ohio during 2006, more than half the total burden of healthcare-associated CDI cases may have their onset in long-term care facilities, primarily nursing homes [23]. In the report by Campbell et al [23], the burden in Ohio was extrapolated to the entire US population, suggesting that 333 000 initial and 145 000 recurrent healthcare facility–onset CDI cases occur annually nationwide. Meanwhile, CDI is increasingly recognized in the community, where both outpatient antimicrobial exposure and proton pump inhibitor use have been associated with increased risk for disease [24, 25]. Data from the United States, Canada, and Europe suggest that approximately 20%–27% of all CDI cases are community associated, with an incidence of 20–30 per 100 000 population [24–26].
Preliminary data from US vital records indicate that the number of death certificates with enterocolitis due to C. difficile listed as the primary cause of death increased from 793 in 1999 to 7483 in 2008. The age-adjusted death rate for C. difficile increased from 2.0 deaths per 100 000 population in 2007 to 2.3 deaths per 100 000 population in 2008, representing a 15% increase. In 2008, a total of 93% of deaths from C. difficile occurred in persons ≥65 years of age, and C. difficile was reported as the 18th leading cause of death in this age group [27].
CDI in Special Populations: Children and Peripartum Women
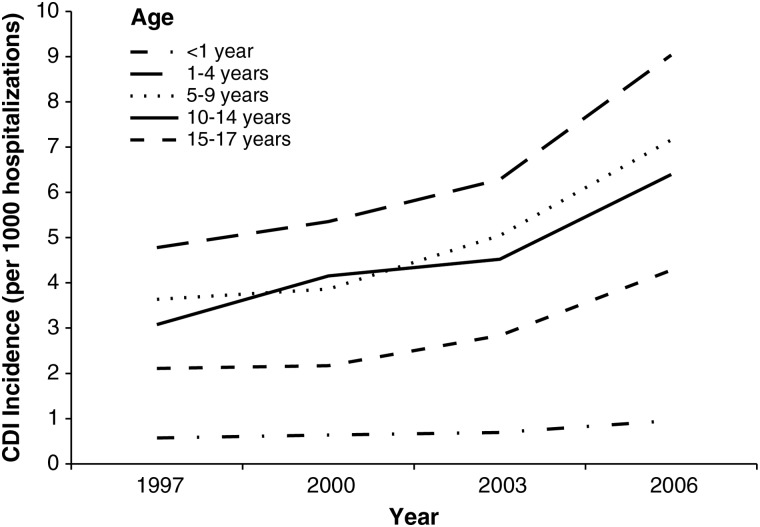
Rates of pediatric CDI-related hospitalizations increased substantially between 1997 and 2006, from 0.724 to 1.28 per 1000 hospitalizations. The highest incidence was reported in children 1–4 years of age, and the lowest in infants <1 year of age (Figure 2). However, when newborns were excluded from the analysis, the rate of CDI-related hospitalization in infants <1 year of age who were not newborn was the third highest [28]. Given the high rate (∼70%) of C. difficile colonization in infants <1 year of age [29], it is difficult to determine whether CDI-related hospitalizations in this age group represent true infection or colonization. Current recommendations are not to routinely test infants <1 year of age for C. difficile. However, studies are urgently needed to determine how often C. difficile in a nonnewborn population <1 year of age causes true disease. Although the epidemiology of C. difficile in the pediatric population remains poorly understood, recent studies suggest that children ≥1 year of age with comorbid conditions, such as cancer, solid organ transplant, gastrostomy, or jejunostomy, are at increased risk for CDI [30, 31].
Figure 2.
Incidence of Clostridium difficile infection per 1000 hospitalizations by age (Healthcare Utilization Project Kids’ and Inpatient Database, United States, 1997–2006). Modified from Zilberberg et al [28]. Abbreviation: CDI, Clostridium difficile infection.
In 2005, severe cases of CDI in peripartum women were reported across 4 US states [32], calling attention to the risk for this disease in this patient population. More recently, analysis of data from the Nationwide Inpatient Sample revealed that the rate of US hospital discharges in peripartum women for whom CDI was listed as any diagnosis was stable until 2004, when it increased from 0.04 to 0.07 per 1000 discharges in 2006. Of the 1706 peripartum women who received a diagnosis of CDI, 67% underwent cesarean delivery, which is probably a surrogate for antimicrobial exposure in this patient population [33].
