Abstract
Chronic hepatitis C virus (HCV) infection is complicated by hepatic fibrosis. Hypothesizing that early fibrogenic signals may originate in cells susceptible to HCV infection, hepatocyte gene expression was analyzed from persons with chronic HCV at different stages of liver fibrosis. Four HCV-infected subjects with pre-cirrhotic liver fibrosis (Ishak fibrosis 3–5) were matched for age, race, and gender to five HCV-infected subjects with no evidence of fibrosis (Ishak fibrosis 0). Hepatocytes from each subject were isolated from liver biopsies using laser capture microdissection. Transcriptome profiling was performed on hepatocyte RNA using hybridization arrays. We found that hepatocytes in pre-cirrhotic fibrosis were depleted for genes involved in small molecule and drug metabolism, especially butyrylcholinesterase (BCHE), a gene involved in the metabolism of drugs of abuse. Differential expression of BCHE was validated in the same tissues and cross-sectionally in an expanded cohort of 143 HCV-infected individuals. In a longitudinal study, serum BCHE activity were already decreased at study inception in 19 fibrosis progressors compared to 20 fibrosis non-progressors (p<0.05). Non-progressors also had decreased BCHE activity over time compared to initial values, but these evolved a median (range) 8.6 (7.8–11.4) years after the study period inception(p<0.05). Laser captured portal tracts were enriched for immune related genes when compared to hepatocytes but pre-cirrhotic livers lost this enrichment.
Conclusion
Overall, we found that chronic HCV is associated with hepatocyte BCHE loss years before hepatic synthetic function is impaired. These results indicate that BCHE may be involved in the pathogenesis of HCV-related fibrosis among injection drug users.
Keywords: butyrylcholinesterase, Hepacivirus, chronic HCV infection, laser capture microdissection, portal tract
Hepatitis C virus (HCV) affects approximately 170 million people worldwide.(1),(2) Nearly 85% of these persons develop chronic infection; the natural history of chronic infection in many cases leads to complications that are the leading reasons for liver transplantation in the U.S. Complications usually begin with hepatic fibrosis, lead to cirrhosis, and can ultimately result in hepatocellular carcinoma.(3) Progression to hepatic fibrosis in chronic HCV infection has been previously associated with common downstream mechanisms such as TGFβ(4, 5) and PDGF signaling pathways(6); however, the earliest molecular mechanisms underlying HCV-induced hepatic fibrosis are largely unknown. Therefore, determining how HCV induces hepatic fibrosis is crucial for identifying targetable biological determinants of progressive HCV infection.
Progressive hepatic fibrosis is orchestrated by several cellular constituents; however, it is not well understood how hepatocytes, the sites of HCV replication, contribute to the ensuing fibrosis separately from abundant inflammation and stellate cells which ultimately produce collagen. To understand the direct link between HCV infection and fibrosis, we hypothesized that hepatocyte transcriptomes could be separated from bulk liver tissue and studied. Human liver biopsies deliver only a limited amount of material, and cell separation has not been attempted to enrich transcriptomes. In addition, the absence of a representative animal model of progressive liver disease has compelled researchers to use in situ methods in studies of HCV pathogenesis. In this context, laser capture microdissection (LCM) is an emerging technology that allows isolation of specific cell types while preserving key anatomic relationships of the tissue.
The present study was structured into three phases: in the first (discovery) phase, gene expression arrays were used to explore potential markers of fibrosis progression from laser captured hepatocytes and portal tracts. In the second (validation) phase, differential expression of the lead gene, butyrylcholinesterase (BCHE), a critical enzyme in cocaine and heroin metabolism, was confirmed in an expanded cross-sectional cohort of HCV-infected IDUs by measuring a surrogate of BCHE protein expression in archived serum samples. In the third (longitudinal) phase, BCHE expression over time was measured in liver disease progressors and non-progressors. BCHE, therefore, is a potential pathogenic node that may link drugs of abuse with the development of liver disease in persons with chronic HCV.
Materials and Methods
Study populations
The AIDS Link to Intravenous Experience (ALIVE) cohort is a community-based study that enrolled participants to study the natural history of HCV and HIV infections. Between 1996 and 1998, 210 of 1625 participants were randomly selected from ALIVE to participate in a cross-sectional study designed to determine the severity and correlates of liver disease.(7) Liver biopsies were obtained from participants (details below). A subset of 116 subjects had a second liver biopsy with careful interval follow-up, and were the focus of subsequent study.(8) Pre-cirrhotic (PC) liver tissues were chosen for the discovery cohort from five subjects with chronic HCV infection and Ishak fibrosis stage 3–5 who had sufficient tissue stored in OCT. Tissues that were stored in Trizol® or other lysis buffers were excluded to avoid homogenization of transcriptomes between cellular constituents. Five control tissues with baseline Ishak fibrosis score of 0 and no evidence of fibrosis (NF) were selected from persons matched for HCV status, age, race and gender. All subjects in the discovery cohort were HIV negative. One PC tissue was later excluded because the subject was found to be HBsAg positive, leaving nine subjects.
