Abstract
Background
Xeroderma pigmentosum complementation group F (XPF or ERCC4) plays a key role in DNA repair that protects against genetic instability and carcinogenesis. A series of epidemiological studies have examined associations between XPF polymorphisms and cancer risk, but the findings remain inconclusive.
Methodology/Principal Findings
In this meta-analysis of 47,639 cancer cases and 51,915 controls, by searching three electronic databases (i.e., MEDLINE, EMBASE and CNKI), we summarized 43 case-control studies from 29 publications on four commonly studied polymorphisms of XPF (i.e., rs1800067, rs1799801, rs2020955 and rs744154), and we did not find statistical evidence of any significant association with overall cancer risk. However, in stratification analyses, we found a significant association of XPF-rs1799801 with a reduced cancer risk in Caucasian populations (4,845 cases and 5,556 controls; recessive model: OR = 0.87, 95% CI = 0.76–1.00, P = 0.049, P = 0.723 for heterogeneity test, I2 = 0). Further genotype-phenotype correlation analysis showed that the homozygous variant CC genotype carriers had higher XPF expression levels than that of the TT genotype carriers (Student’s t test for a recessive model: P = 0.046). No publication bias was found by using the funnel plot and Egger’s test.
Conclusion
This meta-analysis suggests a lack of statistical evidence for the association between the four XPF SNPs and overall risk of cancers. However, XPF-rs1799801 may be associated with cancer risk in Caucasian populations, which needs to be further validated in single large, well-designed prospective studies.
Introduction
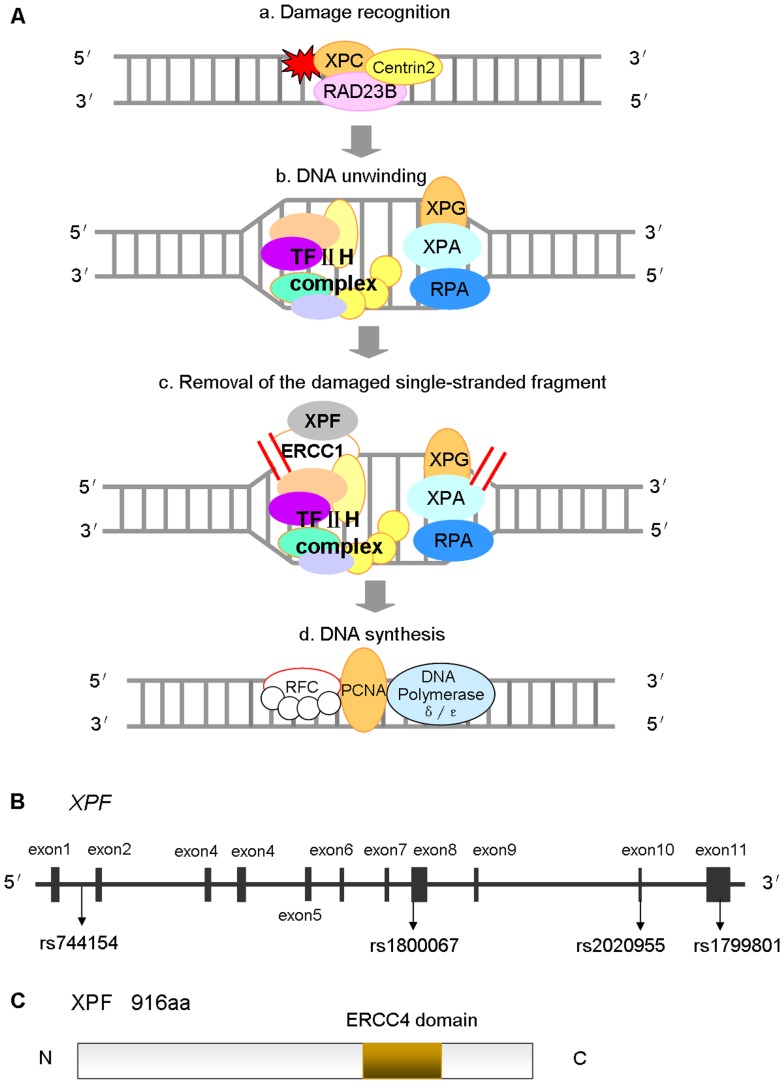
Nucleotide excision repair (NER) is the most versatile, well studied DNA repair mechanism in humans, mainly responsible for repairing bulky DNA damage, such as DNA adducts caused by UV radiation, mutagenic chemicals, or chemotherapeutic drugs [1]. The repair process includes excising and removing damaged nucleotides and synthesizing to fill the resultant gap by using the complementary DNA strand as a template [1]. Therefore, reduced DNA repair capacity (DRC) may lead to genomic instability and carcinogenesis, and genes involved in the NER pathway are candidate cancer susceptibility genes [1]–[3]. NER involves at least four steps ( Figure 1A ): (a) damage recognition by a complex of bound proteins including XPC; (b) unwinding of the DNA by the TFIIH complex that includes XPD; (c) removal of the damaged single-stranded fragment by molecules including an ERCC1/XPF complex; and (d) synthesis by DNA polymerases [4].
Figure 1. Structural characteristics and function of XPF as modified from KEGG and GeneBank database.
(A) NER involves at least four steps: (a) damage recognition by a complex of bound proteins including XPC, (b) unwinding of the DNA by the TFIIH complex that includes XPD, (c) removal of the damaged single-stranded fragment by molecules including an ERCC1/XPF complex, and (d) synthesis by DNA polymerases; (B) The XPF gene map labeled with 11 exons, and four polymorphisms that have been commonly studied for their associations with cancer risk (i.e., rs1800067, rs1799801, rs2020955 and rs744154); (C) The XPF protein consists of 916 amino acids, containing an ERCC4 domain. Abbreviation: NER, nucleotide excision repair; KEGG, Kyoto Encyclopedia of Genes and Genomes.
One of the NER genes, xeroderma pigmentosum complementation group F (XPF), also called excision repair cross-complimentary group 4 (ERCC4), is located on chromosome 16p13.12, contains 11 exons and spans approximately 28.2 kb ( Figure 1B ) [5]. It is a key component involved in the 5′ incision made during NER [2]. The XPF protein consists of 916 amino acids, containing an ERCC4 domain ( Figure 1C ) that is one of the nuclease family, in which essential meiotic endonuclease 1 (EME1) acts as an essential component of a Holliday junction resolvase to interact with MUS81 [6], [7]. The ERCC4 domain is also necessary for forming a tight complex with ERCC1 as a structure-specific DNA repair endonuclease responsible for the 5′-primer incision during DNA excision repair ( Figure 1C ) [8], [9]. In addition to NER, this complex is suggested to play a role in removal of DNA interstrand cross-links (ICL) [10] and DNA double-strand breaks (DSB) as well [11].
To date, a total of 580 single nucleotide polymorphisms (SNPs) in the XPF gene have been reported according to the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?chooseRs=all&go=Go&locusId=2072), some of which have been shown as susceptibility loci for several kinds of cancer, including those of the breast, endometrium, and colorectum [12]–[15]. For example, an important and frequent XPF polymorphism – rs1800067 (Arg415Gln), which results in an arginine-to-glutamine transition at codon 415 ( Figure 1B ), may affect protein interactions, diminish the activity of the ERCC1/XPF complex and alter genetic susceptibility to cancer [16]. The XPF-rs1799801 (Ser835Ser) polymorphism ( Figure 1B ), though not altering amino acids, was reported to be a risk factor for cancer [17]. Another commonly studied XPF SNP, (rs2020955) is a serine-to-proline transition at codon 662, which is less frequent but potentially affecting the function of the gene. Interestingly, another commonly studied XPF SNP (rs744154) is located at intron 1, and its functionality is unknown ( Figure 1B ). To date, associations of these four SNPs with cancer risk have been investigated by a number of reported studies [12]–[15], [17]–[41], but the results are inconclusive, partially because of a possible weak effect of the polymorphisms on cancer risk or study design with a relatively small sample size to detect such weak associations in each of the published studies. Therefore, we performed a meta-analysis that assemblies a large sample size to derive a more precise risk estimate for the commonly studied XPF polymorphisms (each investigated at least by four published studies) with an improved statistic power to detect their associations with cancer risk.
