Abstract
Activation of the unfolded protein response sensor PKR-like endoplasmic reticulum kinase (Perk) attenuates endoplasmic reticulum (ER) stress levels. Conversantly, if the damage is too severe and ER function cannot be restored, this signaling branch triggers apoptosis. Bcl-2 homology 3-only family member Bim is essential for ER stress-induced apoptosis. However, the regulatory mechanisms controlling Bim activation under ER stress conditions are not well understood. Here, we show that downregulation of the miR-106b-25 cluster contributes to ER stress-induced apoptosis and the upregulation of Bim. Hypericin-mediated photo-oxidative ER damage induced Perk-dependent cell death and led to a significant decrease in the levels of miRNAs belonging to miR-106b-25 cluster in wild-type (WT) but not in Perk−/− MEFs. Further, we show that expression of miR-106b-25 and Mcm-7 (host gene of miR-106b-25) is co-regulated through the transcription factors Atf4 (activating transcription factor 4) and Nrf2 (nuclear factor-erythroid-2-related factor 2). ER stress increased the activity of WT Bim 3′UTR (untranslated region) construct but not the miR-106b-25 recognition site-mutated Bim 3′UTR construct. Overexpression of miR-106b-25 cluster inhibits ER stress-induced cell death in WT but did not confer any further protection in Bim-knockdown cells. Further, we show downregulation in the levels of miR-106b-25 cluster in the symptomatic SOD1G86R transgenic mice. Our results suggest a molecular mechanism whereby repression of miR-106b-25 cluster has an important role in ER stress-mediated increase in Bim and apoptosis.
Keywords: miR-106b-25, unfolded protein response, apoptosis, Bim, ATF4, NRF2
The endoplasmic reticulum (ER) is a multifunctional signaling organelle that controls a wide range of cellular processes. The major physiological functions of the ER include folding of membrane and secreted proteins, synthesis of lipids and sterols, and storage of free calcium.1 In mammals, three ER transmembrane proteins Ire1 (inositol-requiring enzyme 1), Atf6 (activating transcription factor 6), and Perk (PKR-like endoplasmic reticulum kinase), respond to the accumulation of unfolded proteins in the ER lumen.2 Activation of Ire1, Atf6, and Perk initiates an ER-to-nucleus intracellular signaling cascade collectively termed as the unfolded protein response (UPR).3 The most salient feature of the UPR is to increase the transactivation function of a gamut of basic leucine zipper transcription factors, such as Atf6, Atf4, and Xbp1 (X-box binding protein 1).2 Once activated, these transcription factors coordinate transcriptional induction of ER chaperones and genes involved in ER-associated degradation to enhance the protein folding capacity of the cell and to decrease the unfolded protein load of the ER, respectively.1 However, if the damage is too severe and ER homeostasis cannot be restored, apoptosis ensues. The role of B-cell the lymphoma 2 (Bcl-2) family in ER stress-induced apoptosis is emphasized by concurrent repression of Bcl-2 and upregulation of BCL2L11 (Bim) by the ER stress-specific transcription factor DDIT3 (Chop, C/EBP-homologous protein), a key determinant of ER stress-induced apoptosis.4, 5 Bcl-2 homology 3 (BH3)-only family member, Bim is essential for ER stress-induced apoptosis.5 A major mechanism of regulation of Bim-dependent apoptosis is the control of its expression. Bim is regulated at the transcriptional,5 post-transcriptional,6, 7 and post-translational5, 8 levels. ER stress activates Bim by Chop-C/EBPα-mediated transcriptional activation and protein phosphatase 2A-mediated dephosphorylation, which prevents its proteasomal degradation.5 Furthermore, expression of BH3 only proteins PMAIP1 (Noxa) and BBC3 (Puma) has been reported to be upregulated in mouse embryonic fibroblasts (MEFs) undergoing ER stress-induced apoptosis.9 However, exact mechanism involved in transition of the UPR from a protective to an apoptotic phase is not clearly understood.
A class of small RNAs, known as microRNAs (miRNAs), have been shown to be critically involved in many cellular processes including the control of cell survival and cell death.10 The main function of miRNAs is to direct post-transcriptional regulation of gene expression, typically by binding to 3′UTR (untranslated region) of cognate mRNAs and inhibiting their translation and/or stability.11 The miR-106b-25 cluster comprises a group of three miRNAs on chromosome 7 and is transcribed as a single polycistronic unit.12 The miR-106b-25 cluster is located within the 13th intron of the protein-coding gene Mcm-7.12 Amplification and overexpression of miR-106b-25 cluster has been documented in B-cell lymphoma, lung cancer and gastric cancer.12 The miR-106b-25 cluster functions as an oncogene by suppressing the expression of negative regulators of cell proliferation such as the pocket proteins pRb and p130, PTEN, the cyclin-dependent kinase inhibitors p21 and p57, and proapoptotic Bcl-2 family protein, Bim.13, 14, 15 Deletion of the miR-106b-25 cluster in mice does not result in any obvious phenotype6 but loss of both miR-106b-25/miR-17-92 clusters results in embryonic death associated with severe cardiac developmental abnormalities, consisting of defective ventricular and atrial septation and thinner ventricles.6 However, in contrast to the wealth of information about the biological effects of the miR-106b-25 cluster, little is known about its regulation. Only a few studies have recently related ER stress with miRNA biology. miRNAs have been recently shown to be controlled by ER stress through CHOP, modulating the expression of proteins with a high tendency to misfold, thus alleviating ER stress.16 In addition, XBP1 modulates the expression of miRNAs related to regulation of the immune system.17 Finally, a recent study indicated that ER stress downregulates miRNAs that are important for tumor survival.18
In this study, we report the role of miR-106b-25 cluster in ER stress responses. We show that miR-106b-25 cluster is repressed by the Perk-dependent transcription factors Atf4 and nuclear factor-erythroid-2-related factor 2 (Nrf2). Ectopic expression of Atf4 or Nrf2 leads to reduced expression of miR-106b-25 cluster, which is accompanied by increased expression of Bim. We provide evidence that repression of miR-106b-25 cluster contributes to the ER stress-mediated increase of Bim and apoptosis. Taken together, our results suggest a role for miR-106b-25 cluster in ER stress-mediated increase in Bim and apoptosis. This new level of control may fine tune UPR-induced apoptosis in pathologic states such as hypoxia, ischemia/reperfusion injury, neurodegeneration, atherosclerosis, and diabetes.
Results
Role of miR-106b-25 cluster in ER stress-induced induction of Bim
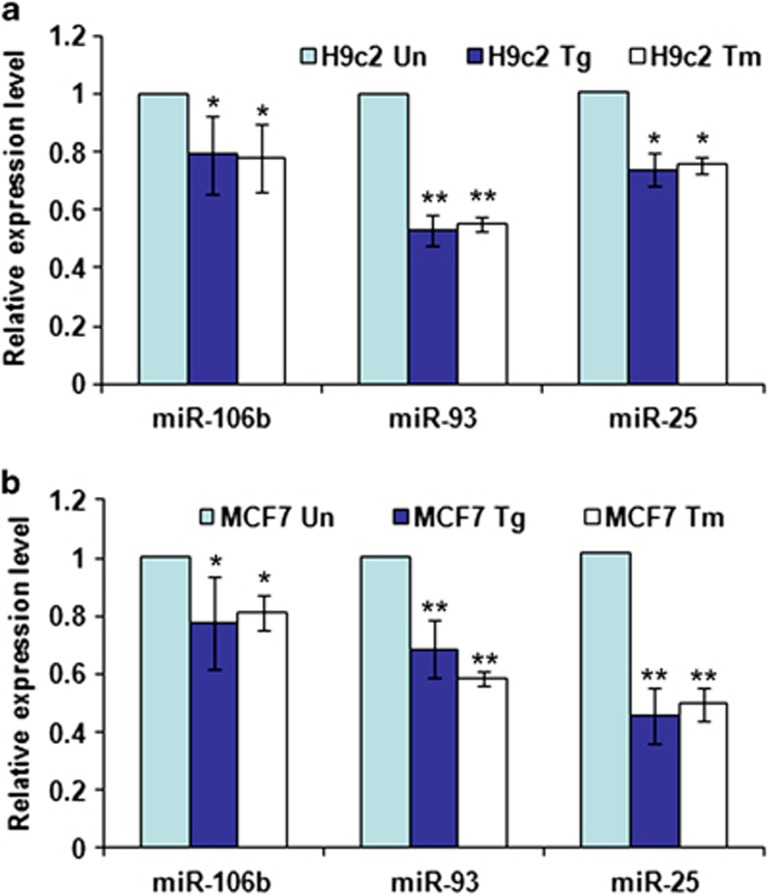
Given that miRNAs can influence the expression of multiple targeted transcripts, we reasoned that miRNAs may have an important regulatory role during the ER stress response. In preliminary experiments, the relative abundance of miRNAs comprising the Sanger miRBase database (Release 11.0) were analyzed by microarray (LC sciences, Houston, TX, USA) using RNA from H9c2 cells during conditions of ER stress. We observed a significant downregulation of miRNAs belonging to the miR-106b-25 cluster in H9c2 cells treated with thapsigargin (Tg) or tunicamycin (Tm). We validated the downregulation of miRNAs comprising miR-106b-25 cluster in H9c2 cells during conditions of ER stress by quantitative real-time PCR (Figure 1a). Recently, it has been shown that Dicer is specifically cleaved by caspase-3 at two sites STTD1476 and CGVD1538 within the RNase IIIa domain during apoptosis.19 Caspase-3-mediated cleavage of Dicer1 during apoptosis reduces its catalytic activity, and reduces the production of mature miRNAs.19 No cleavage of Dicer1 was observed in caspase-3-deficient, MCF-7 cells during staurosporine or actinomycin-induced apoptosis.19 Therefore, to exclude the possibility that reduced levels of miR-106b, miR-25, and miR-93 observed during conditions of ER stress is due to the proteolytic inactivation of Dicer1 we determined the levels of miR-106b, miR-25, and miR-93 in MCF-7 cells upon exposure to ER stress. A reduction in expression levels of miR-106b, miR-25, and miR-93 was observed during ER stress conditions in MCF-7 cells (Figure 1b).