Molecular Epidemiology
Increased CDI incidence and severity have been attributed largely to the emergence of a new strain of C. difficile, designated restriction endonuclease analysis type BI, North American pulsed-field gel electrophoresis type 1 (NAP1), polymerase chain reaction (PCR) ribotype 027 (ie, BI/NAP1/027) [4]. Several characteristics found in BI/NAP1/027 may contribute to its hypervirulence, including polymorphisms in an important toxin production downregulatory gene, tcdC; increased toxin production [34]; presence of the gene encoding binary toxin (ctdA and ctdB); high-level fluoroquinolone resistance (leading to its being selected over other strains in settings of high fluoroquinolone use); and polymorphisms in tcdB that could result in improved toxin binding [35]. Moreover, recent whole-genome comparative analyses have demonstrated a number of genetic rearrangements that BI/NAP1/027 has undergone in the past 20 years. The comparative genome and phenotypic analysis performed by Stabler et al [36] showed that the genome of the epidemic BI/NAP1/027 strain has 5 additional genetic regions compared with its historic counterpart (ie, nonepidemic 027 strain). These additional genes may have contributed to the phenotypic differences between these strains relating to motility, survival, antimicrobial resistance, and toxicity. The multiple mechanisms C. difficile has to modify its genetic content may offer further clues to why BI/NAP1/027 has emerged as a globally dominant strain.
Although the BI/NAP1/027 strain has been associated with the current CDI epidemic, some countries in Europe are starting to see decreases in the prevalence of this strain [37, 38]. However, it is likely that other epidemic strains of C. difficile will emerge in the future. As demonstrated by Belmares et al [39], the molecular epidemiology of C. difficile is both diverse and dynamic, with some strains causing large clusters during certain periods and then becoming endemic; for example, the restriction endonuclease analysis group J strain was associated with multiple outbreaks in the United States and United Kingdom in the 1990s but currently is less common [38, 40].
DIAGNOSIS OF CDI
The enzyme immunoassay (EIA) for detection of toxins A and B has been the most widely used diagnostic test for CDI because of its rapid turnaround, low cost, and simplicity. However, EIAs for toxins A and B are known to have low sensitivity (60%–80%) compared with toxigenic stool culture [5]. Alternative approaches to enhancing the sensitivity of detecting toxigenic C. difficile have been developed, and currently 5 US Food and Drug Administration–cleared PCR assays are available in the United States. These PCR assays have a short turnaround time, and their sensitivities range from 84% to 94% compared with toxigenic stool culture, making their use by clinical laboratories very attractive [5]. In a recent multicenter study, Tenover et al [6] demonstrated that the overall sensitivity of PCR assay for tcdB was significantly greater than that of routine toxin EIA testing (93% vs 60%; P < .001), with toxigenic culture used as the final arbiter.
The use of more sensitive and rapid testing for CDI diagnosis is critical for the clinical management of patients. However, healthcare facilities should expect an increase in CDI rates when transitioning to a PCR-based assay and also should emphasize appropriate testing practices to avoid detection of asymptomatic carriers. A recent study by Belmares et al [41] reported an almost 2-fold increase in C. difficile hospital rates and monthly C. difficile case load after the transition to PCR-based testing for CDI.
ONGOING C. DIFFICILE SURVEILLANCE IN THE UNITED STATES
The increased CDI incidence and severity during recent years [16–19] have highlighted the need for standardized definitions and surveillance methods to detect outbreaks, assess disease trends, and facilitate comparison of CDI rates across healthcare institutions. Therefore, interim recommendations for surveillance, including case definitions, were published in 2007 by the Centers for Disease Control and Prevention (CDC) [42]. These recommendations were incorporated into 2 CDC surveillance systems to monitor CDI rates in the United States: the National Healthcare Safety Network (NHSN) and the CDI surveillance system of the Emerging Infections Program (EIP).
The NHSN is a secure Internet-based surveillance that integrates and supersedes 3 legacy surveillance systems at CDC: the National Nosocomial Infections Surveillance system, the Dialysis Surveillance Network, and the National Surveillance System for Healthcare Workers. The NHSN has a module in its Patient Safety Component that enables US healthcare facilities to monitor CDI by performing either infection surveillance or laboratory-identified (LabID) event reporting. The infection surveillance option relies on trained infection preventionists to perform active, patient-based, prospective surveillance of healthcare–associated CDI. In contrast, the LabID event option relies almost exclusively on data obtained from the laboratory. Data from the LabID event reporting will enable healthcare facilities to calculate proxy measures of C. difficile events, exposures, and healthcare acquisition [43]. Moreover, the LabID event option is well suited for electronic data capture and transmission to the NHSN; definitions for health record vendors to use in facilitating electronic reporting of LabID events to the NHSN are now available [44]. Both preliminary LabID event data in the NHSN (CDC; unpublished data) and other reports of electronically collected hospital surveillance data [45] suggest that current rates of hospital-onset CDI in US hospitals average in the range of 6–8 CDI cases per 10 000 patient-days.
