Living organisms are highly organized adaptive systems. Whereas the maintenance of organization requires conservation, adaptation requires variation. Evolution, the fundamental strategy of Life, is an interplay of genetic variation and phenotypic selection. The wide variety of spontaneous mutation rates among natural isolates of bacteria show that the capacity for producing genetic variation is itself heritable (1, 2). Genetic variation results from somewhat imprecise reproduction due to a less-than-perfect fidelity of the transmission of genomic sequences. Could it be that the dialectic of conservation and change is reflected in the elementary act of DNA replication such that one strand is replicated with higher fidelity (conservation) than the other strand (change)? Fijalkowska et al. (3) suggest, in this issue of the Proceedings, that there is unequal fidelity in the copying of two complementary DNA template strands in the course of replication of the Escherichia coli chromosome.
Fijalkowska et al. (3) base their conclusion on the observation that the frequencies of specific point mutations change up to 5-fold when the mutational site is inverted. The mutational site is always the same codon in the lacZ gene, and mutation is measured as reversion frequency for a set of lac− mutants that are known to revert only by mutation to the original sequence (4). The authors justifiably argue that the two orientations of the DNA segment containing the lacZ gene, at the same integration site (att lambda), would “preserve all aspects of DNA metabolism, with the direction of replication fork movement relative to the gene as the only exception.” Because this assumption is fundamental to the interpretation of the results, it is important to think of all alternatives. There are at least two other parameters that can potentially affect the precision of DNA replication upon inversion: direction of transcription relative to replication (discussed by the authors) and the distance of the mutational site from some fixed chromosomal points. If spontaneous mutations occur in a polarized way with fixed points of increased mutation rates distributed all along the chromosome, then changing the distance from such points could change the mutation frequency (5). Such sites could be as frequent as initiation sites for Okazaki fragments. Because the effect of inversion on mutation frequency was tested only on one mutational site (codon) and at a single chromosomal location, one cannot dismiss this possibility. However, the interpretation of the data is compelling, and it is supported by previous findings of radically different mutagenicity of some DNA lesions (6) and sequences (7) when residing in one or the other complementary strand. DNA sequences such as inverted repeats and G+C-rich trinucleotide repeats (involved in several human hereditary neuromuscular diseases) can form hairpin-like secondary structures (7). Such secondary structures in the template strand can act as barriers to elongation of DNA synthesis and thus cause deletions in the newly synthesized strand through a replicative bypass of such structures. Alternatively, expansions of trinucleotide repeats can occur by replicative idling—i.e., repeated polymerase slippage at the site of the secondary structure. Obviously, polymerase slippage cannot account for all base substitution mutagenesis.
What aspect of the asymmetry of the replication fork is relevant to the unequal replicational fidelity of the two strands and at which biochemical step of DNA replication, or its editing, does the inequality occur? Is the asymmetry of leading (continuous synthesis) versus lagging (discontinuous synthesis) strand responsible for the asymmetry in fidelity? A little bit of mystery still lingers as to the nature and extent of discontinuity of DNA synthesis in vivo. The original paper by Okazaki et al. (8) reported discontinuous synthesis on both strands, with the lagging strand being far more discontinuous. Furthermore, ligase-deficient mutants of both E. coli and yeast revealed similarly high discontinuity on both strands (9–11). Therefore, it is not clear whether the strand difference in (dis)continuity is replicational or ligational. Could it be that both strands are replicated equally discontinuously after strand separation in front of the replication fork, but the ligation of the leading strand is, as would be expected, faster? DNA strand discontinuities (nicks) are required for mismatch repair in all organisms tested (12), such that error correction by mismatch repair could be more efficient on the lagging strand. However, although Fijalkowska et al. suggest that lagging strand synthesis is at least 5 times more accurate than leading strand synthesis, they exclude that either mismatch repair or polymerase-associated exonucleolytic proofreading is responsible for the asymmetry in accuracy, because mutants deficient in the former (mutL) and/or latter (mutD5) editing mechanism showed similar asymmetry. The authors therefore consider that the asymmetry in fidelity occurs in the elementary act of DNA synthesis, which is nucleotide insertion and elongation.