Figure 1.
Downregulation of miRNAs of the miR-106b-25 cluster during conditions of ER stress. (a) H9c2 cardiomyocytes were either untreated (Un) or treated with (1.0 μM) Tg and (1.0 μg/ml) Tm for 24 h. The expression level of indicated miRNAs was quantified by real-time RT-PCR, normalizing against snoRNA. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (b) MCF-7 cells were either untreated (Un) or treated with (2.0 μM) Tg and (2.0 μg/ml) Tm for 24 h and the expression level of indicated miRNAs was quantified by real-time RT-PCR, normalizing against U6 snRNA. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired t-test compared with untreated cells)
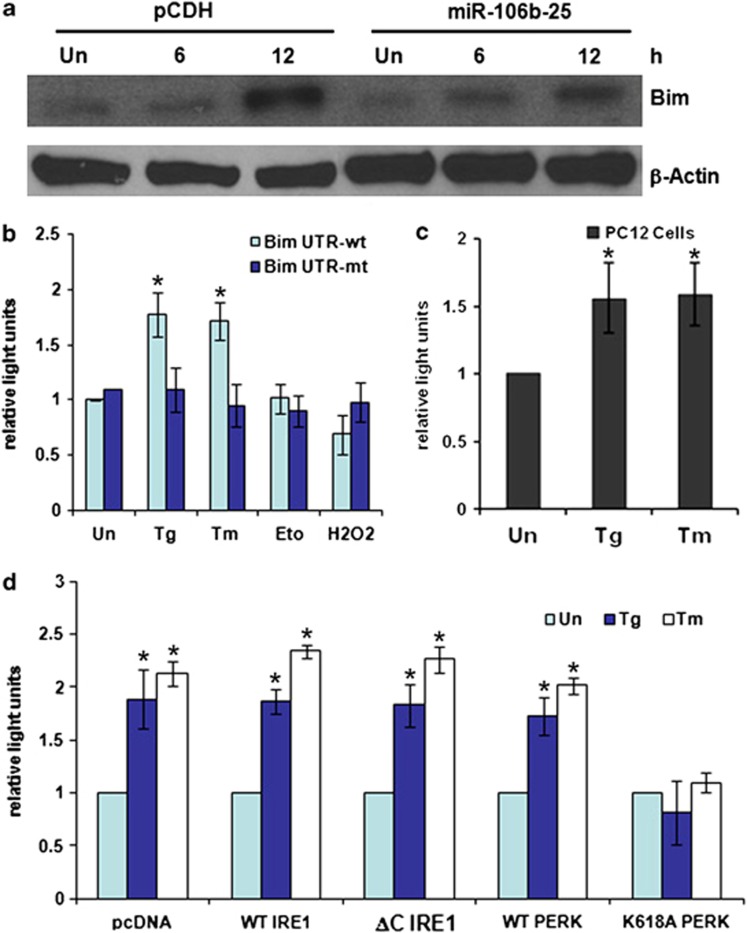
Next, we evaluated the role of the miR-106b-25 cluster in regulation of ER stress-induced expression of Bim. To recapitulate natural conditions, we expressed the three miRNAs of miR-106b-25 cluster (miRs-106b/93/25), which are naturally co-transcribed, in pheochromocytoma 12 (PC12) cells. We found that ectopic expression of miR-106b-25 cluster attenuated the ER stress-mediated upregulation of Bim in PC12 cells (Figure 2a). To determine the role of miR-106b-25 in regulating the ER stress-mediated increase in Bim levels, we used a reporter plasmid with wild-type (WT) Bim 3′UTR (Bim UTR-wt) and mutated miR-106b-25 recognition site in Bim 3′UTR (Bim UTR-mut) cloned into the psiCHECK2 vector.7 We observed that treatment with ER stress-inducing agents, Tg and Tm caused an increase in the luciferase activity of Bim UTR-wt construct in MCF-7 cells (Figure 2b). This increase was not observed in cells transfected with the Bim UTR-mut construct, thus demonstrating that mutation of the binding site for members of the miR-106b-25 cluster in the Bim 3′UTR abolishes its regulation during ER stress (Figure 2b). Further, we did not observe any significant changes in the luciferase activity of Bim UTR-wt construct in MCF-7 cells upon treatment with etoposide (Eto) or hydrogen peroxide (H2O2) (Figure 2b). We observed a similar increase in the luciferase activity of Bim UTR-wt construct during ER stress conditions in PC12 cells (Figure 2c). Ire1 and Perk are two proximal sensors of UPR in the ER membrane.2 To explore the mechanisms behind the upregulation of luciferase activity of Bim UTR-wt construct during ER stress we used dominant-negative constructs of Ire1 and Perk. Co-expression of Perk-K618A completely inhibited the ER stress-mediated upregulation of luciferase activity of Bim UTR-wt (Figure 2d). Co-expression of the dominant-negative Ire1α, (ΔC Ire1α) did not affect the upregulation of luciferase activity of Bim UTR-wt (Figure 2d). These data suggest that the Perk arm of the UPR is required for upregulation of luciferase activity of Bim UTR-wt during conditions of ER stress.
Figure 2.