Methods
Literature Search Strategy
We first used two electronic databases (MEDLINE and EMBASE) to identify all case-control studies published to date on an association between XPF polymorphisms and cancer risk (the last search update on December 16, 2011, using the search terms “XPF” or “ERCC4”; “cancer”, “neoplasia”, “malignancy” or “carcinoma”; “polymorphism” or “variant”). To expand the coverage of our searches, we further searched Chinese National Knowledge Infrastructure (CNKI) database (http://www.cnki.net) (1979–), using the terms “XPF” or “ERCC4”; “cancer” in Chinese. Additional published studies on this topic in the references of each publication were also hand reviewed. We further contacted study investigators to identify some unpublished or submitted studies. Only studies with a full text article were included. The authors of published papers were also contacted directly, if crucial data were not reported in original papers. When more than one of the same patient populations were included in different publications, only the most recent or complete study with the largest sample size was included in this meta-analysis.
Selection Criteria
Studies included in the current meta-analysis had to meet the following criteria: evaluation of XPF polymorphisms and cancer risk; more than three studies were available for a certain SNP; written in English or Chinese; case-control study design; sufficient information needed to estimate odds ratios (ORs) and their 95% confidence intervals (CIs); and independent from other studies to avoid double weighting in the estimates derived from the same study. In addition, investigations in control subjects with cancer patients or departure from Hardy-Weinberg equilibrium (HWE) were also excluded from the final analysis.
Data Extraction
Two authors (STY and HJ) independently extracted data and reached a consensus on all of the items. The following information was extracted from each report: the first author, year of publication, country of origin, ethnicity, cancer type, study type (retrospective and prospective), control source [population-based (PB), hospital-based (HB) and family-based (FB)], DNA source (e.g., blood, lymphocytes, and buccal cells), and genotyping methods, total numbers of cases and controls, minor allele frequency (MAF) and numbers of cases and controls with the wild-type, heterozygous and homozygous genotypes. For studies including subjects of different racial descents and having complete genotyping data for each race, data were extracted separately for each ethnic group (categorized as Caucasian, African American, Asian or others). When a study did not state the detailed genotyping result for each ethnic group or if it was impossible to separate participants according to the data presented, the sample was termed as “mixed”. If the numbers of genotyping methods in a study were more than three and no detailed method information was given, the methods were defined “pooled”. Furthermore, references involving different ethnic groups, different types of cancer and different institutions were divided into multiple single study samples for subgroup analyses.
Quantitative Data Synthesis
The numbers of cases and controls by the wild-type, heterozygous and homozygous genotypes were collected from each study to evaluate the risk of developing cancers (ORs and 95% CIs). We further performed stratification analyses by cancer type (if one cancer type was investigated in less than three studies, it would be merged into the “other cancers” group), study type (retrospective and prospective), ethnicity (Caucasian, African American, Asian or others), control source (HB, PB and FB) and sample size (numbers of cases <500, 500–1000 and >1000).
HWE was evaluated for control subjects of each study, using the goodness-of-fit χ2-test, and P<0.05 was considered representative of departure from HWE. Crude ORs with 95% CIs were used to assess the strength of associations between the XPF polymorphisms and cancer risk. The pooled ORs were calculated by using homozygous model (variant homozygous vs. wild-type) and recessive model (homozygous vs. heterozygous + wild-type). For each study, we estimated statistical power to detect an OR of 1.50 (for a risk effect) or its reciprocal 0.67 (for a protective effect), with an α level equal to the observed P value [42]. The χ2-based Q test was performed to assess between-study heterogeneity and considered significant if P<0.05 [43].
Heterogeneity was also quantified with the I2 statistic, a value that indicates what proportion of the total variation across studies is beyond chance. Specifically, 0% indicates no observed heterogeneity, and larger values show increasing heterogeneity [44]. When P value of the heterogeneity test was ≥0.05, the fixed-effects model, based on the Mantel-Haenszel method was used, which assumes the same homogeneity of effect size across all studies [45]. Otherwise, the random-effects model, based on the DerSimonian and Laird method, was more appropriate, which tends to provide wider 95% CIs as the results of the constituent studies differ among themselves [46]. Subgroup analyses were also performed by cancer type, ethnicity, control source and sample size. To assess the effects of individual studies on the overall risk of cancer, sensitivity analysis was performed by excluding each study at a time individually and recalculating the ORs and 95% CIs. Potential publication bias was estimated by the inverted funnel plot, in which the standard error of log (OR) of each study was plotted against its log (OR) [47], and an asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was assessed by the method of Egger’s linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of the ORs [47]. The significance of the intercept was determined by the t test as suggested by Egger, and P<0.05 was considered representative of statistically significant publication bias [47]. If publication bias existed, the Duval and Tweedie nonparametric “trim and fill” method was used to adjust for it [48].
Genotype-phenotype Correlation Analysis
To evaluate biological plausibility of our findings, we used the data on XPF polymorphism genotypes and XPF transcript (mRNA) expression levels both available for 270 lymphoblastoid cell lines by SNPexp online tool (http://app3.titan.uio.no/biotools/help.php?app=snpexp), which provides a convenient and platform-independent way to calculate and visualize the correlation between the HapMap genotypes in a genomic region of interest and gene expression levels [49]. The genotyping data were from the international HapMap phase II release #23 dataset (http://www.hapmap.org) consisting of 3.96 million SNPs that were genotyped using genomic DNA from the 270 individuals from four worldwide populations [CEU: 90 Utah residents with ancestry from northern and western Europe; CHB: 45 unrelated Han Chinese in Beijing; JPT: 45 unrelated Japanese in Tokyo; YRI: 90 Yoruba in Ibadan, Nigeria] [50], [51]. The data of gene expression levels in the same 270 HapMap individuals were from GENEVAR (GENe Expression VARiation, http://www.sanger.ac.uk/resources/software/genevar/) and were detected by using genome-wide expression arrays (47294 transcripts) from EBV-transformed lymphoblastoid cell lines [52]. Student’s t test and analysis of variance test were used to evaluate the differences in the relative mRNA expression levels among different genotype groups. All analyses were conducted by using STATA version 11.0 (Stata Corporation, College Station, TX) and SAS version 9.1 (SAS Institute, Cary, NC). All P values were two-sided with a significance level of P<0.05.
Results
Flow of Included Studies
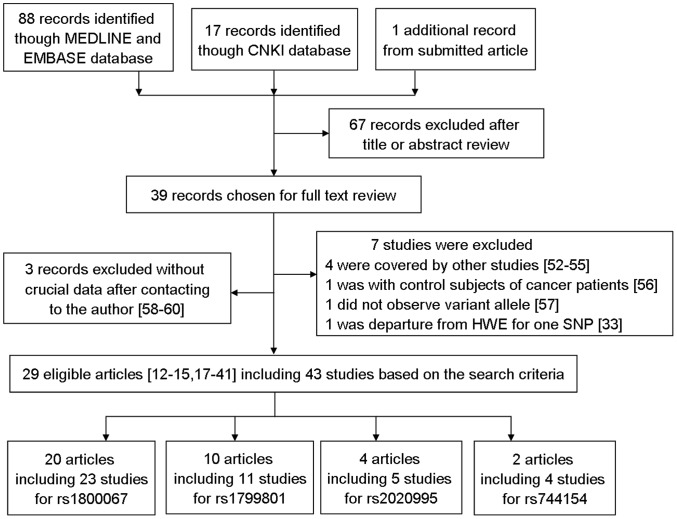
As showed in Figure 2 , a total of 88 published and unduplicated records from the MEDLINE and EMBASE databases, 17 records from the CNKI database and one submitted record were retrieved by using the key words mentioned in the Methods, of which 39 studies examined the association of the commonly studied XPF polymorphisms [i.e., rs1800067 (Arg415Gln, exon 8), rs1799801 (Ser835Ser, exon 11), rs2020955 (Ser662Pro, exon10) and rs744154 (intron 1); Figure 1B ] with cancer risk. Among those 39 publications, four [53]–[56] were excluded because their patient populations were included in other studies [12], [15], [31], one case-control study was excluded because control subjects were of cancer patients [57], one was excluded because no variant allele was observed [58], one study was excluded for departure of the genotype distribution from HWE [33], and three was excluded because of unavailable data to extract ORs and 95% CIs even after having contacted the authors [59]–[61]. The remaining 29 publications of case-control studies contained 43 case-control studies, with a total of 47,639 cancer cases and 51,915 controls of different ethnicities for studying the four polymorphisms.