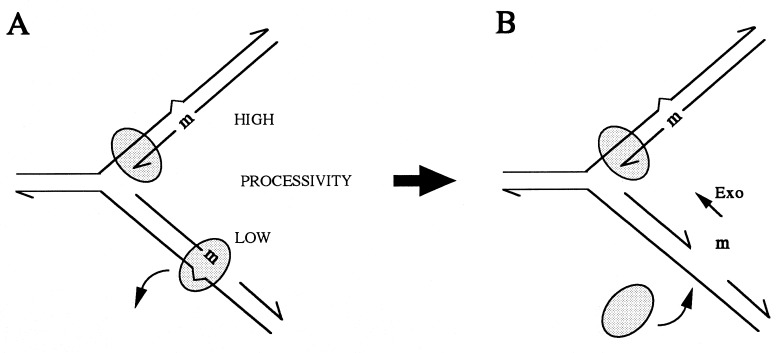
The differences in the fidelity of diverse DNA polymerases are manifested much more in their capacity to elongate DNA after an erroneously inserted nucleotide (error tolerance) than in their propensity to make the mistake (misinsertion) (13). Because only surviving mistakes become mutations, only those nucleotide misinsertions that are followed by elongation of the DNA will become mutations if not removed by mismatch repair. Thus, high-fidelity DNA polymerases do not extend synthesis from a mismatched strand terminus unless the mismatched nucleotide is removed by some 3′-end-specific exonuclease activity (5, 13). Therefore, the authors prefer the hypothesis that the E. coli replicative complex, DNA polymerase III (Pol III) holoenzyme, dissociates from the terminal mistake more readily when on the lagging than when on the leading strand. The abandoned mismatched end can be removed by any 3′-end-specific exonuclease, and the Pol III holoenzyme can resume the lagging strand replication. The presumably higher processivity of DNA synthesis on the leading strand would increase the probability of extension of a mismatched terminus, hence its lower fidelity (Fig. 1). The above hypothesis proposed by Fijalkowska et al. is attractive, although one cannot dismiss the possibility that the nucleotide selection is lower on leading than on lagging strand. For example, if leading strand synthesis is faster (in a stepwise progression mode), its fidelity will be diminished. Along the lines of their hypothesis and of their discovery of dnaE antimutator mutants encoding the DNA-synthesizing subunit of the Pol III holoenzyme (15), it is clear that, in these antimutators, the fidelity of the leading (lower fidelity) strand must have been improved. Perhaps only leading strand replication fidelity has been improved in dnaE antimutators—e.g., by a decrease in processivity or velocity?
Figure 1.
Strandedness, processivity, and fidelity in DNA replication. Leading strand synthesis is probably more processive than lagging strand synthesis. Therefore, according to Fijalkowska et al. (3), replication errors (m) in the leading strand are more readily fixed by the extension of DNA synthesis. Replication errors in the lagging strand are more likely to cause dissociation of the replication complex (A), leaving the mismatched 3′ terminus free for excision by some cellular exonuclease (free m), allowing the replicase to resume synthesis (B). Mutation (m) is a misincorporated nucleotide; the shaded elipsoid is the DNA replication complex.
How would unequal fidelity of replication of the two strands of a single DNA molecule withstand selective evolutionary pressure? Particularly in the case of bacteria, if one strand acted as a more faithful keeper of evolutionary memory, the other providing more mutations for adaptation, they should immediately compete because the progeny of the two strands would occupy the same space (molecular sib competition or sib selection?). When low mutation rates are favored, there should be a strong selection for improved fidelity of the leading (lower-fidelity) strand, whereas when high mutation rates are favored, decreased fidelity of the lagging strand, or of both strands, would be selected for. The persistence of unequal mutation rates of the two strands may be due to frequently alternating selections for low and high mutation rates in the evolutionary history of bacteria. Indeed, modeling of bacterial adaptive evolution (16, 17), experimental evolution of bacteria (18), and studies of mutation rates among natural isolates of E. coli (1, 2) all suggest that there are frequent selective pressures for increased mutation rates during bacterial evolution. Frequent alternations of feast and famine may favor the conservation/change dialectic in the act of bacterial DNA replication.