Validation of the differential expression of BCHE was performed using an expanded group of subjects’ samples derived from the same cohort. Serum BCHE activity (SBA) was measured on 116 well-characterized subjects with serum samples and contemporaneous liver disease assessment as mentioned above; an additional 7 samples from subjects in ALIVE with careful follow-up were added to represent more advanced liver disease.(8) Despite enrichment, the panel had few subjects with biopsy proven cirrhosis (n=2); therefore, 20 subjects from ALIVE who had two consecutive fibroscan values greater than 12 kPa were added to the study.(9) In total, SBA was tested in 143 subjects for cross-sectional validation.
Longitudinal SBA testing was performed on a subset of the validation cohort with regularly sampled serum and contemporaneous fibrosis staging. Cases (n=19), defined as progressors, had two biopsies with Metavir scores ≤ 2 followed by two consecutive fibroscan values averaging greater than 10 kPa with the last fibroscan ≥ 12kPa. Controls, defined as non-progressors, were matched for age, gender and race, and had two biopsies with Metavir score ≤ 2 followed by two consecutive averaging fibroscan values less than 10kPa with the last fibroscan < 12 kPA. Cases and controls were picked for having minimal fibrosis at earliest time points. Samples from cases and controls were chosen at regularly-spaced time points spanning the earliest and the most recent ascertainment of liver disease (11.75±1 years from time point 1 to time point 4) to estimate the natural history of fibrosis progression in the cases.
Tissue Preparation and Laser capture microdissection
Liver biopsies for the discovery cohort were obtained from 1997–1998 using 16-gauge needles, and were divided in two at the bedside; one piece was immediately snap-frozen and stored in a liquid nitrogen repository.(7) The second and larger piece was formalin fixed, stained with H&E and trichrome, and graded and staged for fibrosis (Ishak fibrosis and Metavir) by an experienced hepatopathologist. Tissues that were of poor quality or of inadequate length were excluded before review. Metavir scores from subsequent biopsies and intervening fibroscan values for the same subjects confirmed staging result from the first biopsy. In preparation for LCM, tissues were thawed and sectioned at −24°C. Seven and 10 μm sections were made onto polyethylene naphthalate membrane slides, fixed in 70% ethanol, and hematoxylin-stained for morphologic and nuclear detail. The duration of room temperature exposure before LCM was ≤ 10 minutes. The Leica LMD6000 laser capture microdissecting system was used for LCM. Two portal tracts and six parenchymal segments were captured per subject. Parenchymal segments were captured according to anatomic landmarks and hepatic zonation to include two periportal segments, two midzonal segments, and two centrilobular segments. Segments were captured in Trizol® buffer and stored at −80°C until RNA isolation.
RNA purification and assays
RNA was isolated from each sample separately using the Agencourt®RNAdvance Cell v2 system (Beckman Coulter Genomics, Danvers, MA). Isolated RNA from each sample was divided into three aliquots. The first was tested for housekeeping transcripts using qPCR. The second was tested for HCV RNA using the ABBOT® RealTime HCV Assay. The third was linearly amplified using the NuGEN Ovation Pico WTA System® (NuGEN Technologies, Inc., San Carlos, CA). Attempts to quantify RNA using a NanoDrop® (Thermo Scientific NanoDrop Products, Wilmington, DE) were unsuccessful because of the small numbers of captured cells; therefore, quantitative PCR (qPCR) for GAPDH mRNA was used to estimate the number of captured cells after standardizing to a known quantity of Hepatoma 3B cells in culture, resulting in the estimate of ~20 copies per cell. Samples were submitted to the Johns Hopkins Microarray Core Facility and hybridized to the GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA). The mean slope of 5′ intensity versus 3′ intensity was 0.07±0.06, indicating no preferential mRNA degradation. Results of the microarray were validated by performing qPCR using SyBR Green® and gene-specific primers on 1/8 hepatic parenchyma sections from each subject. Raw data from microarrays is available on NCBI GEO via accession number GSE33650 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=pnmnhuoacgiaglk&acc=GSE33650).
BCHE Protein Expression
Serum BCHE activity (SBA), a surrogate for protein expression, was measured on archived specimens at a dilution of 1:500 using the DetectX Butyrylcholinesterase Fluorescent Activity Kit® (Arbor Assays, Ann Arbor, MI). Using this kit, SBA is quantified by the cleavage of a known and specific substrate for BCHE, butyrylthiocholine, that exposes a free thiol. A proprietary reagent contains a maleimide group that irreversibly reacts with free thiols; the product of this reaction is a conjugated fluorescent compound that can be quantified according to a standard curve. Since BCHE is highly polymorphic, the activity assay allows sensitive detection of SBA and avoids cross-detection of acetylcholinesterase activity. Differences in SBA between independent groups were determined by the Mann-Whitney-Wilcoxon test. Within group differences in the longitudinal analysis were identified using the paired Wilcoxin Signed Rank Test in R with appropriate null hypotheses.
Statistical Analysis
Results from microarray hybridization were analyzed using the Bioconductor package in R. Data were normalized with the “rma” procedure using a custom HGU133Plus2 annotation (CDF: Brainarray Version 13, hgu133plus2hsentrezg), to avoid known problems associated with the affymetrix annotation. (10, 11) The normalized data was then analyzed using the “affy” and “limma” packages in Bioconductor.(12, 13) Genes absent in greater than 95% of the samples were excluded from the analysis, providing annotation for 11170 out of a possible 18185 genes available on the array.(14) Adjusted p-values or False Discovery Rates (FDR) were calculated using the default Benjamini & Hochberg method.(15) Genes with a fold change of >=2 at an adjusted p value of < 0.05 were considered differentially expressed in all comparisons unless mentioned otherwise. Gene functions were found and enriched using DAVID, an online tool.(16) Bonferroni Correction was used to adjust for multiple comparisons under DAVID using the 11170 genes as the background set.