The NHSN's Multidrug-Resistant Organism/CDI module was released in 2009 and, as of 3 April 2012, had 386 facilities monitoring CDI through infection surveillance, 1480 facilities monitoring CDI through LabID event, and 139 monitoring CDI using both methods. Of the total 1866 healthcare facilities reporting CDI data to the NHSN, 40% are located in states with mandatory reporting for C. difficile: California, Illinois, New York, and Tennessee. In these states with mandatory reporting, data will be used to inform state-based prevention initiatives.
The EIP's CDI surveillance is active population laboratory-based surveillance in selected counties in 10 US states [46]. Trained surveillance epidemiologists in the 10 EIP sites investigate all positive C. difficile toxin assay or molecular assay reports from clinical, reference, and commercial laboratories for residents of the surveillance catchment areas. Residents who have a stool specimen positive for C. difficile >8 weeks after the last positive specimen and are >1 year of age are considered to have new cases of CDI. CDI cases are classified as community associated if specimens are collected in the outpatient setting or within 3 calendar days after hospital admission from patients who have no prior healthcare exposure (ie, hospitalization or long-term care facility residency in the 12 weeks before stool specimen collection). The EIP's CDI surveillance was launched in 2009 and has approximately 10.2 million persons under surveillance. As part of this surveillance, a convenience sample of stool specimens is submitted to reference laboratories for culture and molecular characterization. Data from the EIP CDI surveillance will be used to make national estimates of disease burden in both community and healthcare settings, monitor the impact of national prevention efforts or vaccine implementation, and provide information on which strain types are responsible for CDI and their changes in distribution over time.
The NHSN and EIP surveillance systems complement each other: the NHSN measures the burden of CDI in healthcare facilities, whereas the EIP measures the burden of CDI at the population level. Although reporting of CDI through either of these systems has begun only during the past 2 years, initial analysis during the next year should provide insights concerning disease burden and population targets for prevention.
CONCLUSION
Our understanding of C. difficile epidemiology, pathogenesis, and diagnosis continues to evolve. The increases in CDI incidence and severity make C. difficile one of the most important healthcare-associated pathogens. In addition, increases in the detection of CDI in previously low-risk populations pose new challenges that public health officials and healthcare providers will need to face. Clostridium difficile is proving itself to be genetically facile, which can increase its capacity to adapt to new environmental circumstances and lead to the emergence of new epidemic strains. Finally, transition from EIA to PCR will probably improve our ability to detect and manage CDI but will also result in increased CDI rates because of its increased sensitivity. In this era of rapid change in C. difficile epidemiology, national surveillance is crucial to monitor the incidence, identify populations at risk, and characterize the molecular epidemiology of strains causing CDI.
Notes
Acknowledgments. We thank Dawn Sievert, PhD, for providing the numbers of facilities participating in the NHSN Multidrug-Resistant Organism/CDI module; Erik R. Dubberke, MD, for helping with CDI burden estimates using hospital discharge data (Figure 1); and Marya Zilberberg, MD, for helping with the modification of Figure 2 in this article.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Supplement sponsorship. This article was published as part of a supplement entitled “Fidaxomicin and the Evolving Approach to the Treatment of Clostridium difficile Infection,” sponsored by Optimer Pharmaceuticals, Inc.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hall IC, O'Toole E. Intestinal flora in newborn infants with a description of a new pathogenic anaerobe, Bacillus difficile. Am J Dis Child. 1935;49:390–402. [Google Scholar]
- 2.Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med. 1978;298:531–4. doi: 10.1056/NEJM197803092981003. [DOI] [PubMed] [Google Scholar]
- 3.Tedesco FJ, Barton RW, Alpers DH. Clindamycin-associated colitis. A prospective study. Ann Intern Med. 1974;81:429–33. doi: 10.7326/0003-4819-81-4-429. [DOI] [PubMed] [Google Scholar]