Because DNA replication is usually bidirectional and because many organisms have a large number of replication origins, the mutational bias would not affect entire DNA strands in a single block, but the mutational effect would be patchy. Such a mutational bias is expected to affect particularly some error-prone DNA sequences which would be consistently replicated by either leading or lagging strand synthesis. It is remarkable that strand compositional bias found in some bacterial genomes correlates with replicational asymmetry expected from the position of the origin and termination of DNA replication (19). Some particular sequences are expected to evolve much faster than others. Mutation rates at blocks of many kinds of simple repetitive sequences, such as microsatellites, are expected to be very sensitive to the strandedness (20). As for base substitution mutagenesis, one should keep in mind that spontaneous chemical modifications (base losses, oxidations, deaminations, alkylations, etc.) may be at least as frequent sources of mutations as are genuine replication errors (21).
Footnotes
The companion to this commentary is published on pages 10020–10025.
References
- 1.Le Clerc J E, Bauguang L, Payne W L, Cebula T A. Science. 1997;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 2.Matic I, Radman M, Taddei F, Picard B, Bingen E, Denamur E, Eion J. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- 3.Fijalkowska I J, Jonczyk P, Tkaczyk M M, Bialoskorska M, Schaaper R M. Proc Natl Acad Sci USA. 1998;95:10020–10025. doi: 10.1073/pnas.95.17.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cupples C G, Miller J H. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marians K J. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 6.Veaute X, Fuchs R P P. Science. 1993;261:598–600. doi: 10.1126/science.8342022. [DOI] [PubMed] [Google Scholar]
- 7.Sinden R R, Wells R D. Curr Opin Biotechnol. 1992;3:612–622. doi: 10.1016/0958-1669(92)90005-4. [DOI] [PubMed] [Google Scholar]
- 8.Okazaki R, Okazaki T, Sakebe K, Sugimoto K, Sugino A. Proc Natl Acad Sci USA. 1968;59:598–605. doi: 10.1073/pnas.59.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konrad E B, Modrich P, Lehman I R. J Mol Biol. 1973;77:519–529. doi: 10.1016/0022-2836(73)90220-9. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman M M, Hicks M L, Gellert M. J Mol Biol. 1973;77:531–536. doi: 10.1016/0022-2836(73)90221-0. [DOI] [PubMed] [Google Scholar]
- 11.Johnston L H, Nasmyth K A. Nature (London) 1976;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 12.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 13.Echols H, Goodman M F. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 14.Lecomte P, Doubleday O P, Radman M. J Mol Biol. 1986;189:643–652. doi: 10.1016/0022-2836(86)90494-8. [DOI] [PubMed] [Google Scholar]
- 15.Fijalkowska I J, Schaaper R M. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnasco M O, Thaler D S. Phys Lett A. 1996;221:287–292. [Google Scholar]
- 17.Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon P H, Godelle B. Nature (London) 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 18.Sniegowski P D, Gerrish P J, Lenski R E. Nature (London) 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 19.Mrazek J, Karlin S. Proc Natl Acad Sci USA. 1998;95:3720–3725. doi: 10.1073/pnas.95.7.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debrauwere H, Gendrel C G, Lachat S, Dutreix M. Biochimie. 1997;79:577–586. doi: 10.1016/s0300-9084(97)82006-8. [DOI] [PubMed] [Google Scholar]
- 21.Schaaper R M, Dunn R L. Genetics. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]