Abstract
Kinesin-1 motor proteins walk parallel to the protofilament axes of microtubules as they step from one tubulin dimer to the next. Is protofilament tracking an inherent property of processive kinesin motors, like kinesin-1, and what are the structural determinants underlying protofilament tracking? To address these questions, we investigated the tracking properties of the processive kinesin-8, Kip3. Using in vitro gliding motility assays, we found that Kip3 rotates microtubules counterclockwise around their longitudinal axes with periodicities of ∼1 μm. These rotations indicate that the motors switch protofilaments with a bias toward the left. Molecular modeling suggests 1), that the protofilament switching may be due to kinesin-8 having a longer neck linker than kinesin-1, and 2), that the leftward bias is due the asymmetric geometry of the motor neck linker complex.
The founding member of the kinesin superfamily, the cargo-transporting kinesin-1, has been studied in great detail. Dimeric kinesin-1 constructs 1) are mechanical processive, taking ∼100 of 8-nm steps in a hand-over-hand fashion without detaching from the microtubule; and 2), walk parallel to the axis of microtubule protofilaments as they step from one tubulin dimer to the next. The latter was inferred from gliding motility assays, where microtubules propelled by motors bound to a planar substrate surface rotated around their longitudinal axis with periodicities corresponding to the helical course of the protofilaments in supertwisted microtubules (1,2). Interestingly, protofilament tracking of kinesin-1 is lost in nonprocessive, monomeric constructs (3). There, and also for other nonprocessive microtubule motors such as the kinesin-14 Ncd (4) or axonemal dynein (5), significantly shorter pitches of microtubule rotations in gliding motility assays were observed. As suggested previously (6) this may indicate that protofilament tracking is an inherent property of processive microtubule motors.
To explore this idea further, we investigated the rotations of 14-protofilament microtubules (left-handed helical pitch of ∼8 μm (2)) in gliding motility assays using kinesin-8 that has been observed to perform ≈12 μm-long processive runs in vitro (7). Streptavidin-coated quantum dots (QDs), sparsely bound to the microtubules, served as reporters of microtubule rotations (Fig. 1 A). Information on the three-dimensional paths of the QDs—and thus on microtubule rotations—were obtained from 1), two-dimensional tracking of the QDs with nanometer precision in x and y (8), in combination with 2), z information derived from fluorescence-interference contrast (FLIC) (2) (Fig. 1, B–D). FLIC originates from destructive and constructive interference effects close to reflecting surfaces and gives rise to a modulation of the detected intensity of a fluorescent object depending on its height above the surface. Specifically, the microtubule-attached QDs appear dark when they are in close proximity to the surface (i.e., when being located between the microtubule and the surface) but brighten up significantly when being further away (i.e., when on the microtubule lattice pointing away from the surface). In our experiments, we observed counterclockwise rotations (looking from the trailing microtubule plus-end in the direction toward the leading minus-end) with an average pitch of 0.93 μm ± 0.20 μm (mean ± SD, N = 75; N is the number of complete rotations obtained from 15 gliding microtubules). Considering the geometry of the assay, the counterclockwise directionality of the rotations corresponds to the motors stepping with a perpetual bias (∼1 protofilament switch event per forward movement over 10 tubulin dimers) toward the left.
Figure 1.
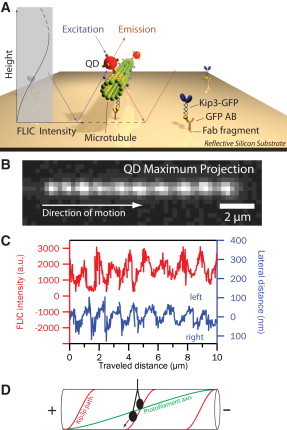
Monitoring Kip3-driven microtubule rotations in gliding motility assays. (A) Schematic of the experimental setup. Imaging is performed on top of a reflective silicon surface using fluorescence interference contrast (FLIC) microscopy (2). (B) Maximum projection of the fluorescence signal of a microtubule-attached quantum dot in the Kip3 gliding motility assay. (C) FLIC intensity (red) and lateral distance from the microtubule path (blue) of the quantum dot shown in panel B versus traveled distance along the microtubule path. The periodic FLIC signal is indicative of repeated up- and down-motion. (D) Schematic of the deduced Kip3 path (red) in comparison to the protofilament axis (green) on a 14-protofilament microtubule.