Figure 2. Flow diagram of included studies for this meta-analysis.
Studies Characteristics
Table 1 lists the essential information for all studies, including first author, year of publication, cancer type, country, ethnicity, study type, source of control, numbers of cases and controls, MAF of controls, statistical power, source of DNA and genotyping methods, grouped by different polymorphisms. For the XPF-rs1800067 SNP, the final analysis included nine breast cancer studies [13], [21], [23], [27], [29], [31], [32], four colorectal cancer studies [14], [22], [24], [28], three cancer studies of head and neck [18], [25], [41], two lung cancer studies [15], [20], and five studies of other cancers [12], [19], [26], [30]. Overall, 17 studies used Caucasians, three used African Americans, one used Latinos, and two used mixed ethnic populations. There were 12, nine, one and one studies using PB, HB, PB/HB and FB design, respectively. For the XPF-rs1799801 SNP, the final analysis included three prostate cancer studies [19], [34], three bladder cancer studies [33], [35], [39], two breast cancer studies [17], [37] and three studies of other cancers [12], [36], [38]. Among them, six studies used Caucasians, two used African Americans, and three used Asians. Six studies were PB design and five HB design. In addition, there were five and four studies having investigated rs2020955 [27], [30], [33], [34] and rs744154 SNPs [39], [40], respectively.
Table 1. Characteristics of studies included in the meta-analysis for an association between XPF-rs1800067, rs1799801, rs2020995, rs744154 SNPs and risk of cancers.
| First author, Year | Cancer type | Country | Ethnicity | Study type | Source of control | Cases/Controls | MAF of controls | P a | Powerb | Source of DNA | Genotyping methods |
| XPF-rs1800067 (Arg415Gln) | |||||||||||
| Yu, submitted | Head neck | USA | Caucasian | Retrospective | HB | 1040/1046 | 0.11 | 0.485 | 0.999 | Blood | SNPlex, PCR-RFLP, Quantitative PCR |
| Doherty, 2011 | Endometrial | USA | Caucasian | Retrospective | PB | 703/714 | 0.07 | 0.184 | 0.925 | Blood, Buccal cells | SNPlex, SNaPshot, Quantitative PCR, PCR-RFLP |
| Gil,2011 | Colorectal | Poland | Caucasian | Retrospective | HB | 133/98 | 0.08 | 0.281 | 0.533 | Lymphocytes | PCR-RFLP |
| Krupa, 2011 | Larynx | Poland | Caucasian | Retrospective | HB | 253/253 | 0.06 | 0.682 | 0.876 | Lymphocytes | PCR-RFLP |
| Agalliu, 2010 | Prostate | USA | Caucasian | Retrospective | PB | 1221/1219 | 0.09 | 0.537 | 1 | Lymphocytes | SNPlex |
| Agalliu, 2010 | Prostate | USA | African American | Retrospective | PB | 144/81 | 0.02 | 0.539 | 0.489 | Lymphocytes | SNPlex |
| Rajaraman, 2010 | Brain | USA | Caucasian | Retrospective | HB | 526/471 | 0.07 | 0.134 | 0.809 | Blood | Quantitative PCR |
| Abbasi, 2009 | Larynx | Germany | Caucasian | Retrospective | PB | 248/647 | 0.07 | 0.163 | 0.747 | Buffy coats | Quantitative PCR |
| Han, 2009 | Breast | USA | Caucasian | Prospective | PB | 1145/1142 | 0.07 | 0.527 | 0.998 | Blood | BeadArray |
| Joshi, 2009 | Colorectal | USA | Caucasian | Retrospective | FB | 305/357 | – | 0.985 | 0.969 | Lymphocytes | Quantitative PCR |
| Chang, 2008 | Lung | USA | Latino | Retrospective | PB | 113/299 | 0.06 | 0.331 | 0.609 | Blood, Buccal cells | BeadArray |
| Hung, 2008 | Lung | Europe | Caucasian, Asia | Retrospective | PB,HB | 2520/2619 | 0.08 | 0.002 | 0.995 | Blood | Pooled |
| McWilliam, 2008 | Pancreatic | USA | Caucasian | Retrospective | HB | 470/596 | 0.10 | 0.003 | 0.418 | Blood | SNPstream, Sequencing |
| Rajaraman, 2008 | Breast | USA | Caucasian | Retrospective | PB | 838/1069 | – | 0.516 | 0.995 | Blood | Quantitative PCR |
| Smith, 2008 | Breast | USA | Caucasian | Retrospective | HB | 324/406 | 0.01 | 0.342 | 0.826 | Blood | MassArray |
| Smith, 2008 | Breast | USA | African American | Retrospective | HB | 53/75 | 0.06 | 0.724 | 0.520 | Blood | MassArray |
| Crew, 2007 | Breast | USA | Caucasian | Retrospective | PB | 1018/1065 | 0.09 | 0.535 | 0.999 | Lymphocytes | FP, Sequenom, Quantitative PCR |
| Jorgensen, 2007 | Breast | USA | Caucasian | Prospective | PB | 259/275 | 0.08 | 0.671 | 0.920 | Buffy coats | Quantitative PCR |
| Huang, 2006 | Colorectal | USA | Mixed | Retrospective | PB | 703/716 | 0.07 | 0.313 | 0.960 | Blood | Pooled |
| Mechanic, 2006 | Breast | USA | African American | Retrospective | PB | 757/673 | 0.02 | 0.034 | 0.351 | Lymphocytes | Quantitative PCR, Sequencing |
| Mechanic, 2006 | Breast | USA | Caucasian | Retrospective | PB | 1246/1133 | 0.07 | 0.113 | 0.979 | Lymphocytes | Quantitative PCR, Sequencing |
| Moreno, 2006 | Colorectal | Spain | Caucasian | Retrospective | HB | 360/323 | 0.11 | 0.693 | 0.969 | Blood | APEX microarray |
| Smith, 2003 | Breast | USA | Caucasian | Retrospective | HB | 253/268 | 0.06 | 0.439 | 0.799 | Blood | PCR-RFLP |
| XPF-rs1799801 (Ser835Ser) | |||||||||||
| Doherty, 2011 | Endometrial | USA | Caucasian | Retrospective | PB | 722/727 | 0.29 | 0.896 | 1 | Blood, Buccal cells | SNPlex, SNaPshot, Quantitative PCR, PCR-RFLP |
| Roberts, 2011 | Breast | USA | Caucasian | Retrospective | PB | 1063/1913 | 0.29 | 0.141 | 1 | Blood, mouthwash samples | MassARRAY |
| Agalliu, 2010 | Prostate | USA | Caucasian | Retrospective | PB | 1259/1243 | 0.29 | 0.937 | 1 | Lymphocytes | SNPlex |
| Agalliu, 2010 | Prostate | USA | African American | Retrospective | PB | 145/79 | 0.16 | 0.603 | 0.806 | Lymphocytes | SNPlex |
| Wang, 2010 | Bladder | China | Asian | Retrospective | HB | 234/250 | 0.25 | 0.412 | 0.920 | Blood | PCR-RFLP |
