Abstract
Recent studies have discovered strong differences between the dynamics of nucleic acids (RNA and DNA) and proteins, especially at low hydration and low temperatures. This difference is caused primarily by dynamics of methyl groups that are abundant in proteins, but are absent or very rare in RNA and DNA. In this paper, we present a hypothesis regarding the role of methyl groups as intrinsic plasticizers in proteins and their evolutionary selection to facilitate protein dynamics and activity. We demonstrate the profound effect methyl groups have on protein dynamics relative to nucleic acid dynamics, and note the apparent correlation of methyl group content in protein classes and their need for molecular flexibility. Moreover, we note the fastest methyl groups of some enzymes appear around dynamical centers such as hinges or active sites. Methyl groups are also of tremendous importance from a hydrophobicity/folding/entropy perspective. These significant roles, however, complement our hypothesis rather than preclude the recognition of methyl groups in the dynamics and evolution of biomolecules.
Electronic supplementary material The online version of this article (doi:10.1007/s10867-012-9268-6) contains supplementary material, which is available to authorized users.
Keywords: Protein dynamics, RNA world, RNA dynamics, Nucleic acid dynamics
Introduction
It is recognized that atomic and molecular motions (dynamics) are crucial to the function of biological macromolecules [1–5]. Yet our understanding of microscopic details of molecular motions, their relationship to chemical structure, and how they translate into the function of biomolecules remain very limited. Earlier analysis revealed that the dynamics of proteins at ambient conditions are faster than dynamics of RNA and DNA. Moreover, dynamics of RNA and DNA slow down with cooling much more rapidly than the dynamics of proteins [6]. This difference appears especially strong at lower hydration levels [6, 7]. Detailed experimental and computational studies ascribed this difference to dynamics of methyl groups, which are abundant in proteins and essentially are absent in tRNA [6, 8–11]. Methyl groups have relatively low energy barriers for rotation and, as a result, methyl group dynamics in proteins remain active, even at low hydration levels or low temperatures. This facilitates dynamics within the entire protein and allows the molecules to access a more diverse set of configurations and to sample various conformational states, thus increasing the conformational entropy.
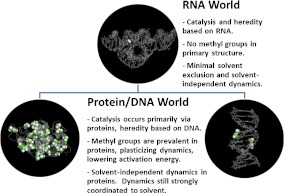
The unexpected observation that methyl groups indeed facilitate the dynamics of proteins, calls our attention to the importance methyl groups might play in facilitating the associated biological functions. In this paper, we formulate a hypothesis (sketched in Fig. 1) based on the implications of these recent studies. We postulate that solvent-independent methyl dynamics were advantageous in the evolution of biomolecules from an RNA-dominated world, lacking methyl dynamics, to the protein/RNA/DNA life we see today. We present experimental evidence showing the difference in dynamics of RNA and proteins, an analysis of methyl group content in biological macromolecules from various classes of proteins and nucleic acids, and analyze positions of the fastest methyl groups in several proteins with respect to the active site. The results of the presented analysis are consistent with the formulated hypothesis. Thus, if true, the hypothesis creates a new way to look at the role of individual structural units (e.g., side groups) in dynamics, activity, and evolution of biological macromolecules.
Fig. 1.
Summary of the hypothesis highlighting the evolutionary trend from RNA-based life, low in methyl content and lacking methyl plasticized dynamics, to protein and DNA-based life that has numerous methyl groups and solvent-independent methyl dynamics
Materials and methods
Neutron scattering data discussed for lysozyme and tRNA were measured with the High-Flux Back-Scattering instrument at the National Institute of Standards and Technology and previously published [7]. Similarly, for DNA measurements, we used the previously-published data obtained using the back-scattering spectrometer at the Research Center Julich in Germany [12]. Details of the measurement and sample preparation can be found in these references. Details of molecular dynamics simulations of lysozyme are presented in Roh et al. [13].
Analysis of methyl content in various classes of biomolecules is based on the number of methyl groups in each molecule, divided by the molecular weight. The content for RNA and DNA was calculated using the average value for each of the primary four nucleotides, whereas the content for protein was calculated as an average of the 78 proteins listed in the database contained in the Supplementary Information. The comparative analysis of methyl content in various classes of proteins was expressed as a ratio of methyl group content to total number of residues. Twenty-three ribosomal proteins, 43 enzymes, and 12 extracellular matrix proteins were analyzed. The exact proteins used are listed in the Supplementary Information.
