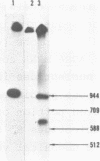
Abstract
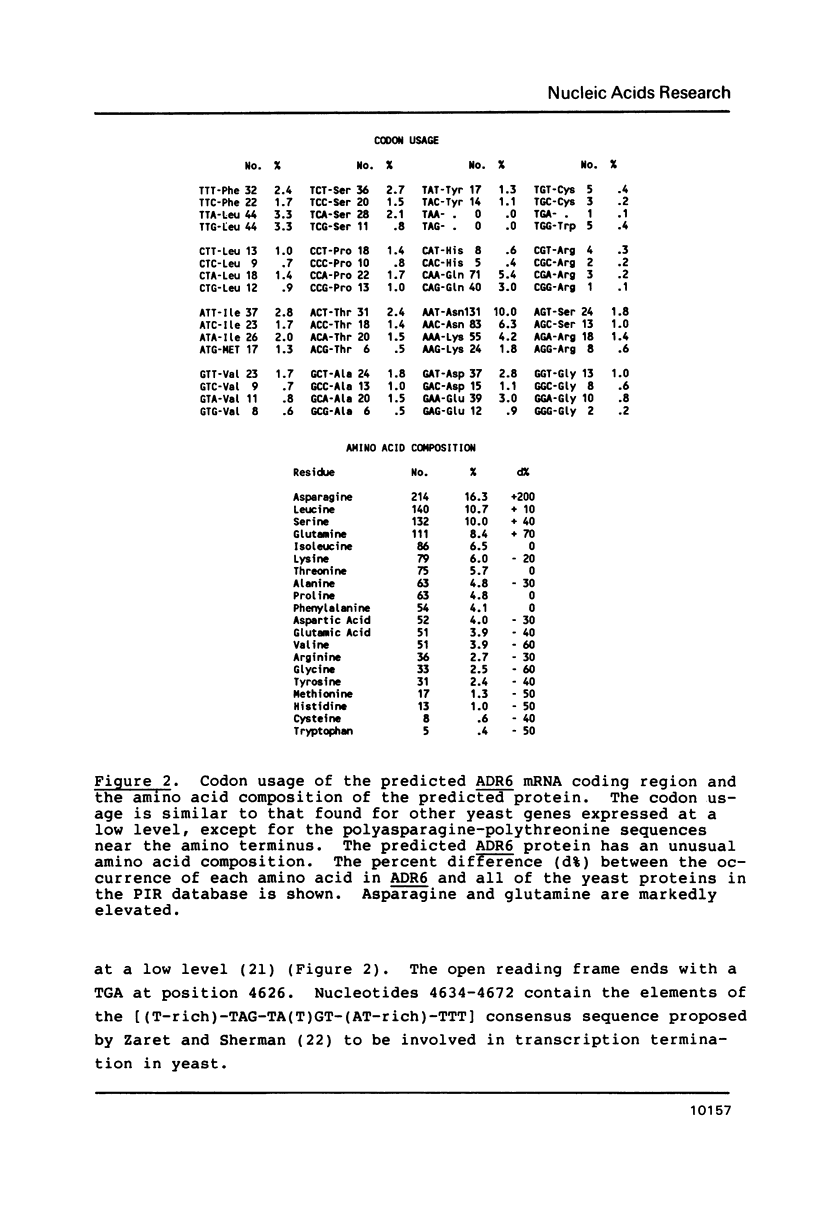
The ADR6 gene of Saccharomyces cerevisiae has an open reading frame which could encode a polypeptide of 1314 amino acids. The predicted mRNA encodes a protein with homopolymeric stretches of asparagine and threonine, particularly near its amino terminus and contains additional sequences consisting of polyglutamine repeats. The predicted protein also contains a potential metal binding (Cys)4-type finger near its carboxy-terminus. An ADR6/beta-galactosidase fusion protein was predominantly nuclear in location, consistent with its role as an activator of ADH2 transcription.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ainley W. M., Macreadie I. G., Butow R. A. var1 Gene on the mitochondrial genome of Torulopsis glabrata. J Mol Biol. 1985 Aug 20;184(4):565–576. doi: 10.1016/0022-2836(85)90303-1. [DOI] [PubMed] [Google Scholar]
- Aline R., Jr, MacDonald G., Brown E., Allison J., Myler P., Rothwell V., Stuart K. (TAA)n within sequences flanking several intrachromosomal variant surface glycoprotein genes in Trypanosoma brucei. Nucleic Acids Res. 1985 May 10;13(9):3161–3177. doi: 10.1093/nar/13.9.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya S. K., Gallo R. C. Three novel genes of human T-lymphotropic virus type III: immune reactivity of their products with sera from acquired immune deficiency syndrome patients. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2209–2213. doi: 10.1073/pnas.83.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J. A., Geever R., Giles N. H. Expression of qa-1F activator protein: identification of upstream binding sites in the qa gene cluster and localization of the DNA-binding domain. Mol Cell Biol. 1987 Mar;7(3):1256–1266. doi: 10.1128/mcb.7.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D. R., Sledziewski A., Young E. T. Deletion analysis identifies a region, upstream of the ADH2 gene of Saccharomyces cerevisiae, which is required for ADR1-mediated derepression. Mol Cell Biol. 1985 Jul;5(7):1743–1749. doi: 10.1128/mcb.5.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis L. T., Denis C. L. Identification of functional regions in the yeast transcriptional activator ADR1. Mol Cell Biol. 1988 May;8(5):2125–2131. doi: 10.1128/mcb.8.5.