Results
Subject and Tissue Characteristics
Cases and controls were well matched by age, gender, and race (Table 1). The median age was 38.6 years, 7/9 were male, and all 9 were African American. Individuals were chronically HCV-infected, and none had received treatment before the time of biopsy. Eight of 9 subjects were infected with genotype 1 (6/8 1a). The median circulating HCV RNA level was 6.5×105 IU/mL (5.8 log10 IU/mL), and was obtained a median (range) 28 (821) days before liver biopsy. Transaminases (ALT and AST) values were available from the nearest visit before biopsy (Table 1).
Table 1.
Characteristics of Discovery Cohort with Four HCV Infected Subjects with Pre-Cirrhotic (PC) Livers and Five HCV Infected Subjects with No Fibrosis (NF) in the Liver.
| Subject ID | Age | Gender | HCV Subtype | HCV Viral Load (Log10) | ALT | AST | MHAI FIB |
|---|---|---|---|---|---|---|---|
| NF 1 | 36 | M | 1b | 5.5 | 31 | 27 | 0 |
| NF 2 | 39 | F | 1a | 5.7 | 28 | 28 | 0 |
| NF 3 | 33 | M | 1a | 7.7 | 33 | 39 | 0 |
| NF 4 | 58 | M | 1a | 6.9 | 17 | 17 | 0 |
| NF 5 | 37 | M | 1b | 5.4 | 35 | 34 | 0 |
| PC 1 | 39 | M | 2 | 6.2 | 35 | 34 | 3 |
| PC 2 | 42 | F | 1a | 6.5 | 39 | 34 | 3 |
| PC 3 | 44 | M | 1a | 5.8 | 43 | 44 | 3 |
| PC 4 | 38 | M | 1a | 5.6 | 42 | 61 | 5 |
The total number of input cells was estimated by qPCR for GAPDH after standardizing to a known quantity of hepatoma cells in culture. RNA was extracted from an estimated median (IQR) of 4535 (1870–5638) portal tract cells and 27,900 (13,800–48,688) hepatic parenchyma cells (Fig. 1), representing 18 and 54 transcriptomes, repectively. Prior to the segregation of hepatic parenchyma and portal tract extracts, no differences in gene expression were observed in the PC tissues versus NF tissues.
Figure 1.
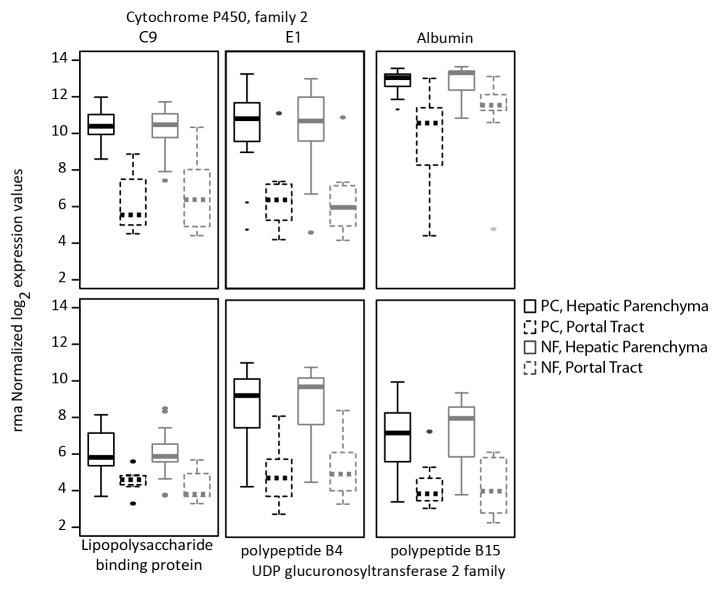
Microanatomic Hepatic Regions Separately Isolated using LCM. (a) Portal tracts (left panel) were identified by the presence of portal veins (PV), bile ducts (BD), and hepatic arteries (HA). After separation, pictures were obtained to confirm isolation and exclusion of hepatocytes (right panel). LM, 20X, hematoxylin. (b) Hepatic parenchyma was identified and categorized by its proximity to portal tracts as denoted by the dashed white line. In this figure, the periportal zone corresponding to the parenchyma surrounding the portal tract is shown pre- (left panel) and post- (right panel) capture. LM, 10X, hematoxylin.
Segregation of portal tracts and hepatic parenchyma
Candidate genes with known or expected differential expression patterns in hepatocytes vs. mononuclear cells (e.g. albumin, cytochrome p450 enzymes) were compared between parenchymal and portal tract segments of all individuals to test the specificity of LCM (Fig. 2). All of the pre-designated hepatocyte-specific genes were more highly expressed in parenchymal segments compared to portal tracts irrespective of fibrosis stage, confirming that LCM of hepatic parenchyma predominantly yielded hepatocytes: albumin expression in the parenchyma was ~5-fold greater than in the portal tract (FDR < 1.67×10−6), while the expression of a representative hepatic microsomal enzyme, CYP2C9, in the parenchyma was ~14 times the expression in portal tracts (FDR < 2×10−13). Due to this higher expression of hepatocyte specific genes in the parenchyma, we assume that the extracted parenchymal section consisted mainly of hepatocytes and hence refer to them as hepatocytes for the rest of the study.
