Abstract
Background
Genomic DNA amplification is a genetic factor involved in cancer, and some oncogenes, such as ERBB2, are highly amplified in gastric cancer. We searched for the possible amplification of other genes in gastric cancer.
Methods and Results
A genome-wide single nucleotide polymorphism microarray analysis was performed using three cell lines of differentiated gastric cancers, and 22 genes (including ERBB2) in five highly amplified chromosome regions (with a copy number of more than 6) were identified. Particular attention was paid to the CRKL gene, the product of which is an adaptor protein containing Src homology 2 and 3 (SH2/SH3) domains. An extremely high CRKL copy number was confirmed in the MKN74 gastric cancer cell line using fluorescence in situ hybridization (FISH), and a high level of CRKL expression was also observed in the cells. The RNA-interference-mediated knockdown of CRKL in MKN74 disclosed the ability of CRKL to upregulate gastric cell proliferation. An immunohistochemical analysis revealed that CRKL protein was overexpressed in 24.4% (88/360) of the primary gastric cancers that were analyzed. The CRKL copy number was also examined in 360 primary gastric cancers using a FISH analysis, and CRKL amplification was found to be associated with CRKL overexpression. Finally, we showed that MKN74 cells with CRKL amplification were responsive to the dual Src/BCR-ABL kinase inhibitor BMS354825, likely via the inhibition of CRKL phosphorylation, and that the proliferation of MKN74 cells was suppressed by treatment with a CRKL-targeting peptide.
Conclusion
These results suggested that CRKL protein is overexpressed in a subset of gastric cancers and is associated with CRKL amplification in gastric cancer. Furthermore, our results suggested that CRKL protein has the ability to regulate gastric cell proliferation and has the potential to serve as a molecular therapy target for gastric cancer.
Keywords: CRKL, Gastric cancer, Cell proliferation, Overexpression, Copy number amplification
Background
Although the overall incidence of gastric cancer is decreasing in many countries, the high incidence of gastric cancer remains a serious health problem, and gastric cancer continues to be the second-leading cause of cancer-related death worldwide [1,2]. Gastric carcinogenesis is a multi-step process in which environmental and genetic factors interact [1-8]. Among the genetic changes observed in cancerous cells, genomic DNA amplification is a well-known alteration that is involved in gastric cancer [4,5,7]. Amplification is often associated with increased expression levels of the genes contained in the amplified loci [5]. Oncogenes in gastric cancer, such as MYC (mapped to chromosome 8q24), KRAS (12p12), and ERBB2 (17q12), are located in such amplified regions [4,5,7,9]. We considered the possibility that there exist genes whose amplification in gastric cancer has not been revealed to date. To uncover such novel gene alterations, we searched for highly amplified genes in gastric cancer using a genome-wide single nucleotide polymorphism (SNP) microarray analysis and found that the CRKLv-crk sarcoma virus CT10 oncogene homolog (avian)-like gene (22q11) is highly amplified in gastric cancer. The CRKL, a member of the CRK family of adapter proteins, consists of an NH2-terminal Src homology 2 (SH2) domain followed by two SH3 domains: SH3n and SH3c [10], and participates in signal transduction in response to growth factors, cytokines, and the oncogenic BCR-ABL fusion protein, resulting in cell proliferation, survival, adhesion, and migration [10,11]. We hypothesized that CRKL might play an important role in gastric carcinogenesis and investigated whether CRKL expression and the function of CRKL protein affect the regulation of cell proliferation in gastric cancer. We also investigated responsiveness of a gastric cancer cell line containing CRKL amplification to a kinase inhibitor, BMS354825, and a CRKL-targeting peptide.
Materials and Methods
Cell lines and surgical specimens
The gastric adenocarcinoma cell lines MKN7, MKN28, MKN74, and AGS were purchased from the Human Science Research Resource Bank (Osaka, Japan) or from American Type Culture Collection (Manassas, VA). Cells were cultured and grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin (100 units/mL), and streptomycin (100 μg/mL) under a 5% CO2 atmosphere at 37°C. Paraffin-embedded gastric tissues obtained from gastric cancer patients who underwent surgery at Toyohashi Municipal Hospital (Japan) were used for the immunohistochemical analysis. Gastric tissue samples obtained from gastric cancer patients who underwent surgery at Hamamatsu University Hospital (Japan) were used for the quantitative reverse-transcription (QRT)-polymerase chain reaction (PCR) analysis. The study design was approved by the Institutional Review Boards (IRBs).
Genome-wide SNP microarray
DNA (250 ng) was digested with NspI restriction enzyme (New England Biolabs, Hertfordshire, UK) and ligated to a universal adaptor sequence. The ligated DNA was PCR-amplified using primers complementary to the universal adaptors, and the PCR products were purified, quantified, and normalized. The products were then fragmented, end-labeled using terminal deoxynucleotidyl transferase, and hybridized to the Affymetrix GeneChip human mapping 250 K NspI arrays (Affymetrix Japan, Tokyo, Japan). After hybridization, the arrays were washed, stained using Affymetrix fluidics station 450, and scanned with a GeneChip Scanner 3000 7 G. Raw SNP call data were extracted using Affymetrix GeneChip Genotyping Analysis software (GTYPE) 4.1. The SNP microarray data were analyzed to determine the total copy number using the CNAG program, as previously described [12,13] (Figure 1).