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C., Fickett J. W., Goad W. B., Lewitter F. I., Rindone W. P., Swindell C. D., Tung C. S. The GenBank genetic sequence databank. Nucleic Acids Res. 1986 Jan 10;14(1):1–4. doi: 10.1093/nar/14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier G., Dressler G. R., Gruss P. Primary structure and developmental expression pattern of Hox 3.1, a member of the murine Hox 3 homeobox gene cluster. EMBO J. 1988 May;7(5):1329–1336. doi: 10.1002/j.1460-2075.1988.tb02948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema W. J., Szeto W. W., Lemley P. V., Orme-Johnson W. H., Ausubel F. M. Nitrogen fixation specific regulatory genes of Klebsiella pneumoniae and Rhizobium meliloti share homology with the general nitrogen regulatory gene ntrC of K. pneumoniae. Nucleic Acids Res. 1985 Jun 25;13(12):4539–4555. doi: 10.1093/nar/13.12.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne B. M., van het Schip A. D., van de Klundert J. A., Arnberg A. C., Gruber M., Ab G. Amino acid sequence of phosvitin derived from the nucleotide sequence of part of the chicken vitellogenin gene. Biochemistry. 1984 Sep 11;23(19):4275–4279. doi: 10.1021/bi00314a003. [DOI] [PubMed] [Google Scholar]
- Bürglin T. R. The yeast regulatory gene PHO2 encodes a homeo box. Cell. 1988 May 6;53(3):339–340. doi: 10.1016/0092-8674(88)90153-5. [DOI] [PubMed] [Google Scholar]
- Campbell D. A., van Bree M. P., Boothroyd J. C. The 5'-limit of transposition and upstream barren region of a trypanosome VSG gene: tandem 76 base-pair repeats flanking (TAA)90. Nucleic Acids Res. 1984 Mar 26;12(6):2759–2774. doi: 10.1093/nar/12.6.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Location of transcriptional control signals and transfer RNA sequences in Torulopsis glabrata mitochondrial DNA. EMBO J. 1985 Feb;4(2):465–473. doi: 10.1002/j.1460-2075.1985.tb03652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colberg-Poley A. M., Voss S. D., Chowdhury K., Gruss P. Structural analysis of murine genes containing homoeo box sequences and their expression in embryonal carcinoma cells. 1985 Apr 25-May 1Nature. 314(6013):713–718. doi: 10.1038/314713a0. [DOI] [PubMed] [Google Scholar]
- Crews S. T., Thomas J. B., Goodman C. S. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell. 1988 Jan 15;52(1):143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- Delmas V., Bastien C., Scherneck S., Feunteun J. A new member of the polyomavirus family: the hamster papovavirus. Complete nucleotide sequence and transformation properties. EMBO J. 1985 May;4(5):1279–1286. doi: 10.1002/j.1460-2075.1985.tb03773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L., Young E. T. Isolation and characterization of the positive regulatory gene ADR1 from Saccharomyces cerevisiae. Mol Cell Biol. 1983 Mar;3(3):360–370. doi: 10.1128/mcb.3.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol Cell Biol. 1987 Aug;7(8):2708–2717. doi: 10.1128/mcb.7.8.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque R. J., Bream G. L., Cannella M. T. Human polyomavirus JC virus genome. J Virol. 