| Hooker, 2008 | Prostate | USA | African American | Retrospective | HB | 252/301 | 0.13 | 0.215 | 0.812 | Blood | MassARRAY |
| Povey, 2007 | Melanoma | UK | Caucasian | Retrospective | PB | 506/441 | 0.28 | 0.041 | 0.856 | Blood | PCR-RFLP |
| Garcia-Closas, 2006 | Bladder | Spain | Caucasian | Retrospective | HB | 1091/1019 | 0.31 | 0.885 | 1 | leukocytes, mouthwash sample | Pooled |
| Lee, 2005 | Breast | Korea | Asian | Retrospective | HB | 386/336 | 0.23 | 0.406 | 0.970 | Blood | DASH Hybaid |
| Matullo, 2005 | Bladder | UK | Caucasian | Retrospective | HB | 204/213 | 0.32 | 0.340 | 0.865 | buffy coats | Pooled |
| Shen, 2005 | Lung | USA | Asian | Retrospective | PB | 117/111 | 0.19 | 0.231 | 0.611 | Sputum | Quantitative PCR |
| XPF-rs2020955 (Ser662Pro) | |||||||||||
| Rajaraman, 2010 | Brain | USA | Caucasian | Retrospective | HB | 514/444 | 0.00 | 0.917 | 0.589 | Blood | Quantitative PCR |
| Hooker, 2008 | Prostate | USA | African American | Retrospective | HB | 254/301 | 0.19 | 0.689 | 0.972 | Blood | MassARRAY |
| Garcia-Closas, 2006 | Bladder | Spain | Caucasian | Retrospective | HB | 1066/1001 | 0.01 | 0.859 | 0.854 | leukocytes, mouthwash sample | Pooled |
| Mechanic, 2006 | Breast | USA | African American | Retrospective | PB | 752/674 | 0.20 | 0.535 | 0.999 | Lymphocytes | Quantitative PCR, Sequencing |
| Mechanic, 2006 | Breast | USA | Caucasian | Retrospective | PB | 249/250 | 0.00 | 0.568 | 0.399 | Lymphocytes | Quantitative PCR, Sequencing |
| XPF-rs744154 (intron) | |||||||||||
| Wang, 2010 | Bladder | China | Asian | Retrospective | HB | 234/250 | 0.27 | 0.004 | 0.252 | Blood | PCR-RFLP |
| Gaudet, 2009 | Breast | Multicenter | Caucasian | Retrospective | PB,HB | 25743/29074 | 0.28 | 0.542 | 1 | Pooled | Quantitative PCR |
| Gaudet, 2009 | Breast | Multicenter | African | Retrospective | PB,HB | 651/571 | 0.25 | 0.870 | 1 | Pooled | Quantitative PCR |
| Gaudet, 2009 | Breast | Multicenter | Asian | Retrospective | PB,HB | 2700/2104 | 0.24 | 0.345 | 1 | Pooled | Quantitative PCR |
Abbreviation: SNP, single nucleotide polymorphism; HB, Hospital based; PB, Population based; FB, Family based; RFLP, Restriction fragment length polymorphisms.
P value for dominant genetic model in the associations with XPF polymorphisms.
Statistical power to detect an OR of 1.5 (or 0.67 = 1/1.5).
Almost all of the cases were histopathologically confirmed, except for six studies [13], [15], [20], [34], [36], [40]. Controls were mainly matched with cases by age and/or other variables except for five studies [22], [24], [26]–[28]. All the studies reached 50% power to detect the associations between XPF polymorphisms and cancer risk, except for five studies [19], [26], [27], [39]. Blood and lymphocytes were the most common source of DNA, and other sources included buccal cells, buffy coat and mouthwash samples. PCR-based methods were most commonly used in genotyping among these studies.
Meta-analysis Results
Table 2 lists the main results of the meta-analysis for the four polymorphisms in the XPF gene. Given that the xeroderma pigmentosum (XP) syndromes caused by XP germ-line mutations fit a recessive genetic model, in which heterozygotes are unaffected [62], we tested the hypothesis that the XPF polymorphisms were associated with overall cancer risk, assuming a recessive genetic model (i.e., only the variant homozygous genotype was considered the risk genotype).
Table 2. Meta-analysis of the associations between XPF polymorphisms and cancer risk under the XP recessive genetic model.
| Variables | No. of | Cases/Controls | Homozygous model | Recessive model | Powerb | ||||||
| studies | OR (95%CI) | P | P het a | I2 | OR (95%CI) | P | P het a | I2 | |||
| rs1800067 (Arg415Gln) | AA vs. GG | AA vs. (AG + GG) | |||||||||
| All | 23 | 14632/15545 | 1.21 (0.73–1.99) | 0.467 | 0.020 | 45.2 | 1.20 (0.73–1.98) | 0.466 | 0.022 | 44.6 | 0.981 |
| Cancer type | |||||||||||
| Breast | 9 | 5893/6106 | 2.27 (0.81–6.35) | 0.119 | 0.040 | 54.6 | 2.26 (0.81–6.29) | 0.118 | 0.041 | 54.1 | 0.422 |
| Colorectal | 4 | 1501/1494 | 0.51 (0.06–4.25) | 0.535 | 0.073 | 68.8 | 0.51 (0.06–4.16) | 0.532 | 0.076 | 68.2 | 0.679 |
| Head neck | 3 | 1541/1946 | 1.47 (0.42–5.18) | 0.551 | 0.240 | 29.9 | 1.47 (0.41–5.23) | 0.550 | 0.238 | 30.4 | 0.651 |
| Others | 7 | 5697/5999 | 0.84 (0.44–1.63) | 0.617 | 0.222 | 28.4 | 0.85 (0.44–1.64) | 0.629 | 0.226 | 27.8 | 0.930 |
| Lung | 2 | 2633/2918 | 0.63 (0.32–1.25) | 0.185 | 0.817 | 0 | 0.95 (0.56–1.61) | 0.841 | 0.092 | 64.8 | 0.891 |
| Prostate | 2 | 1365/1300 | 2.57 (0.91–7.23) | 0.074 | / | / | 0.95 (0.77–1.17) | 0.607 | 0.482 | 0 | 0.579 |
| Brain | 1 | 526/471 | 0.70 (0.16–3.15) | 0.642 | / | / | 1.30 (0.92–1.84) | 0.134 | / | / | 0.193 |
| Endometrial | 1 | 703/714 | 0.63 (0.15–2.64) | 0.524 | / | / | 1.22 (0.91–1.65) | 0.184 | / | / | 0.266 |
| Pancreatic | 1 | 470/596 | 0.13 (0.01–2.42) | 0.172 | / | / | 0.60 (0.43–0.84) | 0.003 | / | / | / |
| Ethnicity | |||||||||||
| Caucasian | 17 | 10342/11082 | 1.46 (0.83–2.54) | 0.186 | 0.043 | 43.1 | 1.45 (0.83–2.53) | 0.192 | 0.043 | 43.1 | 0.829 |
| African American | 3 | 954/829 | 2.61(0.11–64.2) | 0.557 | / | / | 2.67 (0.11–65.67) | 0.548 | / | / | / |
| Latino | 1 | 113/299 | 0.92(0.04–22.6) | 0.956 | / | / | 0.88 (0.04–21.68) | 0.936 | / | / | / |
| Mixed | 2 | 3223/3335 | 0.42 (0.12–1.50) | 0.182 | 0.192 | 41.2 | 0.43 (0.12–1.57) | 0.201 | 0.186 | 42.9 | 0.645 |
| Source of controls | |||||||||||
| PB | 12 | 8395/9033 | 1.13 (0.56–2.28) | 0.726 | 0.079 | 41.9 | 1.13 (0.56–2.26) | 0.740 | 0.078 | 42.0 | 0.945 |