The behavior observed in our experiments is in stark contrast to kinesin-1, for which counterclockwise rotations with an average pitch of 7.9 μm were previously observed using the same experimental technique (2). Consequently, the question arises: which structural determinants decide whether a kinesin acts as a strict protofilament tracker (and—if it does not—from where the directional bias of the off-axis stepping originates)? Assuming motility in a hand-over-hand fashion, it will matter which binding sites on the microtubule lattice are within reach of the forward swinging motor head. This reach is primarily set by the neck linkers, the structural elements that connect the two motor heads to the coiled-coil neck domain. More precisely, the reach is a function of the length of the neck linkers and their three-dimensional path dictated by the volumes that are occupied by the motor heads when bound to the microtubule. Based on primary sequence alignment between Kip3 with other members of the kinesin-8 family and prediction of the start of the coiled-coil dimerization domain with the program PCOILS, we assigned the neck linker region to the amino acids K436–H452 (i.e., 17 amino acids). Accounting for neck linker docking of the rear motor head (K436–Q447) (9), the corresponding length of the neck linkers between both heads, composed of five amino acids from the undocked part of the rear-head neck linker and 17 amino acids from the front-head neck linker, is estimated to be 85 Å (see the Supporting Material).
We then modeled all configurations of Kip3 with both heads bound simultaneously to adjacent tubulin dimers (Fig. 2, A and B, and see the Supporting Material). The estimated three-dimensional distances between the positions where the neck linkers protrude from the motor heads, respecting the volumes of the heads (Fig. 2 C, gray column; see also the Supporting Material), are measures for the minimally required neck linker lengths for each two-head bound configuration. Comparison between the three-dimensional distances obtained from the model and the available neck linker length (85 Å) suggests that a forward-swinging Kip3 head can most readily reach the tubulin dimer in the front (53 Å needed) and can switch to the protofilament on the left (79 Å for left and 93 Å for front-left needed), but it has difficulties in stepping to the protofilament on the right, which would require a longer neck-linker than it actually exhibits (103 Å for front-right and 105 Å for right needed). The main reason why these long neck-linker distances are required (i.e., >100 Å) is that, to reach the tubulin dimer on the right (or front-right), the neck linker has to bend over the humpy back of the front head (see Fig. 2 B, right and front-right). On the contrary, to reach the tubulin dimer on the left (or front-left), this detour is avoided (see Fig. 2 B, left and front-left). The model-derived preference for left-stepping over right-stepping is in agreement with our experimental observations.
Figure 2.
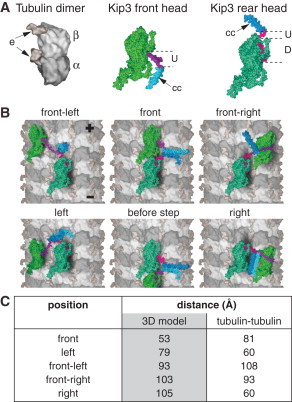
Virtual three-dimensional reconstruction of Kip3 stepping. (A) Tubulin dimer: composed of alpha-tubulin (α) and beta-tubulin (β) monomers, with the unstructured surface-exposed E-Hooks (e). Kip3 front head: Shown with undocked neck linker (U) and following coiled-coil (cc) dimerization domain. Kip3 rear head: Shown with docked (D) and undocked (U) neck linker parts. (B) Illustration of different Kip3 configurations bound with both heads to adjacent tubulin dimers (first heptad repeat of the coiled coil region is artificially unfolded to illustrate all binding configurations). (C) Estimated three-dimensional distances between the positions where the neck linkers protrude from the motor heads, respecting the volumes of the heads (nomenclature as in panel B). For comparison, the direct distances between the tubulin dimers are given.
Modeling as described can be applied for kinesin-1, whose neck linkers are three-amino-acids shorter than the neck linkers of Kip3. Whereas the modeled minimally required neck linker lengths for each two-head-bound configuration are almost identical to the values for Kip3, we estimate an available neck linker length of 63 Å (see the Supporting Material), which explains the strict forward stepping of kinesin-1.