1984 Aug;51(2):458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse A., Fujita T., Yasumitsu H., Kashima N., Hasegawa K., Taniguchi T. Organization and structure of the mouse interleukin-2 gene. Nucleic Acids Res. 1984 Dec 21;12(24):9323–9331. doi: 10.1093/nar/12.24.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel M. D., Pruitt R. E., Meyerowitz E. M. DNA sequences, gene regulation and modular protein evolution in the Drosophila 68C glue gene cluster. J Mol Biol. 1983 Aug 25;168(4):765–789. doi: 10.1016/s0022-2836(83)80074-6. [DOI] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecki M., Zeelon E. P. Cell-free synthesis of rat parotid preamylase. J Biol Chem. 1979 Jan 25;254(2):525–529. [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hahn S., Pinkham J., Wei R., Miller R., Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988 Feb;8(2):655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Hamilton R., Watanabe C. K., de Boer H. A. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987 Apr 24;15(8):3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm G. H., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1986 Jan 10;14(1):5–9. doi: 10.1093/nar/14.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne T. A., Blumberg H., Young E. T. Sequence homology of the yeast regulatory protein ADR1 with Xenopus transcription factor TFIIIA. Nature. 1986 Mar 20;320(6059):283–287. doi: 10.1038/320283a0. [DOI] [PubMed] [Google Scholar]
- Haynes S. R., Rebbert M. L., Mozer B. A., Forquignon F., Dawid I. B. pen repeat sequences are GGN clusters and encode a glycine-rich domain in a Drosophila cDNA homologous to the rat helix destabilizing protein. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1819–1823. doi: 10.1073/pnas.84.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth M. E., Ainley W. M., Shumard D. S., Butow R. A., Grossman L. I. Location and structure of the var1 gene on yeast mitochondrial DNA: nucleotide sequence of the 40.0 allele. Cell. 1982 Sep;30(2):617–626. doi: 10.1016/0092-8674(82)90258-6. [DOI] [PubMed] [Google Scholar]
- Icho T., Wickner R. B. Metal-binding, nucleic acid-binding finger sequences in the CDC16 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1987 Oct 26;15(20):8439–8450. doi: 10.1093/nar/15.20.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemura S., Saitoh E., Sanada K. Isolation and amino acid sequences of proline-rich peptides of human whole saliva. J Biochem. 1979 Jul;86(1):79–86. [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Kidd S., Lockett T. J., Young M. W. The Notch locus of Drosophila melanogaster. Cell. 1983 Sep;34(2):421–433. doi: 10.1016/0092-8674(83)90376-8. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene K. C., Distel R. J., Hecht N. B. Nucleotide sequence of a cDNA clone encoding mouse protamine 1. Biochemistry. 1985 Jan 29;24(3):719–722. doi: 10.1021/bi00324a027. [DOI] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Lin Y., Gross J. K. Molecular cloning and characterization of winter flounder antifreeze cDNA. Proc Natl Acad Sci U S A. 1981 May;78(5):2825–2829. doi: 10.1073/pnas.78.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Ingham P., Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986 Dec 5;47(5):721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986 May 15;157(1):77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987 Nov 6;238(4828):797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- Miller D. L., Folse J. R., Benson P. J., Martin N. C. Identification and consequences of a guanosine-15 to adenosine-15 change in the yeast mitochondrial tRNASerUCX gene. Biochemistry. 1983 Mar 29;22(7):1709–1714. doi: 10.1021/bi00276a029. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M., Gehring W. J. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell. 1987 Feb 13;48(3):465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- Moore J., Engelberg A., Bairoch A. Using PC/GENE for protein and nucleic acid analysis. Biotechniques. 1988 Jun;6(6):566–572. [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Olesen J. T., Guarente L. P. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol. 1987 Feb;7(2):578–585. doi: 10.1128/mcb.7.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V., Manet E., Hardon E., Bickel S. E., Benson M. Structure and sequence of the Drosophila zeste gene. EMBO J. 1987 Mar;6(3):791–799. doi: 10.1002/j.1460-2075.1987.tb04821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Rafalski J. A., Scheets K., Metzler M., Peterson D. M., Hedgcoth C., Söll D. G. Developmentally regulated plant genes: the nucleotide sequence of a wheat gliadin genomic clone. EMBO J. 1984 Jun;3(6):1409–1415. doi: 10.1002/j.1460-2075.1984.tb01985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Regulski M., McGinnis N., Chadwick R., McGinnis W. Developmental and molecular analysis of Deformed; a homeotic gene controlling Drosophila head development. EMBO J. 1987 Mar;6(3):767–777. doi: 10.1002/j.1460-2075.1987.tb04819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuwly S., Kuroiwa A., Baumgartner P., Gehring W. J. Structural organization and sequence of the homeotic gene Antennapedia of Drosophila melanogaster. EMBO J. 1986 Apr;5(4):733–739. doi: 10.1002/j.1460-2075.1986.tb04275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. The