- 4.McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 5.Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211–7. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenover FC, Novak-Weekley S, Woods CW, et al. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol. 2010;48:3719–24. doi: 10.1128/JCM.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaughnessy MK, Micielli RL, Depestel DD, et al. Evaluation of hospital room assignment and acquisition of Clostridium difficile infection. Infect Control Hosp Epidemiol. 2011;32:201–6. doi: 10.1086/658669. [DOI] [PubMed] [Google Scholar]
- 8.McFarland LV, Mulligan ME, Kwok RY, Stamm WE. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320:204–10. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 9.Kuijper EJ, Coignard B, Tüll P ESCMID Study Group for Clostridium difficile; EU Member States; European Centre for Disease Prevention and Control. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12(Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 10.Kelly CP, LaMont JT. Clostridium difficile—more difficult than ever. N Engl J Med. 2008;359:1932–40. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 11.Johnson S, Gerding DN. Clostridium difficile–associated diarrhea. Clin Infect Dis. 1998;26:1027–34. doi: 10.1086/520276. [DOI] [PubMed] [Google Scholar]
- 12.Gould CV, McDonald LC. Bench-to-bedside review: Clostridium difficile colitis. Crit Care. 2008;12:203. doi: 10.1186/cc6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clabots CR, Johnson S, Olson MM, Peterson LR, Gerding DN. Acquisition of Clostridium difficile by hospitalized patients: evidence for colonized new admissions as a source of infection. Infect Dis. 1992;166:561–7. doi: 10.1093/infdis/166.3.561. [DOI] [PubMed] [Google Scholar]
- 14.Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, Donskey CJ. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis. 2007;45:992–8. doi: 10.1086/521854. [DOI] [PubMed] [Google Scholar]
- 15.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhea. Lancet. 1998;351:633–6. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 16.McDonald LC, Owings M, Jernigan DB. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerg Infect Dis. 2006;12:409–15. doi: 10.3201/eid1203.051064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redelings MD, Sorvillo F, Mascola L. Increase in Clostridium difficile-related mortality rates, United States, 1999–2004. Emerg Infect Dis. 2007;13:1417–9. doi: 10.3201/eid1309.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burckhardt F, Friedrich A, Beier D, Eckmanns T. Clostridium difficile surveillance trends, Saxony, Germany. Emerg Infect Dis. 2008;14:691–2. doi: 10.3201/eid1404.071023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gravel D, Miller M, Simor A, et al. Canadian Nosocomial Infection Surveillance Program. Health care-associated Clostridium difficile infection in adults admitted to acute care hospitals in Canada: a Canadian Nosocomial Infection Surveillance Program study. Clin Infect Dis. 2009;48:568–76. doi: 10.1086/596703. [DOI] [PubMed] [Google Scholar]
- 20.Miller BA, Chen LF, Sexton DJ, Anderson DJ. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol. 2011;32:387–90. doi: 10.1086/659156. [DOI] [PubMed] [Google Scholar]
- 21.Lucado J, Gould C, Elixhauser A. Clostridium difficile infections (CDI) in hospital stays. Healthcare Cost and Utilization Project, Agency for Healthcare Research and Quality. Statistical brief #124. Available at: http://www.hcup-us.ahrq.gov/reports/statbriefs/sb124.pdf. Accessed 3 April 2012. [PubMed] [Google Scholar]
- 22.Agency for Healthcare Research and Quality. National and regional estimates on hospital use for all patients from the HCUP Nationwide Inpatient Sample (NIS) Available at: http://www.hcup-us.ahrq.gov/db/nation/nis/nissummstats.jsp . Accessed 3 April 2012. [Google Scholar]
- 23.Campbell RJ, Giljahn L, Machesky K, et al. Clostridium difficile infection in Ohio hospitals and nursing homes during 2006. Infect Control Hosp Epidemiol. 2009;30:526–33. doi: 10.1086/597507. [DOI] [PubMed] [Google Scholar]
- 24.Wilcox MH, Mooney L, Bendall R, Settle CD, Fawley WN. A case-control study of community-associated Clostridium difficile infection. Antimicrob Chemother. 2008;62:388–96. doi: 10.1093/jac/dkn163. [DOI] [PubMed] [Google Scholar]
- 25.Kutty PK, Woods CW, Sena AC, et al. Risk factors for and estimated incidence of community-associated Clostridium difficile infection, North Carolina, USA. Emerg Infect Dis. 2010;16:197–204. doi: 10.3201/eid1602.090953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert PJ, Dyck M, Thompson LH, Hammond GW. Population-based surveillance of Clostridium difficile infection in Manitoba, Canada, by using interim surveillance definitions. Infect Control Hosp Epidemiol. 2009;30:945–51. doi: 10.1086/605719. [DOI] [PubMed] [Google Scholar]