Figure 2. Laser captured hepatic parenchyma is enriched for hepatocyte-specific genes.
LCM was used to isolate separate hepatic regions that are based on microanatomic distinctions. Hepatic parenchyma (solid lines) was separated from portal tracts (dashed lines) in pre-cirrhotic (PC; black lines) and no fibrosis (NF; gray lines) livers, and RNA was extracted and hybridized to expression arrays. Six genes on the array were identified a priori that were anticipated to be specific for hepatocytes. Absolute expression of each gene confirmed enrichment of hepatocytes in the hepatic parenchyma, and exclusion from portal tracts. All comparisons between portal tracts and hepatic parenchyma have adjusted p values <0.01. In each case, the box itself contains the middle 50% of the data. The upper edge of the box indicates the 75th percentile of the data set, and the lower edge indicates the 25th percentile. The line in the box indicates the median value of the data.
Transcriptome Profiling of HCV-infected Hepatocytes
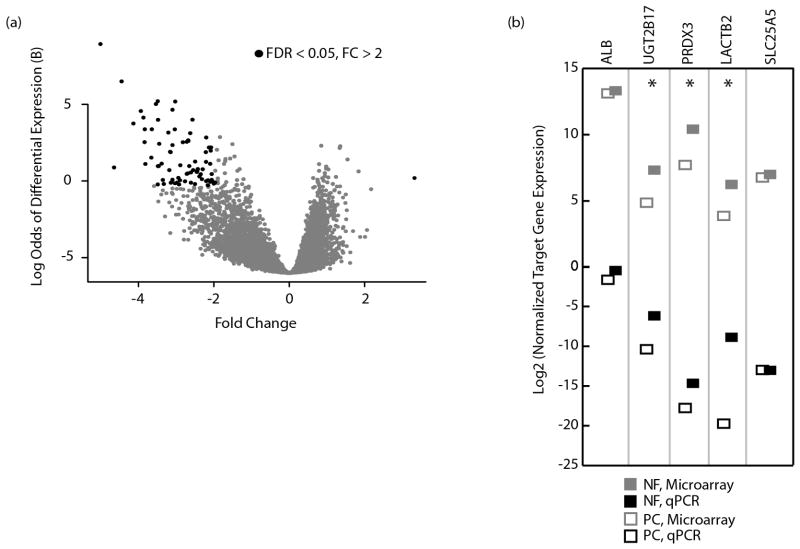
On comparison between hepatocytes in PC and NF groups, 74 genes were found to be differentially expressed. (Fig. 3a; Suppl. Table 1): 73 genes were downregulated and and only one gene was upregulated in hepatocytes from PC tissues. Using GO analysis, oxidative-reductive processes were found to be the most down-regulated processes in PC tissues, with 8/73 (p>0.05, due to small n) belonging to this category. Other genes with decreased expression were associated with metabolic processes, such as steroid and alcohol metabolism genes and genes involved in small molecule and drug metabolism. Hepatic parenchyma from PC tissues had only one upregulated gene in contrast, ubiquitin D (UBD), which has been described earlier.(17) Notably, HCV RNA was detected in 51/54 hepatocyte segments; hence, differences in gene expression between captured hepatocytes from PC and NF liver tissue were more likely to reflect fibrosis rather than HCV replication.
Figure 3. Hepatocytes from PC and NF livers reveal asymmetric transcriptomes.
(a) Shown is a volcano plot of differentially expressed genes between hepatocytes from PC and NF liver tissue, defined by log2 absolute expression of genes in the PC group minus the absolute expression of genes in the NF group. Genes in black have an FDR < 0.05 and show > 2 fold differences in expression (b) Validation of the microarray is shown for selected genes. Median normalized expression levels for the respective microarray values (gray) and qPCR amounts (black) are given for a parenchymal specimen from each subject. The qPCR data was normalized to 7SL while the microarray data was normalized using rma. *p<0.05.
For validation, genes were randomly selected at different positions in the rank list so as not to bias the validation towards outlier genes and tested by qPCR using gene-specific primers. Representative captured material was tested from each subject, showing close agreement between microarray and qPCR results (Fig. 3b). Importantly, albumin was not differentially expressed between PC and NF hepatocytes, indicating that hepatic synthetic function was preserved in PC hepatocytes. BCHE had the most suppressed expression in PC tissues, showing 5-fold lower expression by microarray (FDR=2×10−4), and a log2 (FC) of 13.51 by qPCR (Fig. 4a), and was still significant after exclusion of the PC tissue with Ishak Stage 5.
Figure 4. Validation of differential BCHE expression in tissue and serum.
(a) Normalized log2 expression levels of BCHE by qPCR in representative hepatocytes (left) and by hybridization array in all hepatocytes (right) is shown in NF (solid squares) and PC (open squares) liver tissue. Downregulation of BCHE is shown in hepatocytes in PC compared to NF liver tissue (p<0.05 for both methods, Mann-Whitney-Wilcoxon test). (b, c, d & e) Subjects in the ALIVE cohort were selected for having staged liver disease by liver biopsy or transient elastography and available contemporaneous serum samples. SBA was tested (b) and compared to contemporaneous serum albumin (c), APRI (d) and, bilirubin (e) levels. Median values are shown with first and third quantiles in different groups of Ishak fibrosis stage. Adjusted p values shown were calculated using a pairwise Mann-Whitney-Wilcoxon test with a BH correction in R.