Results and discussion
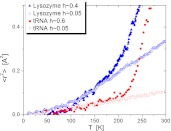
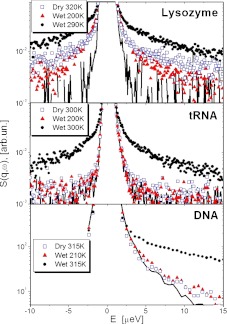
The mean-squared atomic displacements (<r2 >) on the ~1 ns time scale, measured with neutron scattering, demonstrate that <r2 > at biological temperatures (~300 K) are approximately three times larger in dehydrated proteins than in dehydrated tRNA (Fig. 2). The relaxation dynamics of these biomolecules, seen in Fig. 3, appear in scattering measurements as a quasielastic intensity, or a broadening of the central peak. The direct comparison of the neutron scattering spectra of proteins, RNA and DNA shows significant differences at low hydration levels and low temperatures. While protein spectra exhibit significant quasielastic intensity, the spectra of RNA and DNA under the same conditions present just the resolution function. The latter means that the dynamics of RNA and DNA on the ns time scale appear essentially frozen at these conditions [6, 7, 9]. These data show the presence of low activation-energy dynamic processes in the protein molecules, and not in the nucleic acids.
Fig. 2.
Differences in the dynamics of RNA and proteins. Mean squared atomic displacements on a ~1 ns time scale. Comparing hydrated and dry lysozyme (filledandopen triangles, respectively) to hydrated and dry tRNA (filled and open circles). Hydration levels, h, are in g of water per 1 g of biomolecules. The solvent-independent, methyl-group dynamic onset in protein appears at ~100 K and the subsequent hydration-dependent onset at ~200 K in both RNA and proteins (data from [7])
Fig. 3.
Quasielastic neutron scattering spectra of lysozyme, tRNA and DNA (symbols); lines show the resolution function of spectrometers. Data for lysozyme and tRNA are from [7], for DNA from [20]. Lysozyme spectra exhibit relaxation contribution even in the dry state and at T = 200 K, while no quasielastic scattering is observed for dry tRNA and DNA, or for hydrated tRNA and DNA below 210 K
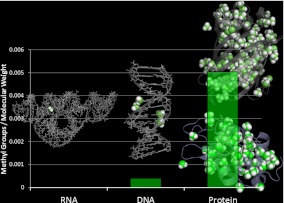
These processes are the rotation and fluctuations of methyl groups, the lowest activation-energy dynamic processes in biological macromolecules. Our observation on methyl group dynamics suggests another evolutionary advantage of proteins—their greater diversity of dynamical processes and the lower energies required for conformational changes. Our initial investigation of this hypothesis is based on the direct comparison of the number of methyl groups in different classes of biomolecules. We will not consider here post-transcriptional methylations of RNA and DNA, although it is conceivable that methylations of nucleic acids such as the cap of mRNA will also have important functions in solvent-excluded regions (such as within the ribosome). There are no methyl groups in the primary structure of RNA while one of DNA’s four nucleic acids contains a single methyl group. Six of the 20 standard amino-acids found in proteins, however, contain methyl groups. Clearly, the number of methyl groups in proteins is significantly higher than in DNA or RNA (Fig. 4).
Fig. 4.
Methyl content in different classes of biomolecules. It is presented in number of methyl groups per molecular weight. Post-translational and post-transcriptional modifications have been disregarded
It is interesting therefore to consider that the amount of methyl groups in biological macromolecules might reflect the evolutionary process. The RNA world hypothesis [14] proposes that RNA preceded proteins and DNA as the molecule of life. This was spawned by the discovery of enzymatic RNAs in the 1980s [15], leading to Gilbert’s theory [14]. RNA-based organisms would have fulfilled the fundamental requirements for life by being both self-replicating and metabolically/catalytically active. The prevailing thought is that the hereditary fidelity of DNA was superior to RNA and proteins are better catalytic agents than ribozymes, and recent work supports this assertion [16]. Why then did RNA also lose its role as primary catalytic agent? The diversity of chemical groups is one of the obvious benefits of proteins over RNA as catalysts. Chemically and structurally, the ‘repertoire’ of biomolecules was expanded from four bases, and a backbone, with functional groups highly interactive with the solvent and each other, to 20 distinct side chains which contain a far wider array of functionality and interaction with each other, potential ligands, and the aqueous environment. However, this also introduced large regions of solvent exclusion and the molecular motions contained within these regions should not be ignored. Such solvent-independent dynamics are typified by methyl group dynamics, which we again point out, are absent in RNA. One can envision how such flexibility (or lack thereof) could translate into the energy barrier involved in enzymatic action. In material terms, we suggest that methyl groups are plasticizing agents in proteins, facilitating dynamics and, subsequently, activity.