genome of human papovavirus BKV. Cell. 1979 Dec;18(4):963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- Sengstag C., Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 1987 Jan 12;15(1):233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Stahl H. D., Bianco A. E., Crewther P. E., Burkot T., Coppel R. L., Brown G. V., Anders R. F., Kemp D. J. An asparagine-rich protein from blood stages of Plasmodium falciparum shares determinants with sporozoites. Nucleic Acids Res. 1986 Apr 11;14(7):3089–3102. doi: 10.1093/nar/14.7.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert P. M., Parry D. A., Idler W. W., Johnson L. D., Steven A. C., Roop D. R. Amino acid sequences of mouse and human epidermal type II keratins of Mr 67,000 provide a systematic basis for the structural and functional diversity of the end domains of keratin intermediate filament subunits. J Biol Chem. 1985 Jun 10;260(11):7142–7149. [PubMed] [Google Scholar]
- Stillman D. J., Bankier A. T., Seddon A., Groenhout E. G., Nasmyth K. A. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988 Feb;7(2):485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A. K., Young E. T. The cloning and mapping of ADR6, a gene required for sporulation and for expression of the alcohol dehydrogenase II isozyme from Saccharomyces cerevisiae. Genetics. 1987 Aug;116(4):531–540. doi: 10.1093/genetics/116.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A. K., Young E. T. The identification and characterization of ADR6, a gene required for sporulation and for expression of the alcohol dehydrogenase II isozyme from Saccharomyces cerevisiae. Genetics. 1987 Aug;116(4):523–530. doi: 10.1093/genetics/116.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlgren M., Aslund L., Franzén L., Sundvall M., Wåhlin B., Berzins K., McNicol L. A., Björkman A., Wigzell H., Perlmann P. A Plasmodium falciparum antigen containing clusters of asparagine residues. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2677–2681. doi: 10.1073/pnas.83.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A., Johansen K. M., Xu T., Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985 Dec;43(3 Pt 2):567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
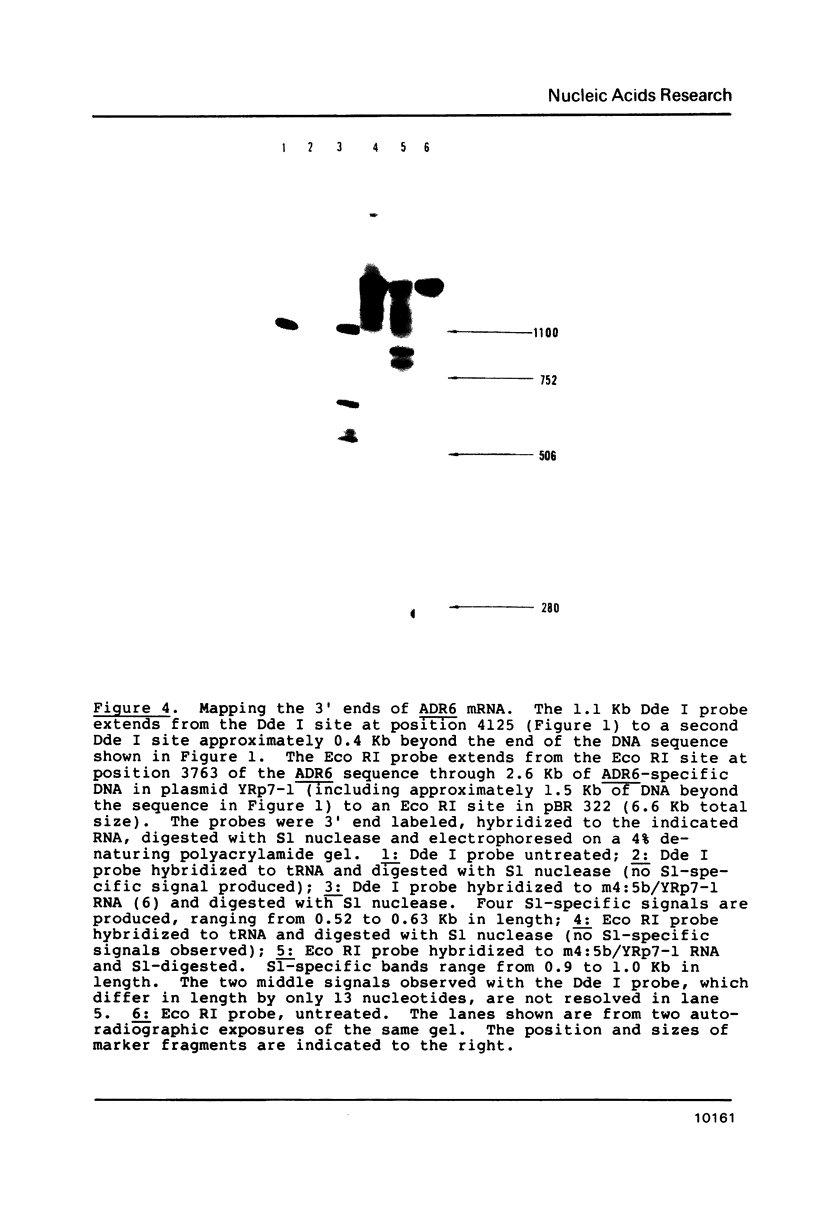
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zassenhaus H. P., Farrelly F., Hudspeth M. E., Grossman L. I., Butow R. A. Transcriptional analysis of the Saccharomyces cerevisiae mitochondrial var1 gene: anomalous hybridization of RNA from AT-rich regions. Mol Cell Biol. 1983 Sep;3(9):1615–1624. doi: 10.1128/mcb.3.9.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]