- 27.Miniño AM, Xu JQ, Kochanek KD. Deaths: preliminary data for 2008. Natl Vital Stat Rep. 2010;59 Hyattsville, MD: National Center for Health Statistics. http://www.cdc.gov/nchs/data/nvsr/nvsr59/nvsr59_02.pdf. Accessed 10 September 2011. [PubMed] [Google Scholar]
- 28.Zilberberg MD, Tillotson GS, McDonald C. Clostridium difficile infections among hospitalized children, United States, 1997–2006. Emerg Infect Dis. 2010;16:604–9. doi: 10.3201/eid1604.090680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Jumaili IJ, Shibley M, Lishman AH, Record CO. Incidence and origin of Clostridium difficile in neonates. J Clin Microbiol. 1984;19:77–8. doi: 10.1128/jcm.19.1.77-78.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandora TJ, Fung M, Flaherty K, et al. Epidemiology and risk factors for Clostridium difficile infection in children. Pediatr Infect Dis J. 2011;30:580–4. doi: 10.1097/INF.0b013e31820bfb29. [DOI] [PubMed] [Google Scholar]
- 31.Tai E, Richardson LC, Townsend J, Howard E, McDonald LC. Clostridium difficile infection among children with cancer. Pediatr Infect Dis J. 2011;30:610–2. doi: 10.1097/INF.0b013e31820970d1. [DOI] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and Prevention (CDC) Severe Clostridium difficile-associated disease in populations previously at low risk—four states, 2005. MMWR Morb Mortal Wkly Rep. 2005;54:1201–5. [PubMed] [Google Scholar]
- 33.Kuntz JL, Yang M, Cavanaugh J, Saftlas AF, Polgreen PM. Trends in Clostridium difficile infection among peripartum women. Infect Control Hosp Epidemiol. 2010;31:532–4. doi: 10.1086/652454. [DOI] [PubMed] [Google Scholar]
- 34.Warny M, Pepin J, Fang A, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–84. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 35.Stabler RA, Dawson LF, Phua LT, Wren BW. Comparative analysis of BI/NAP1/027 hypervirulent strains reveals novel toxin B-encoding gene (tcdB) sequences. J Med Microbiol. 2008;57:771–5. doi: 10.1099/jmm.0.47743-0. [DOI] [PubMed] [Google Scholar]
- 36.Stabler RA, He M, Dawson L, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hensgens MP, Goorhuis A, Notermans DW, van Benthem BH, Kuijper EJ. Decrease of hypervirulent Clostridium difficile PCR ribotype 027 in the Netherlands. Euro Surveill. 2009;14:pii 19402. doi: 10.2807/ese.14.45.19402-en. [DOI] [PubMed] [Google Scholar]
- 38.Bauer MP, Notermans DW, van Benthem BH, et al. ECDIS Study Group. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 39.Belmares J, Johnson S, Parada JP, et al. Molecular epidemiology of Clostridium difficile over the course of 10 years in a tertiary care hospital. Clin Infect Dis. 2009;49:1141–7. doi: 10.1086/605638. [DOI] [PubMed] [Google Scholar]
- 40.Pear SM, Williamson TH, Bettin KM, Gerding DN, Galgiani JN. Decrease in nosocomial Clostridium difficile-associated diarrhea by restricting clindamycin use. Ann Intern Med. 1994;120:272–7. doi: 10.7326/0003-4819-120-4-199402150-00003. [DOI] [PubMed] [Google Scholar]
- 41.Belmares J, Pua H, Schreckenberger P, Parada J. The paradox of accuracy—C. difficile (CD) polymerase chain reaction (PCR) increases nosocomial CD rates while decreasing CD-related isolation days [abstract 150] Presented at: SHEA 2011 Annual Scientific Meeting, 1–4 April 2011; Dallas, Texas. [Google Scholar]
- 42.McDonald LC, Coignard B, Dubberke E, Song X, Horan T, Kutty PK Ad Hoc Clostridium difficile Surveillance Working Group. Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol. 2007;28:140–5. doi: 10.1086/511798. [DOI] [PubMed] [Google Scholar]
- 43.The National Healthcare Safety Network Manual. Patient Safety Component. Protocol Multidrug-Resistant Organism (MDRO) and Clostridium difficile-Associated Disease (CDAD) Module. Available at: http://www.cdc.gov/nhsn/PDFs/MDRO_CDADprotocolv41Dec08final.pdf . Accessed 10 September 2011. [Google Scholar]
- 44.The National Healthcare Safety Network Clinical Document Architecture (CDA) Available at: http://www.cdc.gov/nhsn/CDA_eSurveillance.html . Accessed 3 April 2012. [Google Scholar]
- 45.Benoit SR, McDonald LC, English R, Tokars JI. Automated surveillance of Clostridium difficile infections using BioSense. Infect Control Hosp Epidemiol. 2011;32:26–33. doi: 10.1086/657633. [DOI] [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention. Healthcare-associated infections (HAIs) Available at: http://www.cdc.gov/HAI/surveillance/monitorHAI.html . Accessed 10 September 2011. [Google Scholar]