| HB | 9 | 3412/3536 | 1.71 (0.66–4.43) | 0.268 | 0.062 | 50.0 | 1.71 (0.66–4.40) | 0.268 | 0.063 | 49.7 | 0.639 |
| PB/HB | 1 | 2520/2619 | 0.62 (0.31–1.24) | 0.179 | / | / | 0.64 (0.32–1.28) | 0.210 | / | / | 0.570 |
| Sample size | |||||||||||
| <500 | 12 | 2915/3678 | 1.99 (0.72–5.48) | 0.181 | 0.164 | 33.1 | 1.98 (0.72–5.43) | 0.182 | 0.168 | 32.6 | 0.781 |
| 500–1000 | 5 | 3527/3643 | 0.56 (0.23–1.36) | 0.199 | 0.453 | 0.0 | 0.54 (0.22–1.33) | 0.181 | 0.461 | 0.0 | 0.466 |
| >1000 | 6 | 8190/8224 | 1.23 (0.60–2.51) | 0.566 | 0.016 | 64.1 | 1.24 (0.62–2.50) | 0.545 | 0.019 | 63.1 | 0.731 |
| rs1799801 (Ser835Ser) | CC vs. TT | CC vs. (CT + TT) | |||||||||
| All | 11 | 5979/6633 | 0.91 (0.79–1.04) | 0.166 | 0.783 | 0.0 | 0.89 (0.78–1.01) | 0.080 | 0.764 | 0.0 | 1 |
| Cancer type | |||||||||||
| Prostate | 3 | 1656/1623 | 1.09 (0.82–1.45) | 0.554 | 0.643 | 0.0 | 1.09 (0.83–1.43) | 0.540 | 0.705 | 0.0 | 0.994 |
| Bladder | 3 | 1529/1482 | 0.88 (0.68–1.13) | 0.305 | 0.790 | 0.0 | 0.85 (0.66–1.08) | 0.179 | 0.793 | 0.0 | 0.989 |
| Breast | 2 | 1449/2249 | 0.83 (0.63–1.08) | 0.155 | 0.219 | 33.9 | 0.85 (0.65–1.09) | 0.200 | 0.297 | 8.2 | 0.989 |
| Others | 3 | 1345/1279 | 0.88 (0.66–1.19) | 0.412 | 0.512 | 0.0 | 0.81 (0.61–1.08) | 0.157 | 0.458 | 0.0 | 0.959 |
| Melanoma | 1 | 117/111 | 0.84 (0.52–1.34) | 0.462 | / | / | 1.31 (1.01–1.69) | 0.041 | / | / | 0.298 |
| Lung | 1 | 506/441 | 1.72 (0.54–5.50) | 0.363 | / | / | 1.39 (0.81–2.39) | 0.231 | / | / | 0.314 |
| Endometrial | 1 | 722/727 | 0.85 (0.57–1.26) | 0.417 | / | / | 1.01 (0.83–1.25) | 0.896 | / | / | 0.986 |
| Ethnicity | |||||||||||
| Caucasian | 6 | 4845/5556 | 0.88 (0.76–1.02) | 0.087 | 0.792 | 0.0 | 0.87 (0.76–1.00) | 0.049 | 0.723 | 0.0 | 1 |
| African American | 2 | 397/380 | 1.70 (0.64–4.51) | 0.285 | 0.992 | 0.0 | 1.62 (0.61–4.27) | 0.331 | 0.986 | 0.0 | 0.444 |
| Asian | 3 | 737/697 | 1.07 (0.67–1.71) | 0.784 | 0.408 | 0.0 | 0.99 (0.62–1.57) | 0.963 | 0.374 | 0.0 | 0.968 |
| Source of controls | |||||||||||
| PB | 6 | 3812/4514 | 0.89 (0.76–1.06) | 0.185 | 0.595 | 0.0 | 0.88 (0.75–1.04) | 0.131 | 0.541 | 0.0 | 1 |
| HB | 5 | 2167/2119 | 0.94 (0.74–1.19) | 0.598 | 0.632 | 0.0 | 0.90 (0.72–1.13) | 0.372 | 0.645 | 0.0 | 0.999 |
| Sample size | |||||||||||
| <500 | 6 | 1338/1290 | 1.12 (0.79–1.59) | 0.518 | 0.759 | 0.0 | 1.03 (0.73–1.44) | 0.877 | 0.705 | 0.0 | 0.993 |
| 500–1000 | 2 | 1228/1168 | 0.84 (0.62–1.14) | 0.274 | 0.969 | 0.0 | 0.78 (0.58–1.05) | 0.097 | 0.616 | 0.0 | 0.921 |
| >1000 | 3 | 3413/4175 | 0.88 (0.75–1.05) | 0.153 | 0.357 | 3.0 | 0.90 (0.76–1.06) | 0.185 | 0.388 | 0.0 | 1 |
| rs2020955 (Ser662Pro) | CC vs. TT | CC vs. (CT + TT) | |||||||||
| All | 5 | 2835/2670 | 1.04 (0.70–1.57) | 0.835 | 0.903 | 0.0 | 1.07 (0.72–1.60) | 0.729 | 0.897 | 0.0 | 0.968 |
| rs744154 (intron) | GG vs. CC | GG vs. (CG + CC) | |||||||||
| All | 4 | 29328/31999 | 0.98 (0.92–1.04) | 0.507 | 0.088 | 54.1 | 0.98 (0.92–1.04) | 0.547 | 0.140 | 45.2 | 1 |
Abbreviation: HB, Hospital based; PB, Population based; FB, Family based.
The results were in bold, if the 95% CI excluded 1 or P<0.05.
P value for heterogeneity test.
Statistical power to detect an OR of 1.5 (or 0.67 = 1/1.5).
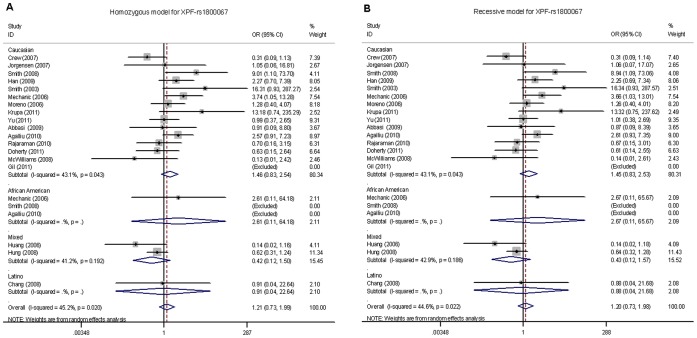
For the XPF-rs1800067 SNP, we obtained genotyping data from 20 publications consisting of 14,632 cancer cases and 15,545 controls. As showed in Table 2 , when all eligible studies were pooled into the meta-analysis, we found that the XPF-rs1800067 polymorphism was not significantly associated with overall cancer risk, with a statistical power of 98% (homozygous model: OR = 1.21, 95% CI = 0.73–1.99, P = 0.020 for heterogeneity test, I2 = 45.2%; recessive model: OR = 1.20, 95% CI = 0.73–1.98, P = 0.022 for heterogeneity test, I2 = 44.6%). In stratification analyses by cancer type, ethnicity, source of controls or sample size, there was no significant association of XPF-rs1800067 SNP with cancer risk in any of the subgroups ( Table 2 , Figure 3A, B ).
Figure 3. Forest plot of cancer risk associated with the XPF-rs1800067 polymorphism stratified by ethnicity.
(A) AA vs. GG in a homozygous model and (B) AA vs. (AG+GG) in a recessive model by the random-effects for each of the 23 published studies. For each study, the estimates of OR and its 95% CI were plotted with a box and a horizontal line. The symbol filled diamond indicates pooled OR and its 95% CI. No significant association between the XPF-rs1800067 polymorphism and cancer risk was found.
For the XPF-rs1799801 SNP, genotyping data of 5,979 cancer cases and 6,633 controls were obtained from 10 publications. Overall, the XPF-rs1799801 polymorphism was not significantly associated with cancer risk (homozygous model: OR = 0.91, 95% CI = 0.79–1.04, P = 0.783 for heterogeneity test, I2 = 0; recessive model: OR = 0.89, 95% CI = 0.78–1.01, P = 0.764 for heterogeneity test, I2 = 0; Table 2 ). However, in stratification analyses, we found a significant association of the XPF-rs1799801 SNP with a reduced cancer risk in Caucasian populations, with a statistical power of 100% (4,845 cases and 5,556 controls; recessive model: OR = 0.87, 95% CI = 0.76–1.00, P = 0.049, P = 0.723 for heterogeneity test, I2 = 0; Table 2 , Figure 4A, B ). After stratified by cancer type, source of controls or sample size, no additional significant association of the XPF-rs1799801 SNP with overall cancer risk was found in any of the subgroups.
Figure 4. Forest plot of cancer risk associated with the XPF-rs1799801 polymorphism stratified by ethnicity.