Validation of differential BCHE expression
BCHE protein is synthesized in the liver and widely distributed in the body, including plasma, brain, and lung. Notably, BCHE is the predominant enzyme that metabolizes cocaine and plays a role in heroin metabolism.(18–22) SBA, a surrogate of the highly polymorphic protein, was measured in an expanded sample of chronic HCV-infected participants to confirm and validate that gene expression differences were manifest at the level of protein expression. The features of the subcohort were characteristic of an urban drug-using population (Suppl. Table 2). Demographic characteristics in each fibrosis stage group were evenly distributed, including race, gender, HIV status, and HBsAg status, although persons with more severe fibrosis were generally older (p<0.01). We observed that SBA (mu/mL) decreased with increasing liver disease stage (Fig. 4b). For reference, serum albumin, bilirubin, and APRI levels were compared at contemporaneous time points (Fig 4c–e) and were similarly found to have significant changes with advanced liver disease.
Longitudinal BCHE expression
The role of BCHE in the pathogenesis of liver fibrosis is contingent on its decreased expression before the onset of advanced liver fibrosis. To test the temporal relationship of BCHE expression with liver fibrosis, SBA was measured in fibrosis progressors and non-progressors at four regularly-spaced time points that spanned the progression of fibrosis from minimal disease to cirrhosis in the progressors (Table 2). The median (range) duration of follow-up in progressors was 4 (1.5–6) years, and in non-progressors was 4.4 (1.6–6.2) years. Median SBA was lower in progressors compared to non-progressors at time point 1 (adj. p=0.04), time point 3 (adj. p=0.04), and time point 4 (adj. p= 0.00057). Indeed, all intermediate time points showed steady and significant decreases of SBA except between time points 1 and 2 (Fig. 5a). These results were compared with serum albumin, a well-characterized clinical markers of impaired liver synthetic function in persons with advanced liver disease. Serum albumin was only significantly lower in progressors compared to non-progressors after the establishment of cirrhosis (Fig. 5b). To confirm that earlier decreases of SBA compared to albumin were not simply due to methodologic differences in the performance of those assays, serum bilirubin was tested in the longitudinal cohort; bilirubin was not different between and within the two groups at any stage (data not shown). Interestingly, SBA in non-progressors was siginificantly lower at time point 3 (adj. p=0.007) and time point 4 (adj. p=0.0001) compared to time point 1, confirming that decreased BCHE expression occurs before the onset of significant fibrosis. In contrast, serum albumin levels were not different between any time points in the non-progressors confirming that its decrease is a result of impaired liver function.
Table 2.
Characteristics of Longitudinal Validation Cohort
|
Progressors N=19 |
Non-progressors N=20 |
||
|---|---|---|---|
| Male, n (%) | 4 (21) | 5 (25) | |
| Black, n (%) | 18 (95) | 20 (100) | |
| HIV Positive, n (%) | 9 (47) | 6 (30) | |
| HBVsAg Positive, n(%) | 0 (0) | 0 (0) | |
| Biopsy 1 | Median Age | 40.8 | 42.0 |
| Median Metavir score | 1.0 | 1.0 | |
| Biopsy 2 | Median Age | 45.3 | 45.9 |
| Median Metavir score | 1.0 | 0.5 | |
| Fibroscan 1 | Median Age | 50.6 | 50.9 |
| Median Transient Elastography Score (kPa) | 16.8 | 5.6 | |
| Fibroscan 2 | Median Age | 53.3 | 55.3 |
| Median Transient Elastography Score (kPa) | 17.1 | 6.7 |
Figure 5. BCHE protein expression is decreased before the onset of hepatic fibrosis.
(a) Median SBA and (b) serum ALB levels during 4 visits spanning an average of 11.75 years for progressors (gray) and non-progressors (black). Median values are depicted with first and third quantiles. The asterisks depict p < 0.05 in a Mann-Whitney-Wilcoxon test between progressors and non-progressors at each time point. The horizontal brackets depict p < 0.05 by paired Wilcoxin Signed Rank Test between time points within the progressor group and non-progressor group with significant differences in SBA and serum ALB levels.
Comparison of Hepatocytes and Portal Tracts
In the setting of chronic viral hepatitis, the portal tracts become chronically inflamed and contain mononuclear cells including B cells, T cells, and monocytes. The hepatic lobules also have chronic inflammation in chronic HCV, but generally less so. To confirm enrichment of cellular inflammation in portal tracts, eighteen segments from portal tract transcriptomes of the original 9 subjects were compared to 54 hepatocyte transcriptomes from the same subjects, yielding 801 differentially expressed genes: 71 genes were upregulated in portal tracts while 730 genes were upregulated in hepatocytes. Portal tracts had low overall expression, but the 71 upregulated genes were categorized into major biological processes under gene ontogeny (GO) using DAVID(16) to confirm the exclusion of contaminating mRNA from hepatic parenchyma. Portal tracts were enriched for immune system related GO categories such as immune system process (26% genes, adjusted p value = 1.8×10−5), immune response (17.1%, adjusted p value = 4.1×10−3), cell adhesion (18.6% genes, adjusted p value = 4.9×10−4) and locomotion (11.4%, adjusted p value = 0.03) In particular, the portal tract was enriched for the expression of chemokine genes and their receptors (CCL2, CCL19, CKLF, CCR5, CXCR4). Consistent with the role of chemokines in trafficking inflammatory cells, genes found in innate immune phagocytic cells (e.g., lysozyme) were upregulated in portal tracts, as were genes important in antigen-presenting cells (e.g., HLA-DQA1, HLA-DPA1). Genes associated with B-cells function (EBF1, BCL11B, PBXIP1, BCL2, BANK1) were differentially regulated with FDR < 0.05 but had a FC < 2. Molecules that form part of several downstream signaling cascades, such as IKBKB, and cell adhesion molecules such as ICAM1 were also upregulated in the portal tracts at FDR<0.05 but FC < 2.