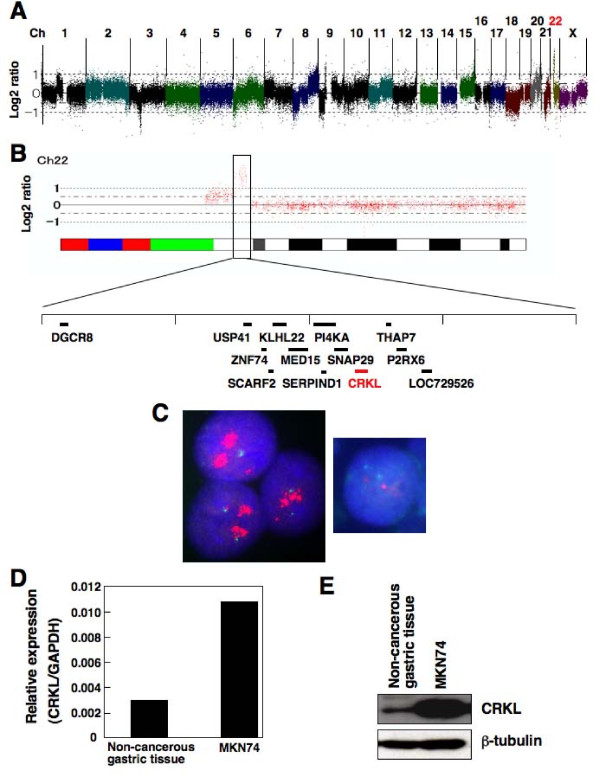
Figure 1.
Identification of highly amplified chromosome regions containing theCRKLgene and the detection of CRKL overexpression in gastric cancer. (A) Genome-wide detection of copy number alterations using a high-density SNP microarray in the MKN74 gastric cancer cell line. The copy number status for the whole genome of MKN74 is shown. DNA (250 ng) was analyzed using an Affymetrix GeneChip 250 K NspI array, and the total copy numbers were determined by analyzing the microarray data using the CNAG program. The chromosome number is shown above the panel. Chromosome 22 is highlighted in red. (B) The copy number status of chromosome 22 of the MKN74 cells is shown. A highly amplified region of chromosome 22 is enlarged, and the genes located in this region are indicated. The CRKL gene is highlighted in red. (C) Detection of CRKL amplification in MKN74 cells using a FISH analysis. The left panel shows the CRKL signal (red) in MKN74 cells, while the right panel shows the CRKL (red) in non-cancerous gastric tissue cells. An extreme increase in the CRKL copy number was observed in the MKN74 cells, while a normal copy number (2) was seen in non-cancerous cells. Nuclei are stained with DAPI. (D) Detection of the increased expression of CRKL mRNA transcript in MKN74 cells using real-time QRT-PCR analysis. The amounts of CRKL transcripts normalized to the amount of GAPDH transcripts are shown in the graph. The average expression level of eight normal gastric mucosa samples was measured as a control. (E) Detection of the increased expression of CRKL protein in MKN74 cells using a western blot analysis. The expression of CRKL was examined using anti-CRKL monoclonal antibody (Y244; 1:500 dilution), horseradish peroxidase-coupled secondary antibody (1:5,000 dilution), and enhanced chemiluminescence detection reagents. The expression of β-tubulin protein was analyzed as an internal control.
WST-8 assay
Cell proliferation and viability were quantified using a Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions [14]. The assay was based on the extracellular reduction of the tetrazolium salt WST-8 by NADH produced in the mitochondria of living cells. The cells were incubated with the WST-8 reagent for 1 hr at 37°C, and the absorbance was measured at 450 nm using an EL340I microplate reader (BIO-TEK Instruments, Winooski, VT) (Figure 2).
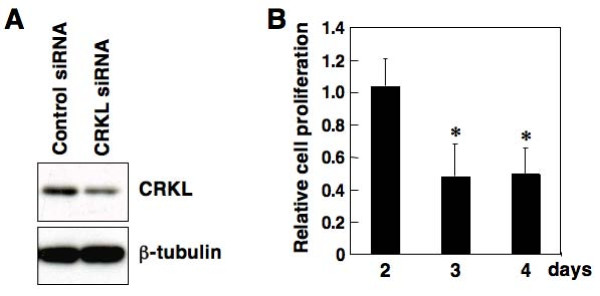
Figure 2.