(A) CC vs. TT in a homozygous model and (B) CC vs. (CT+TT) in a recessive model by the fixed-effects for each of the 11 published studies. For each study, the estimates of OR and its 95% CI were plotted with a box and a horizontal line. The symbol filled diamond indicates pooled OR and its 95% CI. A significant association of the XPF-rs1799801 SNP with a borderline cancer risk in Caucasian populations was found (4845 cases and 5556 controls; recessive model: OR = 0.87, 95% CI = 0.76–1.00, P = 0.049, P = 0.723 for heterogeneity test, I2 = 0).
For XPF-rs2020955 and rs744154 SNPs, a total of 2,835 cancer cases and 2,670 controls and a total of 29,328 cancer cases and 31,999 controls were included, respectively. No significant association of these two SNPs with cancer risk was found in recessive models (OR = 1.07, 95% CI = 0.72–1.60, P = 0.897 for heterogeneity test, I2 = 0%, statistical power = 97%; and OR = 0.98, 95% CI = 0.92–1.04, P = 0.140 for heterogeneity test, I2 = 45.2%, statistical power = 100%, respectively; Table 2 ). Because a limited number of published studies for these two polymorphisms were included, no further stratification analysis was performed.
Heterogeneity and Sensitivity Analyses
Substantial heterogeneities were observed among studies for the association between the XPF-rs1800067 polymorphism and cancer risk (homozygous model: χ2 = 31.02, df = 17, P = 0.020; recessive model: χ2 = 30.66, df = 17, P = 0.022). Therefore, we used the random-effects model that generated wider CIs. For the other three SNPs of the XPF gene (i.e., rs1799801, rs2020955 and rs744154), no heterogeneity was found among studies or in stratification analyses in recessive models (χ2 = 6.58, df = 10, P = 0.764; χ2 = 0.02, df = 1, P = 0.897; and χ2 = 5.47, df = 3, P = 0.140, respectively), and the fixed-effects model was performed. The leave-one-out sensitivity analysis indicated that no single study changed the pooled ORs qualitatively (data not shown).
Publication Bias
The shapes of the funnel plots seemed symmetrical, and Egger’s test suggested that there was no publication bias for studies of XPF-rs1800067, rs1799801, rs2020955 and rs744154 SNPs’ associations with cancer risk in the current meta-analysis [recessive model: P = 0.445, 0.205, no value (i.e., Only two studies were included when assumed a recessive genetic model, which caused no value for the Egger’s test) and 0.663, respectively]. These findings indicated that bias from publications, if any, might not have a significant effect on the results of our meta-analysis for the association between the four commonly studied XPF polymorphisms and overall cancer risk.
Correlation Between XPF-rs1799801 Genotypes and XPF Transcript Expression Levels
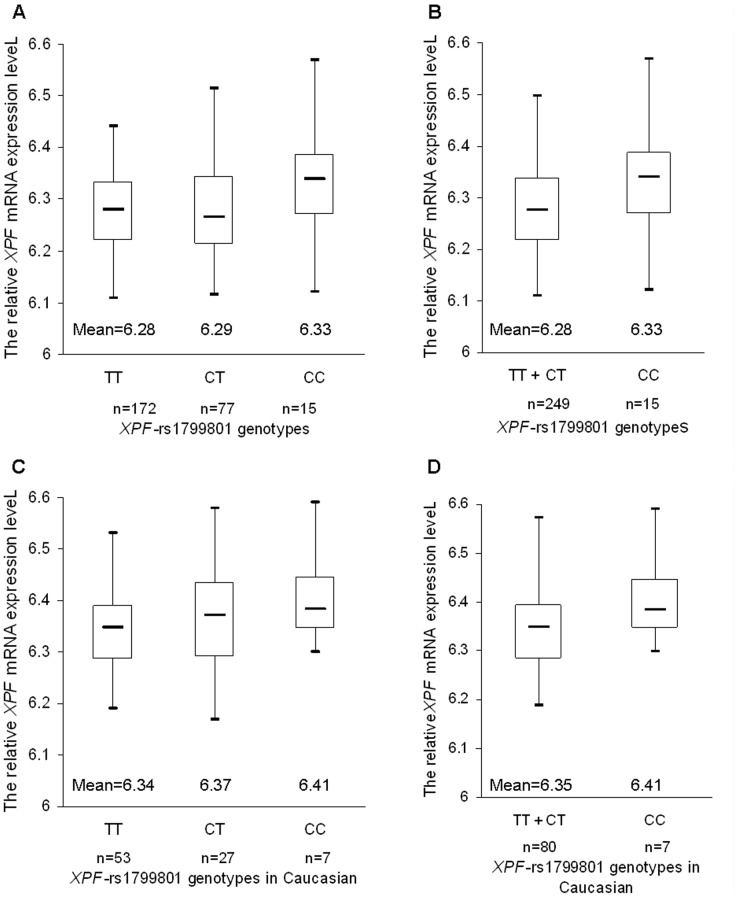
Given that the XPF-rs1799801 SNP, which is located in exon 11, showed a significant association with cancer risk in Caucasian populations, we used the SNPexp online tool to further evaluate biological plausibility underlying the observed association by exploring the correlation between the known XPF-rs1799801 genotypes and the relative expression levels of XPF transcripts. For the 270 individuals whose genotyping and expression data were available for the analysis, there were 172 TT carriers, 77 CT carriers and 15 CC carriers ( Figure 5A ). Homozygous variant CC genotype carriers had significantly higher XPF transcript expression levels than those of wild-type TT carriers and TT+CT carriers (Student’s t test, P = 0.032 and 0.046, respectively; Figure 5A, B ). For the 90 Caucasian subjects, 53 TT carriers, 27 CT carriers and seven CC carriers were observed, but the difference in XPF transcript expression levels between the variant CC genotype, TT and TT+CT genotypes did not reach statistical significance (Student’s t test, P = 0.063 and 0.127, respectively; Figure 5C, D ).
Figure 5. The relative expression levels of XPF transcripts by the known XPF-rs1799801 genotypes in 270 HapMap subjects.
Homozygous variant CC genotype carriers showed a significant increased trend of XPF mRNA expression levels in overall populations, compare to (A) wild-type TT genotype ones, and (B) recessive reference TT+CT genotype ones (Student’s t test, P = 0.032 and 0.046, respectively); but the difference in XPF transcript expression levels between the variant CC genotype and (C) wild-type TT genotype, and (D) TT+CT genotypes did not reach statistical significance (Student’s t test, P = 0.063 and 0.127, respectively).
Discussion
The mechanisms underlying carcinogenesis are multifactorial, and a single genetic variant is usually insufficient to predict risk of cancer, a complex disease phenotype in nature [63]. However, it is likely that suboptimal DNA repair may have a non-specific effect on risk of cancer that originated from DNA damage and subsequent mutation fixation [64]. In this meta-analysis, we summarized all available published data on associations between commonly studied XPF polymorphisms and overall cancer risk. Because germ-line mutations in XP genes cause some rare inherited human syndromes, such as XP, cockayne syndrome (CS) and trichothiodystrophy (TTD) following a recessive genetic model [65]–[67], in which mutant homozygotes manifest the disease but the heterozygotes have a normal phenotype [62]. Therefore, we assessed the associations between XPF polymorphisms and cancer risk by assuming the XP recessive genetic model.
In this meta-analysis of associations between the four commonly studied XPF polymorphisms and cancer risk under the recessive genetic model, we did not find statistical evidence of associations of the XPF-rs1800067, rs2020955 and rs744154 SNPs with cancer risk, nor in stratification analyses. One possible explanation is that these variants, especially of rs1800067 and rs744154, are likely to be low-penetrance SNPs with a very weak effect that needs a much larger sample size to detect [63]. Alternatively, these SNPs may not have any effect on cancer risk, given this meta-analysis of pooling all available studies had included a relatively large sample size. There were two obvious differences between our analysis and another recent meta-analysis of the association between the XPF-rs1800067 SNP and breast cancer risk by Ding [68]. Firstly, Ding et al. presented only one XPF SNP for its association with breast cancer risk, whereas, our analysis included four XPF SNPs for their associations with risk of several cancers with a much larger sample size, which provided a more precise assessment of the associations with risk of cancers, including breast, colorectal and other cancers. Secondly, in the present meta-analysis, we also included one more breast cancer study with 1,145 cases and 1,142 controls of Caucasians for the risk association with XPF-rs1800067 [13]. Furthermore, the subjects from Crew’s study of 1,018 breast cancer cases and 1,065 controls were predominantly of Caucasians [21], leading to a sample size of more than 2,000 Caucasians added to our new analysis, which increased the weight of Caucasians and study power, although we did not find evidence of any association between the XPF-rs1800067 SNP and overall risk of cancers, including breast cancer.