Perk-dependent increases in the activity of Bim 3′UTR reporter construct during conditions of ER stress. (a) pCDH and miR-106b-25 cells were either untreated (Un) or treated with (0.25 μM) Tg for indicated time points. Total protein was isolated and immunoblotting was performed using antibodies against Bim and β-actin. (b) MCF-7 cells were transfected with Bim 3′UTR reporter plasmid containing binding sites for members of miR-106b-25 (Bim UTR-wt) or Bim 3′UTR reporter plasmid with mutated binding sites for members of miR-106b-25 (Bim UTR-mt). At 24 h after transfection cells were left untreated (Un) or treated with (2.0 μM) Tg and (2.0 μg/ml) Tm, (50 μg/ml) Eto, or (600 μM) H2O2. Luciferase activity was measured 48 h after transfection using Dual-Glo assay system and normalized luciferase activity (Renilla/Firefly) is shown. Error bars represent mean±S.D. from three independent experiments performed in duplicate. (c) PC12 cells were transfected with Bim 3′UTR reporter plasmid containing binding sites for members of miR-106b-25 (Bim UTR-wt). At 24 h after transfection cells were left untreated (Un) or treated with (0.25 μM) Tg and (2.0 μg/ml) Tm. Luciferase activity was measured 48 h after transfection using Dual-Glo assay system and normalized luciferase activity (Renilla/Firefly) is shown. Error bars represent mean±S.D. from three independent experiments performed in duplicate. (d) MCF-7 cells were transfected with Bim 3′UTR reporter plasmid containing binding sites for members of miR-106b-25 (Bim UTR-wt) in combination with pcDNA3 (pcDNA), WT Ire1, dominant-negative Ire1 (ΔC Ire1), WT Perk or dominant-negative Perk (K618A Perk) expression plasmids. At 24 h after transfection cells were left untreated (Un) or treated with (2.0 μM) Tg and (2.0 μg/ml) Tm. Luciferase activity was measured 48 h after transfection using Dual-Glo assay system and normalized luciferase activity (Renilla/Firefly) is shown. Error bars represent mean±S.D. from three independent experiments performed in duplicate. (*P<0.05, two-tailed unpaired t-test compared with untreated cells)
Repression of miR-106b-25 cluster during ER stress is Perk-dependent
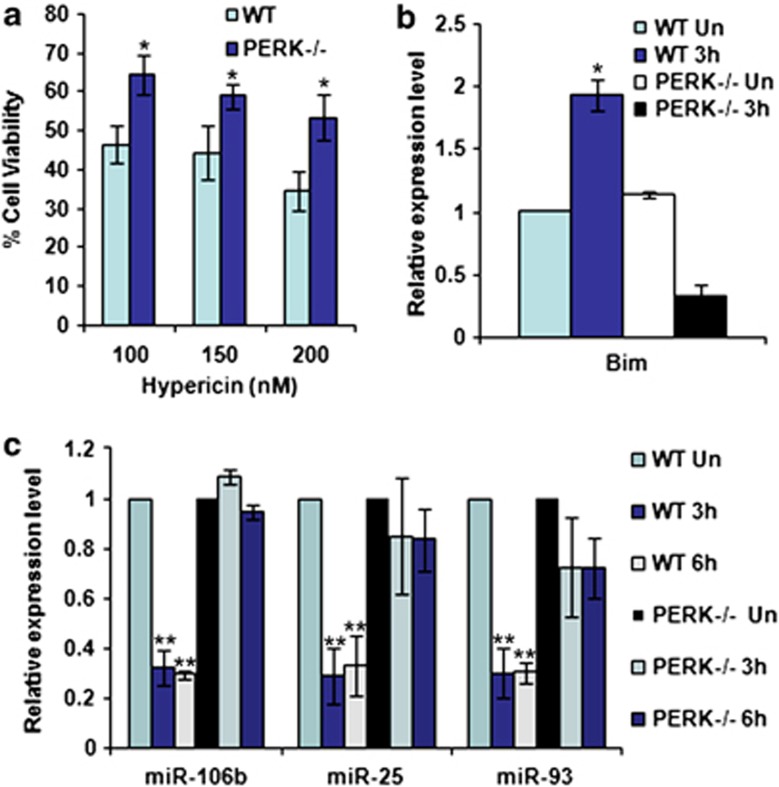
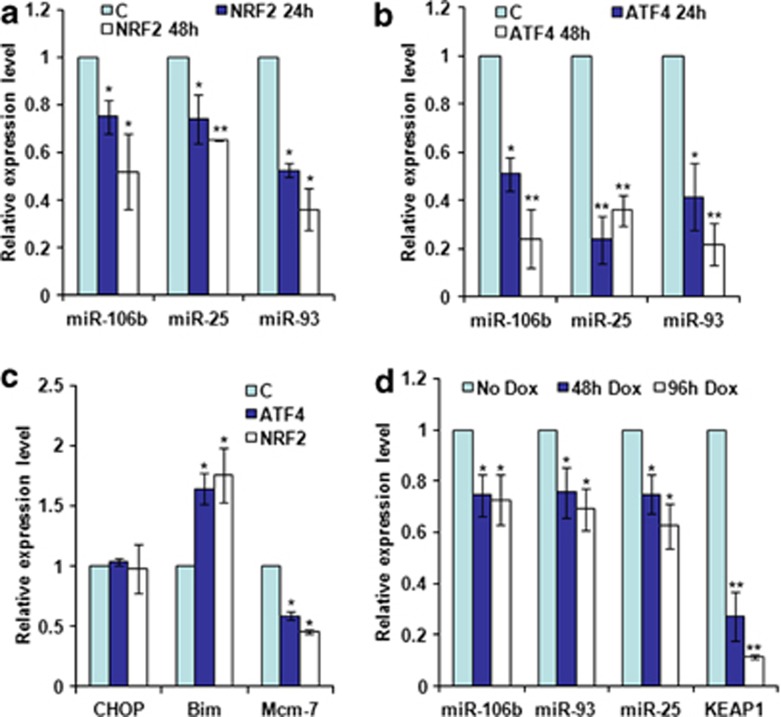
We next used the photosensitizer hypericin that accumulates prevalently in the ER membrane and upon light exposure generates reactive oxygen species, causing a loss-of function of the sarco/endoplasmic reticulum Ca2+–ATPase and consequent ER stress.20 Hypericin-mediated photo-oxidative (phOx) ER damage induces Perk-dependent cell death (Figure 3a). We determined the effect of phOx-mediated ER stress on the expression level of miR-106b-25 cluster in WT and Perk−/− MEFs. We found that phOx-mediated ER stress led to a significant decrease in the levels of miRNAs belonging to miR-106b-25 cluster in WT but not in Perk−/− MEFs (Figure 3c). Furthermore, the expression of Bim was increased in WT but not in Perk−/− MEFs (Figure 3b). The transcription factors Atf4, Nrf2, and Chop are activated downstream of Perk kinase during ER stress.3 As such we determined the effect of ectopic Atf4, Nrf2, and Chop on the expression level of miR-106b-25 cluster. We found that overexpression of both Atf4 and Nrf2 in PC12 cells led to a significant decrease in the levels of miRNAs belonging to miR-106b-25 cluster (Figures 4a and b) whereas expression of Chop had no such effect (Supplementary Figure 1). We found an inverse correlation in the expression of Mcm-7 (host gene for miR-106b-25) and Bim mRNA level upon expression of both Atf4 and Nrf2 in PC12 cells (Figure 4c). However, there was no change in the expression of Chop upon expression of both Atf4 and Nrf2 in PC12 cells (Figure 4c).
Figure 3.
PERK-dependent regulation of miR-106b-25 cluster during conditions of ER stress. (a) WT and Perk knockout (Perk−/−) MEFs were exposed to phOx stress (200 nM hypericin for 2 h, 2.7 J/cm2). The graph shows the surviving fraction of cells 24 h after treatment. Error bars represent mean±S.D. from three experiments performed in duplicate. (*P<0.05, two-tailed unpaired t-test compared with WT MEFs). (b) WT and Perk knockout (Perk−/−) MEFs were exposed to phOx stress (200 nM hypericin for 2 h, 2.7J/cm2) and expression levels of Bim was quantified by real-time RT-PCR, normalizing against Gapdh. Error bars represent mean±S.D. from two independent experiments performed in triplicate. (c) WT and Perk knockout (Perk−/−) MEFs were incubated with 200 nM hypericin for 2 h and irradiated (2.7 J/cm2) (phOx stress). Expression level of miR-106b, miR-25, and miR-93 was quantified by real-time RT-PCR, normalizing against snoRNA. Error bars represent mean±S.D. from two independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired t-test compared with untreated WT MEFs)
Figure 4.
Effect of ectopic Atf4 and Nrf2 on the expression miR-106b-25 cluster and Bim. (a and b) PC12 cells were transfected with pcDNA3 (c), WT Atf4 or Nrf2 expression plasmids and total RNA was isolated at indicated time points. Expression levels of indicated miRNAs were quantified by real-time RT-PCR, normalizing against snoRNA. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired t-test as compared with control transfected cells). (c) PC12 cells were transfected with pcDNA3 (c), WT Atf4, or Nrf2 expression plasmids. Total RNA was isolated and expression levels of Chop, Bim, and Mcm-7 were quantified by real-time RT-PCR, normalizing against Gapdh. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired t-test as compared with control transfected cells). (d) pTRIPZ-shKeap1-293T cells were treated with (500 ng/ml) of doxycycline for indicated time points and expression levels of miR-106b, miR-93, and miR-25 were quantified by real-time RT-PCR, normalizing against snoRNA. Error bars represent mean±S.D. from four independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired t-test as compared with uninduced (No Dox) cells)
Under basal conditions, Nrf2 is localized in the cytoplasm through an interaction with the BTB (broad complex, Tramtrack, and Bric-a-Brac) domain containing protein Kelch-like ECH-associated protein 1 (Keap1).21 Next, we knocked down the levels of Keap1 by introducing the tetracycline-inducible Keap1-targeted short hairpin RNA (shRNAs) into 293T cells and then assessed their effects on expression of miR-106b-25 cluster and Bim. Analysis of Keap1 mRNA levels demonstrated that the system is capable of a dox-dependent knockdown of its target gene (Figure 4d). To address the functionality of the Keap1 knockdown, we analyzed the relative levels of two Nrf2-target genes (HO-1 and NQO-1) in pTRIPZ-shKeap1-293T cells in the absence and presence of dox. We found that levels of two Nrf2-target genes (HO-1 and NQO-1) were increased significantly in the dox-treated cells (Supplementary Figure 2). Hence, the knockdown of Keap1 can be correlated with increase in the levels of Nrf2-target genes. We observed significant downregulation of miRNAs belonging to miR-106b-25 cluster in pTRIPZ-shKeap1-293T cells in presence of dox (Figure 4d). Taken together, these results show Atf4 and Nrf2 can repress expression of the miR-106b-25 cluster independent of ER stress.