Of the 730 genes upregulated in hepatic parenchyma, 725 were associated with GO categories. In contrast to portal tracts, only 43/725 (5.9%, adjusted p-value=1) were associated with the immune system process GO category and 35/725 (4.8%, adjusted p value = 0.82) were associated with immune response. The hepatic parenchyma immune related genes were primarily soluble immune proteins and/or acute phase reactants, such as complement factor B (FC=7, FDR = 1.2×10−10) and mannose-binding lectin (FC=2, FDR=0.004). Of the immune genes that were upregulated in parenchymal sections, several were well-described interferon-stimulated genes (ISGs), such as IFIT2 (FC=2.6, FDR=0.02), IFI6 (log2FC=2.6, FDR=8×10−4), and IFITM3 (FC=7, FDR=4×10−3). Among the genes upregulated in parenchymal sections that were not related to immune function, the majority corresponded largely to GO:0055114, oxidation-reduction processes (19.6%, p = 4.8×10−49) and other metabolic functions.
Comparing portal tracts in PC tissues to those in NF tissues revealed that most upregulation of immune process genes was due to portal tracts in the absence of fibrosis; 98 genes were upregulated in portal tracts from NF tissues while 79 genes were upregulated in portal tracts from PC tissues (Fig. 6). GO categories relating to immune response were enriched in the 26 genes common to both groups. An enrichment of immune related pathways was also observed in the 72 genes that were upregulated only in portal tracts from NF tissues. These 72 included genes involved in T cell activation and differentiation (e.g. NCK2, STAT5A, IL15) as well as cell adhesion molecules (CCL2 and CCL18) and inflammasome related genes (NLRC3, NLRC5). On the other hand, no GO categories were enriched in the 53 genes upregulated in portal tracts from PC tissues, suggesting that progression to fibrosis is associated with loss in immune related genes from the portal tracts.
Figure 6. Gene Ontology (GO) enrichment for biological processes differs between portal tracts in PC and NF liver tissue.
The number of genes upregulated genes in each group is shown. GO categories were assigned using DAVID and an adjusted p < 0.05 (Bonferroni correction) threshold for detection enrichment. The entire set of 11170 genes after removal of absent genes was used as the background for enrichment analysis.
Discussion
In this investigation, we found a novel gene, BCHE, which was downregulated expressed in persons years before the onset of cirrhosis. BCHE is the dominant enzyme that metabolizes cocaine, accounting for 95% of its metabolism. (18–22) Although the natural function of this enzyme remains unclear, it has been proposed as a treatment for cocaine overdose and addiction in humans.(23) This finding was possible because of the use of LCM to differentiate tissue compartments, and was twice-validated in a cohort of HCV-infected persons at different stages of liver disease. If more broadly confirmed, the finding might shed light on the pathogenesis of HCV related fibrosis in IDUs, the largest HCV risk group, and may represent a diagnostically useful disease marker.
The BCHE gene product is an abundant circulating enzyme that has been implicated in liver disease.(24) In an animal model of cocaine intoxication, BCHE −/− knockout mice developed significant hepatic necrosis compared to wildtype mice(25), a finding that is consistent with our detection of lower BCHE expression in human HCV-infected liver tissue taken from IDUs. BCHE is highly polymorphic in its expression, and variants of BCHE have been historically associated with prolonged recovery from anesthetic agents such as succinyl choline.(26) The ALIVE cohort members all have exposure to cocaine and/or heroin, which is the major transmission route of HCV in industrialized countries. Since BCHE is important in the metabolism of cocaine and heroin, its progressive loss with hepatic fibrosis may potentiate the progression of liver disease.
Other studies have examined transcriptional patterns in whole liver from persons with chronic HCV infection. Using expression microarrays, Smith et al. found that liver tissue from persons with chronic HCV and progressive fibrosis had upregulated expression of markers of myofibroblasts and myofibroblasts-like cells compared to HCV-infected persons without liver disease.(27) Similarly, Takahara et al. found that fibrotic liver tissue had higher levels of genes related to inflammation and extracellular matrix compared to normal liver in a cohort of chronically infected persons.(28) A major difference between the present study and earlier ones is the use of LCM to directly compare hepatocytes, rather than pooled hepatic tissue from heterogeneous cellular inputs. This distinction is important since with advancing fibrosis the cellular components of the liver change: inflammatory cells infiltrate the liver while hepatocytes are decreased in number. Indeed, in the present study differences in BCHE expression would not have been detected if high fibrosis transcriptomes were compared in bulk to low fibrosis transcriptomes. Similarly, in earlier studies diseased tissue was obtained from cirrhotic livers, but transcriptional profiles in cirrhosis likely represent the final stages of disease, and do not necessarily reflect the biologic processes that culminate in cirrhosis. In contrast, in the present study tissues from persons with pre-cirrhotic fibrosis (Ishak 3–5) were used to identify host determinants that play an early role in fibrogenesis.