For the XPF-rs1799801 SNP, a total of 5,979 cancer cases and 6,633 controls from 10 independent publications were included. Apparently, studies of these Caucasian populations were quite homogenous, compared with those of XPF-rs1800067 SNP. Although we did not find any significant association with cancer risk, in the stratification analyses, we did find a significant association between the XPF-rs1799801 SNP and cancer risk in Caucasian populations but not in other ethnicities. Further genotype-phenotype correlation analysis showed that homozygous variant CC genotype carriers had significantly increased XPF transcript expression levels in all 270 subjects but not in the 90 Caucasians. This inconsistency is likely due to the reduction in the sample size for Caucasian subjects (n = 90), compared with the overall effect by genotypes of all 270 subjects. Another reason could be the heterogeneity of studies included in the analysis of overall risk, such as different weights of ethnicities included in the overall analysis, which may have confounded the results. For example, for the other two ethnicities, especially African American, less than 500 individuals were included with an insufficient statistical power (44.4%) to detect such an association, which might cause a bias in the combined analysis of the association between XPF-rs1799801 and cancer risk for all populations.
The XPF rs1799801 is highly linked with several other potentially functional SNPs of XPF, such as the rs2276466 SNP, which is located at the 3′-untranslated region (UTR) of XPF. By using the same HapMap and GENEVAR datasets online, in the overall 270 individuals, there were 175 CC carriers, 77 CG carriers and 16 GG carriers for the XPF-rs2276466 SNP. Homozygous variant GG genotype carriers had a significantly higher XPF transcript expression levels than that of wild-type CC carriers and CC+CG carriers (Student’s t test, P = 0.021 and 0.034, respectively). Although the function of the XPF-rs2276466 SNP has not been characterized yet, it is well known that variants, located in the 3?-UTR, particularly at a miRNA binding site, may affect mRNA expression levels [69]. Therefore, additional explanation for the correlation of XPF-rs1799801 SNP with XPF mRNA expression levels may be that some synonymous SNPs appear to act as functional variants in the regulation of gene expression as if they were functional, because of their linkage with other untyped functional SNPs. Additionally, several studies have found that some synonymous SNPs significantly associated with disease phenotypes or traits can be functional by themselves [70], which is because such “silent” polymorphisms may produce a protein product with similar but different structures that may lead to ribosome stalling and alteration of protein folding [71]. Such a hypothesis remains to be tested in future mechanistic studies.
There are several limitations in this meta-analysis, especially for generalization to the general population. First, the quality of the studies included was not optimal. Two studies [14], [15] did not clearly state the ethnicity for genotyping data, and the other two [15], [40] mixed two sources of controls (PB and HB) and did not clearly state the respective genotyping data, which could cause some bias in our estimates. Secondly, obvious heterogeneity across studies for the XPF-rs1800067 SNP, which might result from inclusion of imbalanced ethnic groups and types of cancer, existed in overall and some subgroup analyses. Third, for some SNPs (i.e., XPF-rs2020955 and rs744154) and a certain subgroup (i.e., African Americans and Asians), the numbers of studies and individual sample sizes were relatively small, having no enough statistical power to detect a weak association. Fourth, our results were based on unadjusted estimates, because not all published studies presented with adjusted estimates or when they did, the estimates were not adjusted by the same potential confounders. A more precise analysis should be conducted, if individual data were available, which would allow for the adjustment by other covariants, including age, ethnicity, smoking status, drinking status, environmental factors, and other lifestyles. Finally, although one unpublished study was included in this meta-analysis, many unpublished data may have not been included in the analysis, potentially causing a bias in our meta-analysis.
Overall, our meta-analysis did not provide statistical evidence for an association between the four commonly studied SNPs of the XPF gene and overall risk of several human cancers, but we did find a significant association between the homozygous variant CC genotype of the XPF-rs1799801 SNP and a reduced cancer risk in Caucasian populations. Further genotype-phenotype correlation analysis indicated that the XPF-rs1799801 homozygous variant CC genotype carriers showed an increased trend of XPF expression levels. However, single large, well-designed prospective studies are needed to confirm these findings.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by the funds from China’s Thousand Talents Program at Fudan University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Leibeling D, Laspe P, Emmert S. Nucleotide excision repair and cancer. J Mol Histol. 2006;37:225–238. doi: 10.1007/s10735-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 2.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 3.Shields PG, Harris CC. Molecular epidemiology and the genetics of environmental cancer. JAMA. 1991;266:681–687. [PubMed] [Google Scholar]
- 4.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 5.Liu P, Siciliano J, White B, Legerski R, Callen D, et al. Regional mapping of human DNA excision repair gene ERCC4 to chromosome 16p13.13-p13.2. Mutagenesis. 1993;8:199–205. doi: 10.1093/mutage/8.3.199. [DOI] [PubMed] [Google Scholar]
- 6.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR 3rd, et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 7.Nishino T, Komori K, Ishino Y, Morikawa K. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: similarity between its endonuclease domain and restriction enzymes. Structure. 2003;11:445–457. doi: 10.1016/s0969-2126(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 8.Tsodikov OV, Enzlin JH, Scharer OD, Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc Natl Acad Sci U S A. 2005;102:11236–11241. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripsianes K, Folkers G, Ab E, Das D, Odijk H, et al. The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure. 2005;13:1849–1858. doi: 10.1016/j.str.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, et al. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty JA, Weiss NS, Fish S, Fan W, Loomis MM, et al. Polymorphisms in Nucleotide Excision Repair Genes and Endometrial Cancer Risk. Cancer Epidemiol Biomarkers Prev. 2011;20:1873–1882. doi: 10.1158/1055-9965.EPI-11-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Haiman C, Niu T, Guo Q, Cox DG, et al. Genetic variation in DNA repair pathway genes and premenopausal breast cancer risk. Breast Cancer Res Treat. 2009;115:613–622. doi: 10.1007/s10549-008-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang WY, Berndt SI, Kang D, Chatterjee N, Chanock SJ, et al. Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: XPC polymorphisms modify smoking-related risk. Cancer Epidemiol Biomarkers Prev. 2006;15:306–311. doi: 10.1158/1055-9965.EPI-05-0751. [DOI] [PubMed] [Google Scholar]
- 15.Hung RJ, Christiani DC, Risch A, Popanda O, Haugen A, et al. International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev. 2008;17:3081–3089. doi: 10.1158/1055-9965.EPI-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608. [PubMed] [Google Scholar]
- 17.Lee SA, Lee KM, Park WY, Kim B, Nam J, et al. Obesity and genetic polymorphism of ERCC2 and ERCC4 as modifiers of risk of breast cancer. Exp Mol Med. 2005;37:86–90. doi: 10.1038/emm.2005.12. [DOI] [PubMed] [Google Scholar]
- 18.Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, et al. Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer. 2009;125:1431–1439. doi: 10.1002/ijc.24442. [DOI] [PubMed] [Google Scholar]
- 19.Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA, et al. Genetic variation in DNA repair genes and prostate cancer risk: results from a population-based study. Cancer Causes Control. 2010;21:289–300. doi: 10.1007/s10552-009-9461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, et al. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer. 2008;123:2095–2104. doi: 10.1002/ijc.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crew KD, Gammon MD, Terry MB, Zhang FF, Zablotska LB, et al. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:2033–2041. doi: 10.1158/1055-9965.EPI-07-0096. [DOI] [PubMed] [Google Scholar]