miR-106b-25-dependent induction of Bim is required for ER stress-mediated apoptosis
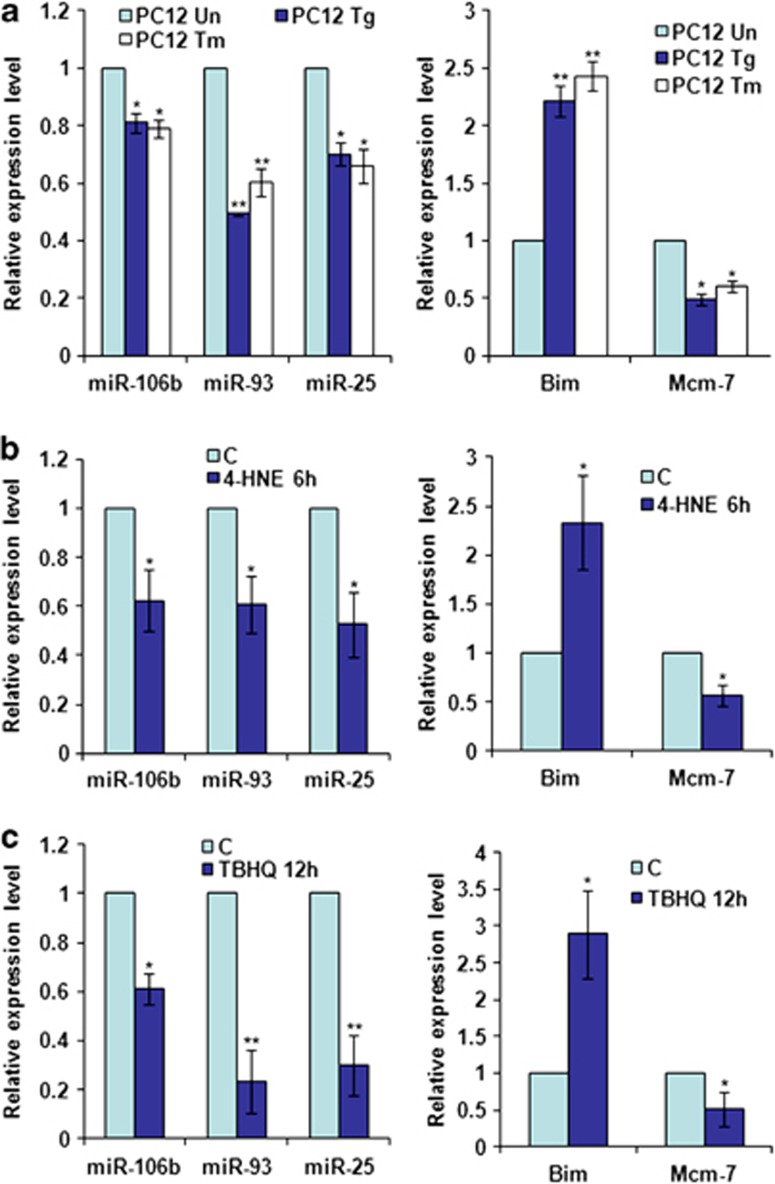
We have previously shown that PC12 cells undergo Bim-dependent apoptosis during conditions of ER stress.22 We therefore treated PC12 cells with Tg and Tm, and evaluated the expression of miR-106b-25 cluster and Bim. We observed significant downregulation of miRNAs belonging to miR-106b-25 cluster in PC12 cells treated with Tg or Tm (Figure 5a). Expression of Mcm-7 (host gene for miR-106b-25) mirrored the expression of miRNAs belonging to miR-106b-25 cluster (Figure 5a). Next, we evaluated the expression of miR-106b-25 cluster upon exposure to 4-hydroxynonenal (4-HNE) and tert-butylhydroquinone (tBHQ). 4-HNE, a lipid peroxidation product is a component of oxidized low-density lipoproteins and is a physiological inducer of ER stress. We found that 4-HNE treatment increased the expression of UPR target genes in PC12 cells (Supplementary Figure 3). tBHQ is a derivative of hydroquinone and has been shown to stabilize Nrf2 by inhibiting its degradation.23 We observed that 4-HNE and tBHQ treatment downregulated the expression of miR-106b-25 cluster and Mcm-7 (Figures 5b and c). Further, we found an inverse correlation between expression of miR-106b-25 cluster and Bim upon exposure of PC12 cells to Tg, Tm, 4-HNE, and tBHQ (Figure 5). Collectively, these results demonstrate that ER stress induced by diverse cytotoxic drugs causes a decrease in miR-106b-25 cluster and increase in Bim mRNA.
Figure 5.
Downregulation of miRNAs belonging to miR-106b-25 cluster during conditions of ER stress. PC12 cells were either untreated (Un) or treated with (0.25 μM) Tg and (2.0 μg/ml) Tm for 24 h (a), (100 μM) 4-HNE for 6 h (b), and (0.5 mM) tBHQ for 12 h (c). The expression levels of miR-106b, miR-93 and miR-25 were quantified by real-time RT-PCR, normalizing against snoRNA. The expression level of Bim and Mcm-7 was quantified by real-time RT-PCR, normalizing against Gapdh. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired t-test as compared with untreated cells)
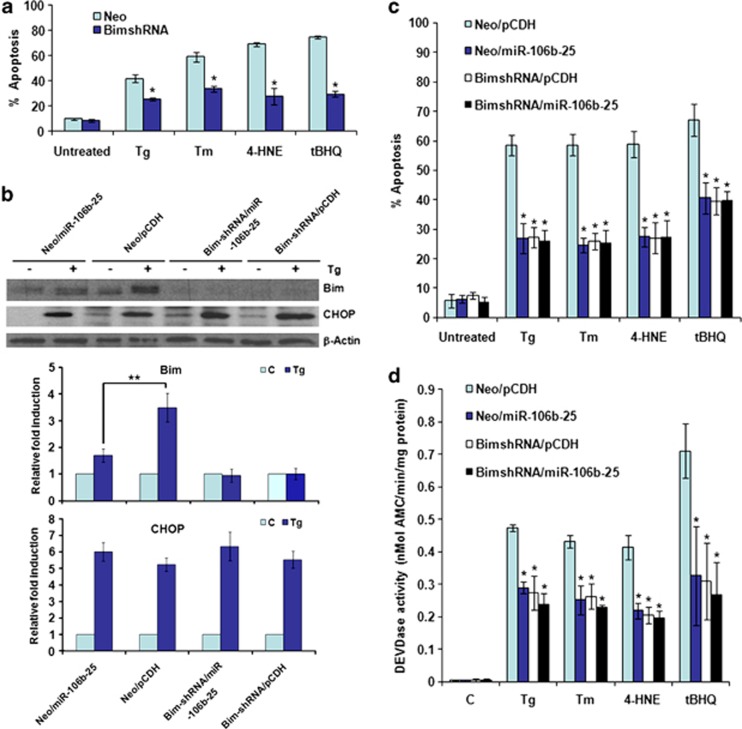
Both 4-HNE and tBHQ have been shown to induce apoptosis in mammalian cells.24, 25 Indeed, we observed cytotoxic effects of 4-HNE and tBHQ in a dose-dependent manner (Supplementary Figure 4). To determine the role of Bim in the cytotoxic effects of Tg, Tm, 4-HNE, and tBHQ, we knocked down Bim levels by introducing Bim-targeted shRNAs into PC12 cells and then assessed their effects on cell survival. Notably, the cytotoxic effect of Tg, Tm, 4-HNE, and tBHQ was attenuated in PC12 cells expressing Bim-targeted shRNAs (Figure 6a). Next we evaluated the role of the miR-106b-25 cluster-dependent regulation of Bim expression and ER stress-induced apoptosis. For this purpose, we expressed the three miRNAs of miR-106b-25 cluster (miRs-106b/93/25) in Neo and Bim-shRNA cells. We found that ectopic expression of miR-106b-25 cluster attenuated the ER stress-mediated upregulation of Bim in PC12 cells (Figure 6b). The effect of miR-106b-25 cluster on ER stress-induced induction of Bim was significant but not as pronounced as Bim-shRNA (Figure 6b). Notably, expression of miR-106b-25 cluster or Bim-shRNA had no effect in Tg-induced expression of Chop (Figure 6b). Next, we used Chop small interfering RNA and evaluated its role in ER stress-induced induction of Bim and apoptosis in PC12 cells. We found that knockdown of Chop expression had no significant effect on the ER stress-induced increase in Bim expression and apoptosis in PC12 cells (Supplementary Figure 5). These results suggest that Bim induction is an important determinant of ER stress-induced cell death in PC12 cells; however, Chop does not have a major role in ER stress-induced induction of Bim in this model. We observed that expression of miR-106b-25 cluster inhibited apoptosis and caspase activity induced by Tg, Tm, 4-HNE, and tBHQ (Figures 6c and d). However, the expression of miR-106b-25 cluster in Bim-shRNA cells did not confer any further protection against Tg, Tm, 4-HNE, and tBHQ-induced apoptosis and caspase activity (Figures 6b and c). These results suggest that Bim is the functionally relevant target of miR-106b-25 cluster in providing resistance to Tg-, Tm-, 4-HNE-, and tBHQ-induced cell death.