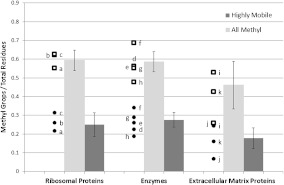
Consideration of this hypothesis suggests that we should look at the role of methyl dynamics in various proteins. For example, traditional model enzymes such as lysozyme and ribonuclease A, and transport proteins such as myoglobin and hemoglobin, have significant numbers of methyl groups in their primary structure, ~0.6 methyl groups per residue, while the structural proteins such as collagen have as few as ~0.3 methyl groups per residue (Fig. 5). This suggests a connection between the required functional flexibility or dynamic activity and the amount of methyl groups in the protein. To investigate this hypothesis further, we have averaged the quantity of methyl groups in a sample of ribosomal, enzymatic, and extracellular proteins (Fig. 5; a detailed list of proteins is presented in the Supplementary Information). Indeed, proteins involved in extracellular matrix roles (collagen, laminin, etc.) have as many as 25% fewer methyl groups per residue. This result is potentially significant: If methyl groups are acting to plasticize proteins and to lower energy barriers for catalytic activity, then it is logical to expect that proteins which function enzymatically would be enriched in plasticizers (methyl groups), and proteins functioning structurally would be depleted in methyl groups.
Fig. 5.
Number of methyl groups and highly mobile (Ile δ, Leu, Met) methyl groups per residue for selected protein classes. Error bars depict the standard deviation. Representative data points have been included; (a) 40S ribosomal protein S11, (b) 40S ribosomal protein S17, (c) 60S ribosomal protein L30, (d) glucose oxidase, (e) calmodulin, (f) adenylate cyclase, (g) dihydrofolate reductase, (h) lysozyme, (i) laminin α-chain, (j) collagen, and (k) plasminogen
More detailed analysis focuses on the fact that not all methyl groups are equal in terms of dynamics. Molecular dynamics simulations and NMR data reveal [11, 17–19] that among the methyl groups the methionine methyl is the most dynamic (i.e., has the lowest energy barrier for rotation), followed by leucine and then the delta methyl group on isoleucine. Of course, their individual energy barriers depend on the position of the residue in the protein 3-D structure. Nevertheless, on average, they remain the fastest, and we will refer to them as ‘highly mobile methyl’ groups. Analysis of the content of highly mobile methyl groups (Fig. 5) reveals that the extracellular matrix proteins have on average ~32% less highly mobile methyl groups than enzymes.
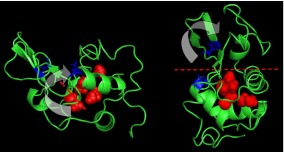
Detailed analysis of distribution of methyl groups in a 3-D structure of biomolecules has only been done for a few proteins. It has been noted that the highly mobile methyl groups in lysozyme are buried near the active site of the enzyme, thus facilitating dynamics in the most functionally important part of the molecule [20]. We illustrate this point in Fig. 6, which shows the spatial distribution of methyl groups in lysozyme. We have superimposed the methyl carbons (red spheres) over the translations associated with binding the substrate (red lines). We can see the abundance of methyl groups in the area centered on this hinge motion (lower right quadrant). Krishnan et al. also analyzed the dynamics of myoglobin using computer simulations [8] and found that the fastest methyl groups are placed in the vicinity of the xenon cavities in the protein. Xenon cavities in myoglobin are crucial for substrate migration to and from the active site. These correlations suggest that the fastest methyl groups might be directly involved in, if not critical to, the function of this protein. Experimental studies of the influence of methyl group deleting mutations on the activity of calmodulin [13] present even more direct evidence of the role of methyl-containing residues in the protein’s function. It was observed that removal of the methyl groups found in the latch region of calmodulin resulted in a significant reduction of activity [20], independent of substrate binding. This observation too is consistent with our hypothesis that methyl groups, and their associated dynamics are critical to protein activity.