- 22.Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, et al. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual’s susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2011;39:527–534. doi: 10.1007/s11033-011-0767-5. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen TJ, Visvanathan K, Ruczinski I, Thuita L, Hoffman S, et al. Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat. 2007;101:65–71. doi: 10.1007/s10549-006-9263-3. [DOI] [PubMed] [Google Scholar]
- 24.Joshi AD, Corral R, Siegmund KD, Haile RW, Le Marchand L, et al. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30:472–479. doi: 10.1093/carcin/bgn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krupa R, Kasznicki J, Gajecka M, Rydzanicz M, Kiwerska K, et al. Polymorphisms of the DNA repair genes XRCC1 and ERCC4 are not associated with smoking- and drinking-dependent larynx cancer in a Polish population. Exp Oncol. 2011;33:55–56. [PubMed] [Google Scholar]
- 26.McWilliams RR, Bamlet WR, Cunningham JM, Goode EL, de Andrade M, et al. Polymorphisms in DNA repair genes, smoking, and pancreatic adenocarcinoma risk. Cancer Res. 2008;68:4928–4935. doi: 10.1158/0008-5472.CAN-07-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mechanic LE, Millikan RC, Player J, de Cotret AR, Winkel S, et al. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis. 2006;27:1377–1385. doi: 10.1093/carcin/bgi330. [DOI] [PubMed] [Google Scholar]
- 28.Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, et al. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- 29.Rajaraman P, Bhatti P, Doody MM, Simon SL, Weinstock RM, et al. Nucleotide excision repair polymorphisms may modify ionizing radiation-related breast cancer risk in US radiologic technologists. Int J Cancer. 2008;123:2713–2716. doi: 10.1002/ijc.23779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajaraman P, Hutchinson A, Wichner S, Black PM, Fine HA, et al. DNA repair gene polymorphisms and risk of adult meningioma, glioma, and acoustic neuroma. Neuro Oncol. 2010;12:37–48. doi: 10.1093/neuonc/nop012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, et al. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–2138. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith TR, Levine EA, Perrier ND, Miller MS, Freimanis RI, et al. DNA-repair genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:1200–1204. [PubMed] [Google Scholar]
- 33.Garcia-Closas M, Malats N, Real FX, Welch R, Kogevinas M, et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:536–542. doi: 10.1158/1055-9965.EPI-05-0749. [DOI] [PubMed] [Google Scholar]
- 34.Hooker S, Bonilla C, Akereyeni F, Ahaghotu C, Kittles RA. NAT2 and NER genetic variants and sporadic prostate cancer susceptibility in African Americans. Prostate Cancer Prostatic Dis. 2008;11:349–356. doi: 10.1038/sj.pcan.4501027. [DOI] [PubMed] [Google Scholar]
- 35.Matullo G, Guarrera S, Sacerdote C, Polidoro S, Davico L, et al. Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol Biomarkers Prev. 2005;14:2569–2578. doi: 10.1158/1055-9965.EPI-05-0189. [DOI] [PubMed] [Google Scholar]
- 36.Povey JE, Darakhshan F, Robertson K, Bisset Y, Mekky M, et al. DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis. 2007;28:1087–1093. doi: 10.1093/carcin/bgl257. [DOI] [PubMed] [Google Scholar]
- 37.Roberts MR, Shields PG, Ambrosone CB, Nie J, Marian C, et al. Single-nucleotide polymorphisms in DNA repair genes and association with breast cancer risk in the web study. Carcinogenesis. 2011;32:1223–1230. doi: 10.1093/carcin/bgr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen M, Berndt SI, Rothman N, Demarini DM, Mumford JL, et al. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer. 2005;116:768–773. doi: 10.1002/ijc.21117. [DOI] [PubMed] [Google Scholar]
- 39.Wang M, Yuan L, Wu D, Zhang Z, Yin C, et al. A novel XPF −357A>C polymorphism predicts risk and recurrence of bladder cancer. Oncogene. 2010;29:1920–1928. doi: 10.1038/onc.2009.484. [DOI] [PubMed] [Google Scholar]
- 40.Gaudet MM, Milne RL, Cox A, Camp NJ, Goode EL, et al. Five polymorphisms and breast cancer risk: results from the Breast Cancer Association Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18:1610–1616. doi: 10.1158/1055-9965.EPI-08-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu H, Liu Z, Huang YJ, Yin M, Wang LE, et al. Association between single nucleotide polymorphisms in ERCC4 and risk of squamous cell carcinoma of the head and neck submitted. 2011. [DOI] [PMC free article] [PubMed]
- 42.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 44.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 45.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 46.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 47.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 49.Holm K, Melum E, Franke A, Karlsen TH. SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics. 2010;11:600. doi: 10.1186/1471-2105-11-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consortium IH. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 51.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Chen C. Nucleotide excision repair genotype and the incidence of endometrial cancer: effect of other risk factors on the association. Gynecol Oncol. 2006;103:891–896. doi: 10.1016/j.ygyno.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 54.Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Voigt LF, et al. Interindividual variation in nucleotide excision repair genes and risk of endometrial cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2524–2530. doi: 10.1158/1055-9965.EPI-05-0414. [DOI] [PubMed] [Google Scholar]
- 55.Smith TR, Liu-Mares W, Van Emburgh BO, Levine EA, Allen GO, et al. Genetic polymorphisms of multiple DNA repair pathways impact age at diagnosis and TP53 mutations in breast cancer. Carcinogenesis. 2011;32:1354–1360. doi: 10.1093/carcin/bgr117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 57.Millikan RC, Hummer A, Begg C, Player J, de Cotret AR, et al. Polymorphisms in nucleotide excision repair genes and risk of multiple primary melanoma: the Genes Environment and Melanoma Study. Carcinogenesis. 2006;27:610–618. doi: 10.1093/carcin/bgi252. [DOI] [PubMed] [Google Scholar]
- 58.Ming-Shiean H, Yu JC, Wang HW, Chen ST, Hsiung CN, et al. Synergistic effects of polymorphisms in DNA repair genes and endogenous estrogen exposure on female breast cancer risk. Ann Surg Oncol. 2010;17:760–771. doi: 10.1245/s10434-009-0802-0. [DOI] [PubMed] [Google Scholar]
- 59.Milne RL, Ribas G, Gonzalez-Neira A, Fagerholm R, Salas A, et al. ERCC4 associated with breast cancer risk: a two-stage case-control study using high-throughput genotyping. Cancer Res. 2006;66:9420–9427. doi: 10.1158/0008-5472.CAN-06-1418. [DOI] [PubMed] [Google Scholar]
- 60.Mort R, Mo L, McEwan C, Melton DW. Lack of involvement of nucleotide excision repair gene polymorphisms in colorectal cancer. Br J Cancer. 2003;89:333–337. doi: 10.1038/sj.bjc.6601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliveira LGJ, Rinck-Junior JA, Moraes AM, Lima CSP. The XPC A2920C, The XPF T30028C and the P53 ARG72pro polymorphisms, Involved in DNA repair, Alter the risk for the Malignant Melanoma. European Journal of Cancer. 2011;47:S654–S655. [Google Scholar]
- 62.Kraemer KH, Lee MM, Andrews AD, Lambert WC. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch Dermatol. 1994;130:1018–1021. [PubMed] [Google Scholar]
- 63.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 64.Manuguerra M, Saletta F, Karagas MR, Berwick M, Veglia F, et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol. 2006;164:297–302. doi: 10.1093/aje/kwj189. [DOI] [PubMed] [Google Scholar]
- 65.Kovacs MS, Hocking DJ, Evans JW, Siim BG, Wouters BG, et al. Cisplatin anti-tumour potentiation by tirapazamine results from a hypoxia-dependent cellular sensitization to cisplatin. Br J Cancer. 1999;80:1245–1251. doi: 10.1038/sj.bjc.6690492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 67.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 68.Ding DP, He XF, Zhang Y. Lack of association between XPG Asp1104His and XPF Arg415Gln polymorphism and breast cancer risk: a meta-analysis of case-control studies. Breast Cancer Res Treat. 2011;129:203–209. doi: 10.1007/s10549-011-1447-9. [DOI] [PubMed] [Google Scholar]
- 69.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 70.Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta. 2009;1794:860–871. doi: 10.1016/j.bbapap.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai CJ, Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM, et al. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol. 2008;383:281–291. doi: 10.1016/j.jmb.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]