Figure 6.
Inhibition of ER stress-induced apoptosis by miR-106b-25 cluster is mediated by repression of Bim. (a) The control (Neo) and Bim-shRNA expressing (Bim-shRNA) PC12 cells were either untreated (Un) or treated with (0.25 μM) Tg, (2.0 μg/ml) Tm, (0.5 mM) tBHQ, and (100 μM) 4-4-HNE for 48 h. Apoptosis was determined with annexin-V/PI staining followed by FACS analysis. Percentages of cells positive for both annexin-V and annexin-V/PI are shown. Average and error bars represent mean±S.D. from three independent experiments. (*P<0.005, two-tailed unpaired t-test as compared with control (Neo) cells). (b) Upper panel, The indicated cells were either untreated (Un) or treated with (0.25 μM) Tg for indicated 24 h. Total protein was isolated and immunoblotting was performed using antibodies against Bim, Chop and β-actin. Lower panel, in the experiment described in (a), induction of Bim and Chop was calculated after densitometric analysis of the autoradiographs with Image J. Average and error bars represent mean±S.D. from three independent experiments.(**P<0.05, two-tailed unpaired t-test as compared with miR-106b-25 expressing (Neo/miR-106b-25) cells). (c) The indicated cells were either untreated or treated with (0.25 μM) Tg, (2.0 μg/ml) Tm, (0.5 mM) tBHQ, and (100 μM) 4-4-HNE for 48 h. Apoptosis was determined with annexin-V/PI staining followed by FACS analysis. Percentages of cells positive for both annexin-V and annexin-V/PI are shown. Average and error bars represent mean±S.D. from three independent experiments. (*P<0.005, two-tailed unpaired t-test as compared with control (Neo/pCDH) cells). (d) The indicated cells were either untreated or treated with (0.25 μM) Tg for 24 h or (2.0 μg/ml) Tm for 24 h, (0.5 mM) tBHQ for 12 h, and (100 μM) 4-4-HNE for 12 h, and DEVDase activity was measured as described in Materials and Methods. Average and error bars represent mean±S.D. from three independent experiments. (*P<0.005, two-tailed unpaired t-test as compared with control (Neo/pCDH) cells)
Downregulation of miR-106b-25 cluster in the SOD1G86Rmice
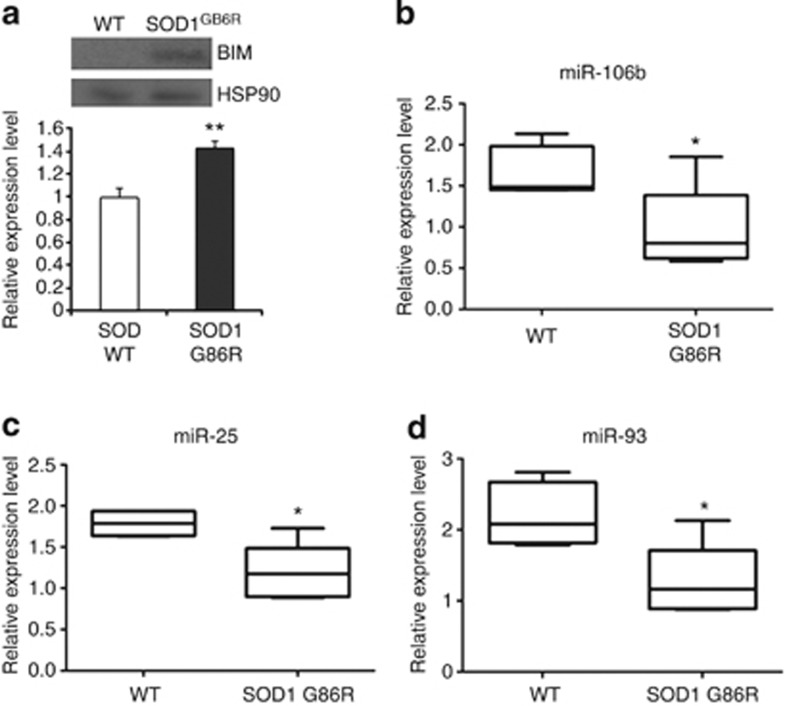
The occurrence of ER stress is associated with disease progression and motoneuron degeneration in amyotrophic lateral sclerosis (ALS) mouse models.26 Many studies have shown that Bim is induced at the protein level in ALS models,26, 27 and deletion of Bim drastically decreases the number of apoptotic cells in the ventral horn of SOD1G93A mice symptomatic mice.28 We therefore determined the expression levels of Bim and miRNA belonging to miR-106b-25 cluster in the post-symptomatic mSOD1G86R transgenic mice. Disease progression was determined as the appearance of abnormal limb-clasping, slight tremor felt in one of the hind limbs, wobbly gait and the first signs of paralysis in one hind limb. We observed significant increase in Bim protein (Figure 7a) and downregulation in the expression of miR-106b-25 cluster in the cortex of mSOD1G86R transgenic mice as compared with the age-matched WT mice (Figures 7b–d). Our results support previous findings, suggesting a functional role of the proapoptotic protein Bim in ALS.28
Figure 7.
Downregulation of miRNAs belonging to miR-106b-25 cluster in the SOD1 mutant mice. (a) The total protein was isolated from the cortex of WT and symptomatic SOD1G86R mice. A total of 40 μg protein per sample used for immunoblotting was analyzed using antibodies against Bim and HSP90. Upper panel, western blot analysis of Bim expression in cortex of WT and symptomatic SOD1G86R mice. Lower panel, average of Bim expression in the cortex of WT (n=2) and symptomatic SOD1G86R (n=7) mice are shown and error bars represent mean±S.D. (**P<0.05, two-tailed unpaired t-test as compared with WT mice). (b–d) The total RNA was isolated from the cortex of WT and symptomatic SOD1G86R mice and expression levels of miR-106b, miR-93, and miR-25 were quantified by real-time RT-PCR, normalizing against U6 snRNA. Box plots showing median and distribution of miR-106b (b), miR-25, (c) and miR-93 (d) in the cortex of WT (n=6) and symptomatic SOD1G86R (n=6) mice are shown. (*P<0.05, Wilcoxon signed rank test as compared with WT mice)
Expression of miR-106b-25 cluster can determine sensitivity to ER stress
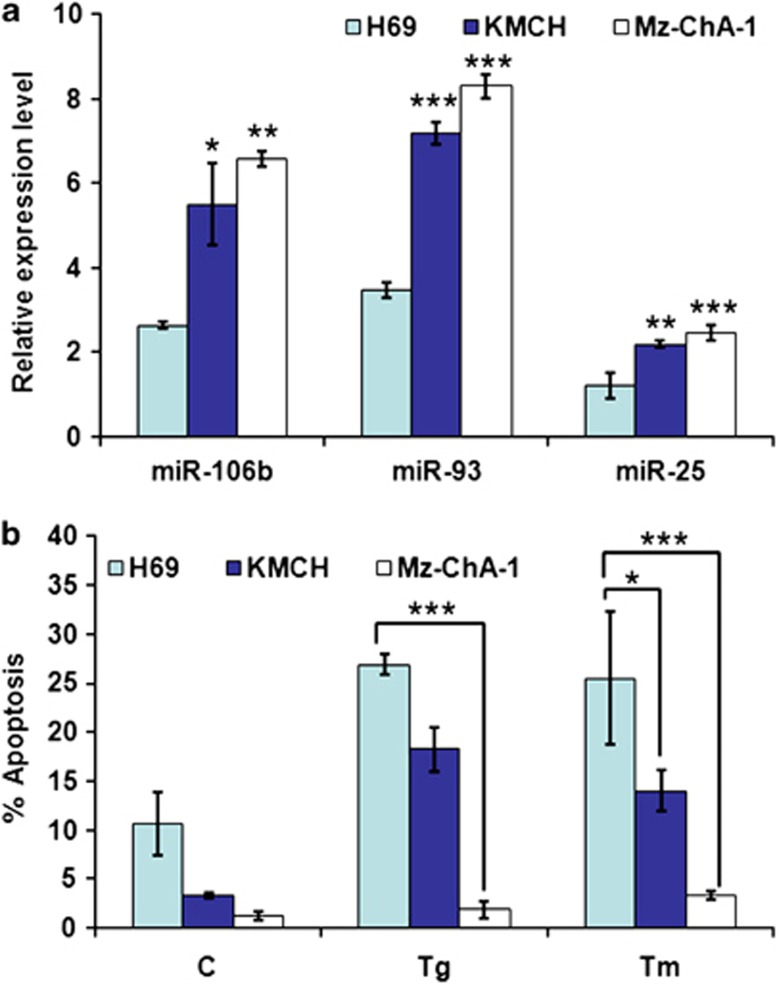
Next, we evaluated whether expression level of miRNAs belonging to miR-106b-25 cluster can affect the sensitivity to ER stress-induced cell death. For this purpose we used human cholangiocarcinoma (H69, Mz-ChA-1, KMCH) cell lines, which have been previously reported to have differential expression of miR-106b-25 cluster.29 We found that Mz-ChA-1 and KMCH cell lines, which have increased basal expression of miR-106b-25, are resistant to ER stress-induced apoptosis as compared with H69 cells (Figures 8a and b). These results suggest that reduced expression level of miR-106b-25 can sensitize the cells to ER stress-induced cell death.