It is important to note that the selection criteria for progressors and non-progressors included staging by biopsy and elastography to sufficiently power the longitudinal analysis and to span a long enough time to track the natural progression of liver disease in both groups. While it is possible that down-regulation of BCHE in tissues with early fibrosis was the result of poor global hepatic synthetic function, it is unlikely since albumin mRNA expression was preserved in the same tissues. Indeed, it is notable that BCHE loss was seen on a per cell basis from these tissues, at a stage of disease in which there was very little compromise of hepatic synthetic function. Moreover, SBA in the longitudinal cohort were decreased at the earliest time points in progressors compared to non-progressors, and at least 4 years before decreases in albumin were seen, indicating that BCHE loss is a predictive marker of future fibrosis progression. Decreased BCHE expression, therefore, is more likely to reflect early events rather than being the result of cirrhosis. Underscoring the early loss of BCHE expression, longitudinal testing confirmed that SBA was also decreased with time in non-progressors despite the lack of significant liver disease, strongly suggesting a role in pathogenesis. Complementary functional studies will be of benefit to confirm a causal role of BCHE in fibrosis progression.
It should be noted that while albumin was significantly down-regulated in portal tracts, its absolute expression was higher than expected. Since albumin is a highly expressed gene, its higher expression in portal tracts may have been due to small numbers of contaminating hepatocytes; however, the overall transcriptional signature of portal tracts was markedly different from hepatocyte transcriptomes. As expected, soluble immune markers were found in hepatocytes, while markers of cellular immunity appeared to be enhanced in portal tracts. We found progression to fibrosis was associated with a loss of expression of immune related genes in portal tracts. Cellular immunologic reprogramming may, therefore, contribute to fibrosis progression, although more detailed study is required to identify the key immunologic signatures that lead to fibrosis.
As part of the LCM strategy, hepatic parenchyma was separately captured by classical lobular zones into periportal, midzonal, centrilobular regions to identify differential patterns of expression in these distinct populations of hepatocytes. Previous work has identified a panel of markers, such as glutamine synthase and urea cycle genes, that allow molecular distinction of lobular zones.(29) In this study, however, a priori selected molecular markers did not discriminate zonal regions identified by LCM (data not shown). In addition, hierarchical clustering did not differentiate regions as assigned by their LCM designation irrespective of fibrosis stage. It is not clear from the present study whether chronic HCV infection itself altered the zonal expression of these molecular markers, or if LCM was of insufficient resolution to distinguish lobular zones. Future studies will include LCM of available uninfected tissues to test the effect of HCV infection on zonation. It will also be important to confirm these findings in other populations, since the discovery and validation cohorts were predominantly comprised of African-Americans.
By employing a novel method for differentiating hepatocytes from other cells, we detected differential expression of a series of genes in hepatocytes from HCV-infected subjects with pre-cirrhotic fibrosis compared to hepatocytes from livers with no fibrosis. Not only was BCHE mRNA expression different in the discovery tissues, but BCHE protein expression was also different years before fibrosis progression was detected. If confirmed, this finding would suggest roles for BCHE in detection of fibrosis and possibly in treatments to prevent fibrosis progression.
Supplementary Material
Acknowledgments
Financial Support: NIH DA 016078 (D.T.), AI 081544 (A.B.) and EY 001765 (Wilmer Core Grant)
We thank Eric Scholten for assistance with image analysis; and Jocelyn Ray for assistance in RNA processing.