Fig. 6.
Distribution of the most dynamically active methyl groups in lysozyme (red spheres), based on simulation and defined as the methyl groups with the lowest activation barrier to rotation. The protein is presented along the hinge axis and rotated 90 degrees. The blue residues are the critical active residues (E35 and D52). We direct the reader’s attention to the proximity of these most dynamic methyl groups and the axis of the hinge bending
The plasticization of proteins by the presence of methyl groups may also affect the existence of stable tertiary structures in proteins compared to those observed in RNA. Dynamically, methyl protons are the largest contributor to the observed amplitude of motion in proteins. One can speculate that methyl groups influence both the folding process through solvent exclusion and the attainment of well-defined, thermodynamically stable structures. The motion of methyl groups in proteins contributes a significant amount of entropy that stabilizes the structure of proteins beyond that which is achievable in RNA. Thus methyl groups provide both the well-recognized hydrophobic contribution to protein folding and an additional entropic contribution to the Gibbs free energy. From an evolutionary perspective, the inclusion of methyl groups may give proteins the dual advantage over RNA of both facilitating dynamically required motions and providing the thermodynamically favorable species to expedite protein folding and/or stabilize the uniquely defined equilibrium structures. We note that stabilization of RNA structure is quite different from that of proteins due to the significant role of charge interactions in RNA.
It is also known that modern nucleic acid biomolecules are often post-transcriptionally methylated. We have no way of knowing if ancient RNA-based life was capable of utilizing methylation, but it does exist in modern organisms. DNA methylation is a common mechanism of gene deactivation in higher animals, and is one of the most common post-transcriptional modifications of RNA products. Methylations have been reported in rRNA, tRNA, and in the 3′ cap of mRNA. The dynamics of these molecules have not been investigated as closely as proteins to this point but we hypothesize that methyl groups act as intrinsic plasticizers in these systems as well. The role of post-transcriptional methylation is an interesting topic; however, it is beyond the scope of the current paper, and is a subject for future work.
Conclusions
We have presented a hypothesis that early life based on RNA for both catalysis and gene storage lacked methyl groups, and subsequently, lacked access to the low-energy dynamic processes provided by methyl groups. These processes plasticize the dynamics of proteins due to their low activation energies and remain active even at low hydrations and temperatures, particularly important for extreme conditions. This is a new perspective with which to consider the progression of biomolecule evolution: A lowering of activation energy required to initiate dynamic processes associated with catalysis provided an advantage to organisms functioning with early proteins. We suggest that this be added to the list of the driving forces of evolution from RNA, for both storage of genetic material and metabolic catalysis, to DNA for storage and proteins for the vast majority of catalysis. Finally, we suggest a comparative study of ribozyme and enzyme dynamics. We hypothesize that the dynamical differences will be strongly connected with the energetic efficiency of their catalytic activity, illustrating what we interpret as an evolutionary advantage.
Electronic Supplementary Material
Funding
JDN and APS acknowledge DOE support through the EPSCoR program (grant DE-FG02–08ER46528) and support from SNS through UT-Battelle. HON acknowledges support of the Center for Structural Molecular Biology (CSMB) funded by the Office of Biological and Environmental Research under FWP ERKP291, using facilities supported by the U.S. Department of Energy. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the U.S. Department of Energy under contract No. DE-AC05–00OR22725.