Figure 8.
Effect of miR-106b-25 expression on sensitivity to ER stress-induced cell death. (a) The expression of miR-106b-25 cluster members was measured by qRT-PCR from total RNA isolated from lysates of the indicated cell lines. Results are presented as relative expression using Z30 as an internal control and employing the delta-delta Ct method. Error bars represent mean±S.E. from three independent experiments performed in triplicate. Statistical significance was determined by ANOVA with Bonferroni post hoc correction; *P<0.05, **P<0.01, ***P<0.001, compared with H69 cells. (b) Indicated cell lines were treated with (2.0 μM) Tg and (2.0 μg/ml) Tm for 24 h. After treatment the apoptotic nuclei were determined as described in Materials and Methods section. Error bars represent mean±S.E. from three independent experiments performed in triplicate. Statistical significance was determined by ANOVA with Bonferroni post hoc correction; *P<0.05, **P<0.01, ***P<0.001, compared with H69 cells
Discussion
ER stress constitutes a physiological as well as pathological stress stimulus, which when overwhelming, can lead to apoptotic death of the damaged cell.1 This study demonstrates that miRNAs belonging to the miR-106b-25 cluster have an important role in ER stress-induced apoptosis through the regulation of proapoptotic BCL2-family members. Moreover, it demonstrates how the miR-106b-25 cluster regulates Bim expression by post-transcriptional mechanisms during ER stress (Figure 9). We have used a variety of cell lines in this study, for example H9c2 (rat cardiomyocytes), MEFs, and the cancer cell lines PC12 (rat pheochromocytoma) and MCF-7 (human breast cancer), which all possess different features and observed reduced expression of miRNAs belonging to miR-106b-25 cluster during UPR. Our results suggest that downregulation of the miR-106b-25 cluster in response to ER stress is a general event. This report provides evidence that during conditions of ER stress expression of Bim is inversely related to miRNAs belonging to miR-106b-25 cluster. Expression of miR-106b-25 in PC12 cells abrogated ER stress-induced increase in Bim, albeit not completely (Figure 2a). These results underscore the importance of post-transcriptional modifications in regulating the levels of Bim during ER stress.
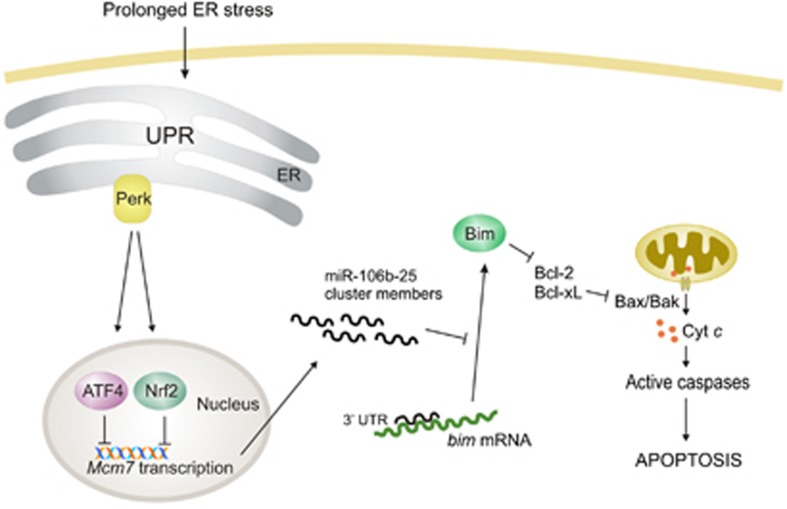
Figure 9.
Schematic summary detailing the main conclusions of this manuscript. During ER stress the transcription factors Atf4 and Nrf2 are activated and downregulate the Mcm7, host gene of miR-106b-25 cluster in a Perk dependent manner, thus contributing to induction of Bim and ER stress-induced apoptosis (see text for details)
Chop is a transcription factor that is induced by several stress stimuli, such as ER stress, amino-acid starvation, DNA damage, or nitric oxide, but how Chop mediates apoptosis has so far been unclear. Recently, a new pathway linking the Chop branch of the UPR to apoptosis was described where Chop, through endoplasmic reticulum oxidoreductin 1 alpha induction, activates inositol trisphosphate receptor-mediated ER calcium release and triggers apoptosis.30 ER stress has been shown to upregulate Bim through Chop-C/ebpα-mediated direct transcriptional induction.5 Although ER stress induced Bim transcription in several cell types, the level of induction was not uniform and there was no apparent transcriptional induction in 293T kidney epithelial cells.5 Chop is clearly important for ER-stress-induced apoptosis in many scenarios; however, Chop is not uniformly essential for cell death induced by ER stress. PERK–/– and AFT4–/– MEFs and eIF2α (Ser51Ala) knock-in MEFs are hypersensitive to ER-stress-induced apoptosis although, they fail to induce Chop during ER stress.31 These data support our results showing that Chop does not have a major role in ER stress-induced induction of Bim in this scenario.
What is the biological significance of transcriptional repression of miR-17-92 cluster by Atf4 and Nrf2? Transcription factors, Atf4 and Nrf2 are activated during ER stress in a Perk-dependent manner. Genetic and pharmacological experiments have demonstrated that Perk signaling can confer both protective and proapoptotic effects in response to ER stress.31, 32 For instance, genetic deletion of Perk sensitizes the cells to ER stress-induced apoptosis.31 However, sustained Perk signaling has been shown to impair cell proliferation and promote apoptosis.33 Atf4 has been shown to act as a pro-death transcriptional activator in the nervous system that propagates death responses to oxidative stress in vitro and to stroke in vivo.34 Atf4 also mediates ER stress-induced cell death of neuroectodermal tumor cells in response to fenretinide or bortezomib.35 In unstressed cells, Nrf2 is maintained in a latent cytoplasmic complex by virtue of its binding to the BTB protein, Keap1.21 As expected, Keap1-deficient mice show increased activity of Nrf2 but die perinatally.36 Further, sustained activation of Nrf2 in autophagy protein 5-deficient murine livers due to p62-mediated stabilization of Nrf2 has been reported to be a major cause of toxicity in autophagy-impaired livers.37 However, the molecular mechanisms by which Atf4 and Nrf2 exert their proapoptotic effects are not clearly understood. The data presented here provide the molecular mechanism underlying Perk-mediated cell death where repression of miR-106b-25 cluster by Atf4 and Nrf2 has an important role in the ER stress-mediated increase in Bim protein and apoptosis.
The occurrence of ER stress is associated with disease progression and motoneuron degeneration in ALS mouse models.38 More importantly, analysis of post-mortem samples derived from sporadic ALS (sALS) patients demonstrated upregulation of the UPR transcription factor Atf4 in tissue from human sALS patients.39 Notably, it has been demonstrated that Bim is upregulated in a familial ALS (fALS) mouse model during the symptomatic stage and ablation of BIM in vivo reduced cellular apoptosis in the ventral horn of a transgenic mouse model of fALS, increasing lifespan.28 Our results suggest an active engagement of the miR-106b-25 cluster in fine tuning the expression of Bim in fALS. Blocking the mitochondrial apoptotic pathway has been shown to preserve motor neuron viability and function in a mouse model of fALS model, suggesting therapeutic benefits of targeting the mitochondrial apoptosis pathway.40 Therefore, it is crucial to identify specific upstream regulators of mitochondrial cell death machinery in motoneurons as potential therapeutic targets. Collectively, our results uncover a new function of miR-106b-25 cluster in vivo, and suggest that small molecules such as Atf4/Nrf2 inhibitors or mimics of miRNAs of miR-106b-25 cluster may be a feasible strategy for the treatment of protein folding disorders in the nervous system.