List of Abbreviations
- LCM
Laser Capture Microdissection
- BCHE
Butyrylcholinesterase
- SBA
Serum BCHE activity
- NF
No Fibrosis
- PC
Pre-cirhottic
References
- 1.World Health Organization. Hepatitis C: global prevalence. Weekly Epidemiological Record. 1997;72:341–348. [Google Scholar]
- 2.Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006 May 16;144(10):705–714. doi: 10.7326/0003-4819-144-10-200605160-00004. [DOI] [PubMed] [Google Scholar]
- 3.Thomas DL, Seeff LB. Natural history of hepatitis C. Clin Liver Dis. 2005 Aug;9(3):383–98. vi. doi: 10.1016/j.cld.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000 Jan 28;275(4):2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 5.Lin W, Tsai WL, Shao RX, Wu G, Peng LF, Barlow LL, et al. Hepatitis C virus regulates transforming growth factor beta1 production through the generation of reactive oxygen species in a nuclear factor kappaB-dependent manner. Gastroenterology. 2010 Jun;138(7):2509–18. 2518. doi: 10.1053/j.gastro.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau DT, Luxon BA, Xiao SY, Beard MR, Lemon SM. Intrahepatic gene expression profiles and alpha-smooth muscle actin patterns in hepatitis C virus induced fibrosis. Hepatology. 2005 Aug;42(2):273–281. doi: 10.1002/hep.20767. [DOI] [PubMed] [Google Scholar]
- 7.Rai R, Wilson LE, Astemborski J, Anania F, Torbenson M, Spoler C, et al. Severity and correlates of liver disease in hepatitis C virus-infected injection drug users. Hepatology. 2002 May;35(5):1247–1255. doi: 10.1053/jhep.2002.33151. [DOI] [PubMed] [Google Scholar]
- 8.Wilson LE, Torbenson M, Astemborski J, Faruki H, Spoler C, Rai R, et al. Progression of liver fibrosis among injection drug users with chronic hepatitis C. Hepatology. 2006 Apr;43(4):788–795. doi: 10.1002/hep.21091. [DOI] [PubMed] [Google Scholar]
- 9.Kirk GD, Astemborski J, Mehta SH, Spoler C, Fisher C, Allen D, et al. Assessment of liver fibrosis by transient elastography in persons with hepatitis C virus infection or HIV-hepatitis C virus coinfection. Clin Infect Dis. 2009 Apr 1;48(7):963–972. doi: 10.1086/597350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003 Apr;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 11.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004 Feb 12;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 13.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 14.Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002 Dec;18(12):1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- 15.Benjamini Y. In: Controlling the False Discovery Rate: A Paractical and Powerful Approach to Multiple Testing. 57. Hochberg Y, editor. 1995. pp. 289–300. [Google Scholar]
- 16.Huang dW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith MW, Yue ZN, Korth MJ, Do HA, Boix L, Fausto N, et al. Hepatitis C virus and liver disease: global transcriptional profiling and identification of potential markers. Hepatology. 2003 Dec;38(6):1458–1467. doi: 10.1016/j.hep.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 18.Gorelick DA. Enhancing cocaine metabolism with butyrylcholinesterase as a treatment strategy. Drug Alcohol Depend. 1997 Dec 15;48(3):159–165. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 19.Kamendulis LM, Brzezinski MR, Pindel EV, Bosron WF, Dean RA. Metabolism of cocaine and heroin is catalyzed by the same human liver carboxylesterases. J Pharmacol Exp Ther. 1996 Nov;279(2):713–717. [PubMed] [Google Scholar]
- 20.Brzezinski MR, Abraham TL, Stone CL, Dean RA, Bosron WF. Purification and characterization of a human liver cocaine carboxylesterase that catalyzes the production of benzoylecgonine and the formation of cocaethylene from alcohol and cocaine. Biochem Pharmacol. 1994 Nov 1;48(9):1747–1755. doi: 10.1016/0006-2952(94)90461-8. [DOI] [PubMed] [Google Scholar]
- 21.Dean RA, Zhang J, Brzezinski MR, Bosron WF. Tissue distribution of cocaine methyl esterase and ethyl transferase activities: correlation with carboxylesterase protein. J Pharmacol Exp Ther. 1995 Nov;275(2):965–971. [PubMed] [Google Scholar]
- 22.Salmon AY, Goren Z, Avissar Y, Soreq H. Human erythrocyte but not brain acetylcholinesterase hydrolyses heroin to morphine. Clin Exp Pharmacol Physiol. 1999 Aug;26(8):596–600. doi: 10.1046/j.1440-1681.1999.03090.x. [DOI] [PubMed] [Google Scholar]
- 23.Gao Y, LaFleur D, Shah R, Zhao Q, Singh M, Brimijoin S. An albumin-butyrylcholinesterase for cocaine toxicity and addiction: catalytic and pharmacokinetic properties. Chem Biol Interact. 2008 Sep 25;175(1–3):83–87. doi: 10.1016/j.cbi.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogunkeye OO, Roluga AI. Serum cholinesterase activity helps to distinguish between liver disease and non-liver disease aberration in liver function tests. Pathophysiology. 2006 May;13(2):91–93. doi: 10.1016/j.pathophys.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Duysen EG, Li B, Carlson M, Li YF, Wieseler S, Hinrichs SH, et al. Increased hepatotoxicity and cardiac fibrosis in cocaine-treated butyrylcholinesterase knockout mice. Basic Clin Pharmacol Toxicol. 2008 Dec;103(6):514–521. doi: 10.1111/j.1742-7843.2008.00259.x. [DOI] [PubMed] [Google Scholar]
- 26.Xie W, Altamirano CV, Bartels CF, Speirs RJ, Cashman JR, Lockridge O. An improved cocaine hydrolase: the A328Y mutant of human butyrylcholinesterase is 4-fold more efficient. Mol Pharmacol. 1999 Jan;55(1):83–91. doi: 10.1124/mol.55.1.83. [DOI] [PubMed] [Google Scholar]
- 27.Smith MW, Walters KA, Korth MJ, Fitzgibbon M, Proll S, Thompson JC, et al. Gene expression patterns that correlate with hepatitis C and early progression to fibrosis in liver transplant recipients. Gastroenterology. 2006 Jan;130(1):179–187. doi: 10.1053/j.gastro.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Takahara Y, Takahashi M, Zhang QW, Wagatsuma H, Mori M, Tamori A, et al. Serial changes in expression of functionally clustered genes in progression of liver fibrosis in hepatitis C patients. World J Gastroenterol. 2008 Apr 7;14(13):2010–2022. doi: 10.3748/wjg.14.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brosnan ME, Brosnan JT. Hepatic glutamate metabolism: a tale of 2 hepatocytes. Am J Clin Nutr. 2009 Sep;90(3):857S–861S. doi: 10.3945/ajcn.2009.27462Z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.