In summary, we have shown what to our knowledge is the first example of a highly processive kinesin motor (run length of several μm) switching between protofilaments of microtubules. Our modeling suggests that protofilament switching may be due to kinesin-8 having a longer neck linker than kinesin-1 so that it is able to reach the extra distance required to change protofilaments. The leftward bias cannot be explained by the geometry of the microtubule lattice alone (Fig. 2 C, last column) but follows from the additional consideration of the asymmetric geometry of the motor neck linker complex. A leftward torque component, which may be present in the powerstrokes of the individual heads (3,4), may further promote the leftward bias but is not strictly necessary. While our results were under review, left-handed spiraling along microtubules of beads coated with a modified kinesin-1 (with extended neck linkers (10)) was reported (11); the handedness of the bead rotations is consistent with the handedness of our microtubule rotations and our model.
Our results may also provide an alternative explanation for the short-pitch, counterclockwise rotations of microtubules gliding on surfaces coated by dimeric kinesin-5 (Eg5) motors (6). The authors of this report attributed the short pitch to the low processivity of Eg5, arguing that during processive episodes the motor follows the protofilament axis, but when detaching generates an off-axis force leading to microtubule rotation. Considering the structure of Eg5 (neck linker length of 18 amino acids (12)), protofilament switching may, however, also be possible during the processive episodes. For kinesin-2 (neck linker length of 17 amino acids, although reduced in length by ∼5 Å due to proline in cis-conformation at position 13 (12,13)), the propensity to switch protofilaments is controversially discussed and may depend on the stability of the neck domain (11,14).
Previously, Kip3, has been found to depolymerize microtubules in a length-dependent manner (7). The underlying mechanism has been described by an antenna model, where Kip3 binds along the entire microtubule lattice and subsequently walks to the microtubule plus-end relying on its high processivity that is ∼20 times the run length of kinesin-1. During such long runs, motors in vivo are expected to frequently encounter obstacles, such as microtubule-associated proteins. In the case of kinesin-1, shown to follow the microtubule's protofilament axis (1), obstacles cause motor stalling or accelerated detachment. It is exciting to speculate that Kip3 uses protofilament switching to bypass obstacles on the microtubule surface avoiding premature motor release or stalling that could reduce the efficiency of targeting and subsequent depolymerization of the microtubule plus-ends.
Footnotes
Volker Bormuth's present address is Institut Curie, UMR168, Paris, France.
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Supporting Material
References and Footnotes
- 1.Ray S., Meyhöfer E., Howard J. Kinesin follows the microtubule's protofilament axis. J. Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nitzsche B., Ruhnow F., Diez S. Quantum-dot-assisted characterization of microtubule rotations during cargo transport. Nat. Nanotechnol. 2008;3:552–556. doi: 10.1038/nnano.2008.216. [DOI] [PubMed] [Google Scholar]
- 3.Yajima J., Cross R.A. A torque component in the kinesin-1 power stroke. Nat. Chem. Biol. 2005;1:338–341. doi: 10.1038/nchembio740. [DOI] [PubMed] [Google Scholar]
- 4.Walker R.A., Salmon E.D., Endow S.A. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- 5.Vale R.D., Toyoshima Y.Y. Rotation and translocation of microtubules in vitro induced by dyneins from Tetrahymena cilia. Cell. 1988;52:459–469. doi: 10.1016/s0092-8674(88)80038-2. [DOI] [PubMed] [Google Scholar]
- 6.Yajima J., Mizutani K., Nishizaka T. A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat. Struct. Mol. Biol. 2008;15:1119–1121. doi: 10.1038/nsmb.1491. [DOI] [PubMed] [Google Scholar]
- 7.Varga V., Helenius J., Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 8.Ruhnow F., Zwicker D., Diez S. Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 2011;100:2820–2828. doi: 10.1016/j.bpj.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice S., Lin A.W., Vale R.D. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 10.Yildiz A., Tomishige M., Vale R.D. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunnbauer M., Dombi R., Okten Z. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol. Cell. 2012;46:147–158. doi: 10.1016/j.molcel.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Hariharan V., Hancock W.O. Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell Mol. Bioeng. 2009;2:177–189. doi: 10.1007/s12195-009-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shastry S., Hancock W.O. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan X.Y., Acar S., Scholey J.M. Torque generation by one of the motor subunits of heterotrimeric kinesin-2. Biochem. Biophys. Res. Commun. 2010;401:53–57. doi: 10.1016/j.bbrc.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikkawa M., Hirokawa N. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J. 2006;25:4187–4194. doi: 10.1038/sj.emboj.7601299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozielski F., Sack S., Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