Materials and Methods
Cell culture and treatment
MEFs, H9c2, and MCF-7 cells were maintained in Dulbecco's modified eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 μ/ml penicillin and 100 mg/ml streptomycin at 37 °C with 5% CO2. PC12 cells were cultured in DMEM supplemented with 10% heat-inactivated horse serum, 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Sigma-Aldrich, Arklow, Ireland) at 37 °C with 5% CO2. Perk+/+ and Perk−/− MEFs were a gift from Dr. David Ron (Skirball Institute, New York, NY, USA). Cells were maintained in DMEM medium supplemented with 10% FBS, 2 mM glutamine, 1 mM sodium pyruvate, MEM non-essential amino-acid solution, 55 μM 2-mercaptoethanol, 50 μ/ml penicillin, and 50 mg/ml streptomycin at 37 °C, 5% CO2 in a humidified incubator. H69 cells are an immortalized non-malignant human cholangiocyte cell line that was cultured as previously described.29 KMCH and Mz-ChA-1 are human cholangiocarcinoma cell lines, grown in DMEM supplemented with 10% FBS, 0.1 μg/ml insulin, penicillin, and streptomycin. Cells were treated with Tg and Tm for the times indicated. All reagents were from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated. Hypericin preparation and cell photosensitization was performed as reported previously.20 Briefly, before irradiation, MEFs were incubated with 200 nM hypericin for 2 h in FBS-free DMEM and 10% FBS was reconstituted after irradiation.
Generation of stable cell lines
Bim shRNA/miR-106b-25 PC12 Cells
We generated stable PC12 cells with reduced levels of Bim using methods previously described,28 by targeting Bim mRNA with shRNA using the lentiviral expression vector pLKO.1 and puromycin selection (3 μg/ml for 7 days). Constructs were generated by Broad Institute (Boston, MA, USA) based on different criteria for shRNA design (see http://www.broad.mit.edu/genome_bio/trc/rnai.html). To further generate subclones of the Neo- and Bim-shRNA PC12 cells expressing miR-106b-25 cluster Neo and Bim-shRNA cells were transduced with pCDH-CMV-miR-106b-25-EF1-RFP or the corresponding control pCDH-CMV-EF1-RFP. PC12 cells were transduced with the lentivirus using polybrene (5 μg/ml) to increase the transduction efficiency. The subclones expressing miR-106b-25 cluster were obtained by sorting on the basis of red fluorescent protein (RFP) using the FACS (fluorescence-activated cell sorting) AriaII cell sorter (BD Biosciences, Oxford, UK) to attain >90% RFP positivity in the selected population.
Keap1-shRNA 293T cells
Lentivirus was generated with tetracycline inducible pTRIPZ Keap1 shRNA (clone shRNA7: 193-0454-D-11; Thermo Scientific, St Leon-Rot, Germany) in 293T cells using jetPEI transfection reagent (Polyplus transfection, VWR International Ltd, Dublin, Ireland) according to manufacturer's instructions. 293T cells were then transduced with the shRNA lentivirus and selection for shRNA-positive cells was performed with 5 μg/ml puromycin. To determine selection efficiency, expression of Keap1 shRNA was induced by 0.5 μg/ml doxycycline for 48 h and the percentage of RFP-positive cells (>80%) determined by flow cytometry (FACS CantoII; BD).
Animal experimentation
We used as an ALS model the SOD1G86R transgenic strain (the equivalent of human SOD1G85R), which was generated in the FVB/N strain (strain FVB-Tg(Sod1-G86R)M1Jwg/J, The Jackson Laboratory, Bar Harbour, Maine, USA). All animal experiments were performed according to procedures approved by the Faculty of Medicine of the University of Chile (approved protocol CBA no. 0208 FMUCH). Disease progression was determined using methods described previously.39 To monitor SOD1 pathogenesis in vivo, animals were euthanized and tissue was collected at different time points, depending on the analysis required. Total RNA was isolated from the cortex tissue and processed for miRNA expression analysis using standard procedures.
RNA extraction, reverse transcription (RT)-PCR and real-time RT-PCR
Total RNA was isolated using RNeasy kit (Qiagen Ltd, West Sussex, UK) according to the manufacturer's instructions. RT was carried out with 2 μg RNA and Oligo dT (Invitrogen, Bio-Sciences, Dublin, Ireland) using 20 U Superscript II Reverse Transcriptase (Invitrogen). Real-time PCR method to determine the induction of UPR target genes has been described previously. Briefly, complementary DNA (cDNA) products were mixed with 2 × TaqMan master mixes and 20 × TaqMan Gene Expression Assays (Applied Biosystems, Life Technologies Ltd, Paisley, UK) and subjected to 40 cycles of PCR in StepOnePlus instrument (Applied Biosystems). Relative expression was evaluated with ΔΔCT method.
Measurement of miRNA levels using TaqMan qRT-PCR assays
Total RNA was reverse transcribed using the TaqMan miRNA Reverse Transcription Kit and miRNA-specific stem-loop primers (Applied BioSystems) in a small-scale RT reaction (comprised of 0.19 μl of H2O, 1.5 μl of 10X Reverse-Transcription Buffer, 0.15 μl of 100 mM deoxyribonucleotide triphosphates, 1.0 μl of Multiscribe Reverse-Transcriptase (50 U/μl), and 5.0 μl of input RNA (20 ng/μl); components other than the input RNA were prepared as a larger volume master mix), using a Tetrad2 Peltier Thermal Cycler (Bio-Rad, Alpha Technologies Ltd, Wicklow, Ireland) at 16 °C for 30 min, 42 °C for 30 min and 85 °C for 5 min. For miR-106b, miR-25, miR-93, snoRNA, and U6 snRNA, 4.0 μl of RT product was combined with 16.0 μl of PCR assay reagents (comprised of 5.0 μl of H2O, 10.0 μl of TaqMan 2X Universal PCR Master Mix, No AmpErase UNG, and 1.0 μl of TaqMan miRNA Assay) to generate a PCR of 20.0 μl of total volume. Real-time PCR was carried out on an Applied BioSystems 7900HT thermocycler at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Data were analyzed with SDS Relative Quantification Software version 2.2.2 (Applied BioSystems.), with the automatic Ct setting for assigning baseline and threshold for Ct determination.
Plasmid constructs
The psiCHECK2 (Promega, MSC Ltd, Dublin, Ireland) reporter plasmid containing the Bim 3′UTR (psiCHECK2-B2) constructs were from Dr. Klaus Rajewsky, Harvard University, USA. The expression vector for WT Ire1α and Ire1αΔC were kind gifts from Dr. Kazunori Imaizumi, University of Miyazaki, Japan and expression plasmids for Atf4, WT Perk and K618A Perk were obtained from Dr. David Ron. The expression vector for WT Nrf2 was from Dr. Alan Diehl, University of Pennsylvania, USA. The expression vector for WT Chop was from Dr. Andreas Strasser, WEHI, Australia. For lentiviral expression of miRs-106b-25, a 1-kb human genomic fragment was amplified using miR-Vec-miRs-106b/93/2514 as a template and cloned in to pCDH-CMV-EF1-RFP. Transient transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Luciferase reporter assays
In Bim 3′UTR reporter assays, MCF-7 or PC12 cells were transfected with 1 μg of psiCHECK2 (Promega) reporter plasmid containing the Bim 3′UTR. At 24 h after transfection cells were treated with Tg or Tm for 24 h. Firefly luciferase and Renilla luciferase activities were measured 48 h after transfection at 560 nm using 10 s luminescence on Wallac plate reader and then normalized for Firefly luciferase activity.
Western blotting
Cells were washed once in ice-cold PBS and lysed in whole-cell lysis buffer (20 mM HEPES pH 7.5, 350 mM NaCl, 0.5 mM EDTA, 1 mM MgCl2, 0.1 mM EGTA, and 1% NP-40) after stipulated time of treatments and boiled at 95 oC with Laemmli's SDS-PAGE sample buffer for 5 min. The proteins were transferred onto nitrocellulose membrane and blocked with 5% milk in PBS-0.05%Tween. The membrane was incubated with the primary antibody for Bim (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA, Cat# sc-793) or β-actin (Sigma-Aldrich, Arklow, Ireland, Cat# A-5060) for 2 h at room temperature or overnight at 4 °C. The membrane was washed three times with PBS-0.05% Tween and further incubated in appropriate horseradish peroxidase-conjugated secondary antibody (Pierce, Rockford, IL, USA) for 90 min. Signals were detected using West pico chemiluminescent substrate (Pierce).