Contributor Information
Jonathan D. Nickels, Phone: +1-865-9743852, Email: jnickels@alumni.nd.edu
Alexei P. Sokolov, Email: sokolov@utk.edu
References
- 1.Beece D, Eisenstein L, Frauenfelder H, Good D, Marden MC, Reinisch L, Reynolds AH, Sorensen LB, Yue KT. Solvent viscosity and protein dynamics. Biochemistry. 1980;19:5147–5157. doi: 10.1021/bi00564a001. [DOI] [PubMed] [Google Scholar]
- 2.Eisenmesser EZ, Bosco DA, Akke M, Kern D.Enzyme dynamics during catalysis Science 20022951520–1523.2002Sci...295.1520E 10.1126/science.1066176 [DOI] [PubMed] [Google Scholar]
- 3.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D.Intrinsic dynamics of an enzyme underlies catalysis Nature 2005438117–121.2005Natur.438..117E 10.1038/nature04105 [DOI] [PubMed] [Google Scholar]
- 4.Henzler-Wildman K, Kern D.Dynamic personalities of proteins Nature 2007450964–972.2007Natur.450..964H 10.1038/nature06522 [DOI] [PubMed] [Google Scholar]
- 5.Tolman JR.Structural biology: protein dynamics from disorder Nature 20094591063–1064.2009Natur.459.1063T 10.1038/4591063a [DOI] [PubMed] [Google Scholar]
- 6.Khodadadi S, Roh JH, Kisliuk A, Mamontov E, Tyagi M, Woodson SA, Briber RM, Sokolov AP.Dynamics of biological macromolecules: not a simple slaving by hydration water Biophys. J. 2010981321–1326.2010BpJ....98.1321K 10.1016/j.bpj.2009.12.4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caliskan G, Briber RM, Thirumalai D, Garcia-Sakai V, Woodson SA, Sokolov AP. Dynamic transition in tRNA is solvent induced. J. Am. Chem. Soc. 2006;128:32–33. doi: 10.1021/ja056444i. [DOI] [PubMed] [Google Scholar]
- 8.Krishnan M, Kurkal-Siebert V, Smith JC. Methyl group dynamics and the onset of anharmonicity in myoglobin. J. Phys. Chem. B. 2008;112:5522–5533. doi: 10.1021/jp076641z. [DOI] [PubMed] [Google Scholar]
- 9.Roh JH, Briber RM, Damjanovic A, Thirumalai D, Woodson SA, Sokolov AP.Dynamics of tRNA at different levels of hydration Biophys. J. 2009962755–2762.2009BpJ....96.2755R 10.1016/j.bpj.2008.12.3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roh JH, Novikov VN, Gregory RB, Curtis JE, Chowdhuri Z, Sokolov AP.Onsets of anharmonicity in protein dynamics Phys. Rev. Lett. 200595381012005PhRvL..95c8101R 10.1103/PhysRevLett.95.038101 [DOI] [PubMed] [Google Scholar]
- 11.Wood K, Tobias DJ, Kessler B, Gabel F, Oesterhelt D, Mulder FAA, Zaccai G, Weik M. The low-temperature inflection observed in neutron scattering measurements of proteins is due to methyl rotation: direct evidence using isotope labeling and molecular dynamics simulations. J. Am. Chem. Soc. 2010;132:4990–4991. doi: 10.1021/ja910502g. [DOI] [PubMed] [Google Scholar]
- 12.Sokolov AP, Grimm H, Kisliuk A. Slow relaxation process in DNA. J. Biol. Phys. 2001;27:313–327. doi: 10.1023/A:1014228824104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vugmeyster L, Ostrovsky D, Ford JJ, Lipton AS. Freezing of dynamics of a methyl group in a protein hydrophobic core at cryogenic temperatures by deuteron NMR spectroscopy. J. Am. Chem. Soc. 2010;132:4038–4039. doi: 10.1021/ja909599k. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert W.Origin of life—the RNA world Nature 1986319618–618.1986Natur.319..618G 10.1038/319618a0 [DOI] [Google Scholar]
- 15.Guerrier-Takada CGK, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease-P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 16.Leu K, Obermayer B, Rajamani S, Gerland U, Chen IA. The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA. Nucleic Acids Res. 2011;39:8135–8147. doi: 10.1093/nar/gkr525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong KB, Daggett V. Barstar has a highly dynamic hydrophobic core: evidence from molecular dynamics simulations and nuclear magnetic resonance relaxation data. Biochemistry. 1998;37:11182–11192. doi: 10.1021/bi980552i. [DOI] [PubMed] [Google Scholar]
- 18.Bajaj VS, Wel PCA, Griffin RG. Observation of a low-temperature, dynamically driven structural transition in a polypeptide by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2009;131:118–128. doi: 10.1021/ja8045926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roh JH, Curtis JE, Azzam S, Novikov VN, Peral I, Chowdhuri Z, Gregory RB, Sokolov AP.Influence of hydration on the dynamics of lysozyme Biophys. J. 2006912573–2588.2006BpJ....91.2573R 10.1529/biophysj.106.082214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su ZH, Fan DJ, George SE. Role of domain-3 of calmodulin in activation of calmodulin-stimulated phosphodiesterase and smooth-muscle myosin light-chain kinase. J. Biol. Chem. 1994;269:16761–16765. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.