Annexin V/PI Staining
Externalization of phosphatidylserine to the outer leaflet of the plasma membrane of apoptotic cells was assessed using annexin V–fluorescein isothiocyanate according to manufacturer's instruction (BD Pharmingen, San Diego, CA, USA).
Apoptotic nuclei measurements
After treatment, 4′,6-diamidino-2-phenylindole hydrochloride (Sigma-Aldrich, St. Louis, MO, USA) was added at a final concentration of 0.5 mg/ml and cells were stained at 37 °C for 20–30 min. Nuclei were visualized by UV epifluorescence microscopy using a Leica DMI 6000B inverted microscope. Apoptotic nuclei (condensed, fragmented, intensely stained) were counted and presented as a percentage of total nuclei. At least 100 cells were counted per well, and all treatments were done in triplicate.
Analysis of caspase activity
Caspase activity was determined using carbobenzoxy-Asp-Glu-Val-Asp-7-amino-4-methyl-coumarin (DEVD-AMC). Briefly, assay buffer (100 mM HEPES, 10% sucrose, 0.1% CHAPS, 5 mM DTT, 0.0001% NP-40, pH 7.25) containing 50 μM DEVD-AMC (Peptide Institute Inc., Osaka, Japan) was added to samples. Cleavage of the substrate DEVD-AMC to liberate free AMC was measured using a Wallac 1420 Multilabel counter (Perkin Elmer Life Sciences, Dublin, Ireland) with 355 nm excitation and 460 nm emission wavelengths over 25 cycles at 1 min intervals.
Statistical analysis
The data are expressed as mean±S.D. for three independent experiments. Differences between the treatment groups were assessed using two-tailed unpaired student's t-tests. Wilcoxon signed rank test was used for assessing the difference between WT and SOD1G86R animals. The values with a P<0.05 were considered statistically significant.
Acknowledgments
We express our gratitude to Aoife O'Reilly, Maria Ryan, and Shirley Hanley for invaluable assistance. We thank Sandra Healy and Richard Jaeger for critical reading of this manuscript. This publication has emanated from research conducted with the financial support of Science Foundation, Ireland (grant number 09/RFP/BIC2371), Health Research Board (grant numbers HRA_HSR/2010/24 and HRA/2009/59). Supported by FONDECYT no. 1100176, Millennium Institute No. P09-015-F, Muscular Dystrophy Association, ALS Therapy Alliance (to CH), Insercion de Capital Humano Avanzado CONICYT (79100007) (to SM). Supported by GOA grant and by FWO grant G.0492.05. (to PA) of the KU Leuven. Supported by grant R03 DK092263 (to JLM) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of NIDDK.
Glossary
- ALS
amyotrophic lateral sclerosis
- ATF
activating transcription factor
- Bcl-2
B-cell lymphoma 2
- BH3
Bcl-2 homology 3
- BTB
Broad Complex, Tramtrack, and Bric-a-brac
- CDK
cyclin-dependent kinases
- cDNA
complementary DNA
- CHOP
C/EBP-homologous protein
- DMEM
Dulbecco's modified eagle's medium
- eIF2α
eukaryotic translation initiation factor 2α
- ER
endoplasmic reticulum
- Eto
etoposide
- FACS
fluorescence-activated cell sorting
- fALS
familial ALS
- FBS
fetal bovine serum
- H2O2
hydrogen peroxide
- 4-HNE
4-hydroxynonenal
- HO-1
heme oxygenase 1
- Ire1
inositol-requiring enzyme 1
- Keap1
Kelch-like ECH-associated protein 1
- Mcm7
minichromosome maintenance complex component 7
- MEF
mouse embryonic fibroblast
- miRNA
microRNA
- NQO-1
NAD(P)H quinone oxidoreductase 1
- Nrf2
nuclear factor-erythroid-2-related factor 2
- PC12
pheochromocytoma 12
- PERK
PKR-like endoplasmic reticulum kinase
- phOx
photo-oxidative
- PTEN
phosphatase and tensin homolog
- RFP
red fluorescent protein
- RT-PCR
reverse transcription PCR
- sALS
sporadic ALS
- shRNA
short hairpin RNA
- SOD1
superoxide dismutase 1
- tBHQ
tert-butylhydroquinone
- Tg
thapsigargin
- Tm
tunicamycin
- UPR
unfolded protein response
- UTR
untranslated region
- UTR-mut
mutated untranslated region
- WT
wild type
- Xbp1
X-box binding protein 1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by M Piacentini
Supplementary Material
References
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, et al. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem. 2006;281:7260–7270. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, et al. p53-repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Behrman S, Acosta-Alvear D, Walter PA. CHOP-regulated microRNA controls rhodopsin expression. J Cell Biol. 2011;192:919–927. doi: 10.1083/jcb.201010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoszewski R, Brewer JW, Rab A, Crossman DK, Bartoszewska S, Kapoor N, et al. The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J Biol Chem. 2011;286:41862–41870. doi: 10.1074/jbc.M111.304956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Wang X, Gong W, Ni L, Chen C, He X, et al. ER stress negatively modulates the expression of the miR-199a/214 cluster to regulates tumor survival and progression in human hepatocellular cancer. PLoS One. 2012;7:e31518. doi: 10.1371/journal.pone.0031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodgaonkar MM, Shah RG, Kandan-Kulangara F, Affar EB, Qi HH, Wiemer E, et al. Abrogation of DNA vector-based RNAi during apoptosis in mammalian cells due to caspase-mediated cleavage and inactivation of Dicer-1. Cell Death Differ. 2009;16:858–868. doi: 10.1038/cdd.2009.15. [DOI] [PubMed] [Google Scholar]
- Buytaert E, Matroule JY, Durinck S, Close P, Kocanova S, Vandenheede JR, et al. Molecular effectors and modulators of hypericin-mediated cell death in bladder cancer cells. Oncogene. 2008;27:1916–1929. doi: 10.1038/sj.onc.1210825. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegezdi E, Herbert KR, Kavanagh ET, Samali A, Gorman AM. Nerve growth factor blocks thapsigargin-induced apoptosis at the level of the mitochondrion via regulation of Bim. J Cell Mol Med. 2008;12:2482–2496. doi: 10.1111/j.1582-4934.2008.00268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza H, John A. 4-hydroxynonenal induces mitochondrial oxidative stress, apoptosis and expression of glutathione S-transferase A4-4 and cytochrome P450 2E1 in PC12 cells. Toxicol Appl Pharmacol. 2006;216:309–318. doi: 10.1016/j.taap.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Okubo T, Yokoyama Y, Kano K, Kano I. Cell death induced by the phenolic antioxidant tert-butylhydroquinone and its metabolite tert-butylquinone in human monocytic leukemia U937 cells. Food Chem Toxicol. 2003;41:679–688. doi: 10.1016/s0278-6915(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Matus S, Glimcher LH, Hetz C. Protein folding stress in neurodegenerative diseases: a glimpse into the ER. Curr Opin Cell Biol. 2011;23:239–252. doi: 10.1016/j.ceb.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Lee K, Matsuoka M. TDP-43-induced death is associated with altered regulation of BIM and Bcl-xL and attenuated by caspase-mediated TDP-43 cleavage. J Biol Chem. 2011;286:13171–13183. doi: 10.1074/jbc.M110.197483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Fisher J, Pasinelli P, Brown RH, Korsmeyer S, et al. The proapoptotic BCL-2 family member BIM mediates motoneuron loss in a model of amyotrophic lateral sclerosis. Cell Death Differ. 2007;14:1386–1389. doi: 10.1038/sj.cdd.4402166. [DOI] [PubMed] [Google Scholar]
- Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, et al. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Mongillo M, Chin KT, Harding H, Ron D, Marks AR, et al. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J Cell Biol. 2009;186:783–792. doi: 10.1083/jcb.200904060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS ONE. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange PS, Chavez JC, Pinto JT, Coppola G, Sun CW, Townes TM, et al. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J Exp Med. 2008;205:1227–1242. doi: 10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JL, Flockhart R, Veal GJ, Lovat PE, Redfern CP. Regulation of endoplasmic reticulum stress-induced cell death by ATF4 in neuroectodermal tumor cells. J Biol Chem. 285:6091–6100. doi: 10.1074/jbc.M109.014092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes NA, Fisher JK, Austgen K, VandenBerg S, Huang EJ, Oakes SA. Blocking the mitochondrial apoptotic pathway preserves motor neuron viability and function in a mouse model of amyotrophic lateral sclerosis. J Clin Invest. 2010;120:3673–3679. doi: 10.1172/JCI42986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.