Ability of CRKL to regulate cell proliferation in the MKN74 gastric cancer cell line. (A) siRNA knockdown for CRKL in MKN74 cells with CRKL amplification. Cells were reverse-transfected with the siRNA oligonucleotides (20 nM) using HiPerFect Transfection Reagent, and 8 × 105 cells were seeded in 60 mm-dishes with 4 ml media. The expression of CRKL was examined 4 days after the reverse transfection of CRKL siRNA or negative control siRNA using a western blot analysis with anti-CRKL monoclonal antibody (Y244; 1:500 dilution). The level of CRKL protein expression was decreased in CRKL siRNA-transfected cells, compared with mock-transfected cells. The expression of β-tubulin protein was analyzed as an internal control. (B) Decrease in the proliferation of MKN74 cells transfected with siRNA for CRKL. Cells were reverse-transfected with the siRNA oligonucleotides (20 nM), and 1.5 × 104 cells were seeded in 96-well microplates containing 100 μL of media. After the reverse transfection of CRKL siRNA or negative control siRNA, the number of viable cells was counted by measuring the reduction in the tetrazolium monosodium salt WST-8. The cell number of CRKL siRNA-transfected cells relative to that of mock-transfected cells is shown. Values are the mean ± standard deviation of three independent experiments. P-values were calculated using the t-test, and * indicates a statistical significance.
Immunohistochemistry
Tissue microarray (TMA) blocks were prepared as previously described [14-16]. TMA block sections were deparaffinized, rehydrated, and boiled in Tris-EDTA buffer (pH 9.0) for antigen retrieval. Endogenous peroxidase activity was blocked by incubation in a hydrogen peroxide solution. Next, the sections were incubated with a rabbit anti-CRKL monoclonal antibody (Y243; Abcam, Cambridge, UK). The antigen-antibody complex was visualized using Histofine Simple Stain Max-Po (Multi) (Nichirei, Tokyo, Japan) and 3,3'-diaminobenzidine tetrahydrochloride. Counterstaining was performed using hematoxylin. The intensity values of the cells were determined using a 4-point scale according to the color of the cell cytoplasm after CRKL immunostaining as follows: 0, blue; 1, blue-brown; 2, light brown; and 3, brown. The percentage of cells with each intensity value was then multiplied by the intensity value, as described previously [14]. The scores obtained for CRKL immunostaining were classified as either a low expression level (0–0.99) or a high expression level (1.00–3.00) (Figure 3) .
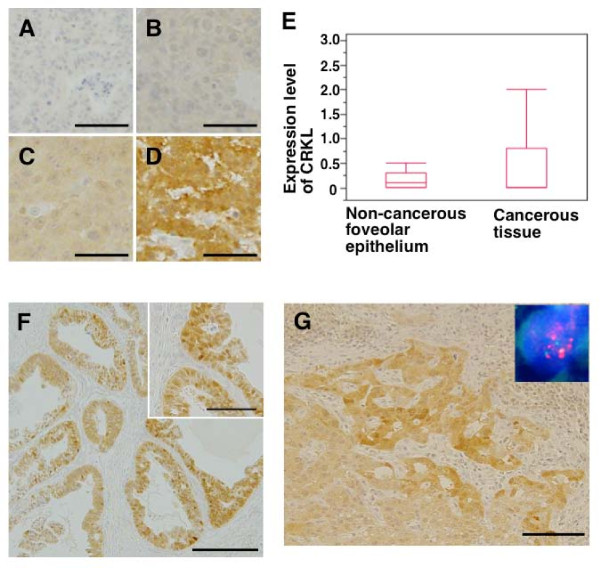
Figure 3.
Immunohistochemical detection of CRKL protein in primary gastric cancer. TMA block sections were subjected to an immunohistochemical analysis using anti-CRKL monoclonal antibody (Y243; 1:100 dilution), Histofine Simple Stain Max-Po (Multi), and 3,3'-diaminobenzidine tetrahydrochloride. Intensity values of 0, 1, 2, and 3 are shown in (A), (B), (C), and (D), respectively. Bar = 50 μm. (E) Box-plot analysis of CRKL protein expression in gastric tissue. A statistically significant difference in the CRKL expression level was detected between non-cancerous gastric foveolar epithelium (n = 41) and gastric cancerous tissue (n = 360). (F) Representative result of the CRKL immunohistochemical analysis. A gastric cancer with a high CRKL expression level is shown. Bar = 500 μm. The inset is a magnified image. Bar = 50 μm. (G) Representative gastric cancer case showing both a high CRKL expression level and CRKL gene amplification. The high CRKL expression level (value = 2.6) was detected using an immunohistochemical analysis. Bar = 100 μm. The inset shows the amplification of CRKL (red) in the cancer cells. The CRKL signal (red) and the control signal for chromosome 22 (green) were detected using a FISH analysis. Nuclei are stained with DAPI.
DNA fluorescence in situ hybridization (FISH)
FISH was performed as previously described [16-19]. Tissue slides were hybridized with a Spectrum Orange-labeled BAC clone (RP11-801O20 and RP11-1058B20) for the CRKL locus (Advanced GenoTechs Co., Tsukuba, Japan) and a Spectrum Green-labeled control probe for the near centromere locus on chromosome 22 (BAC clone: RP11-232E17). 4',6-Diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA) was used for nuclear staining (Figure 3).
MTT assay and direct cell counting
In the experiment involving treatment with the CRKL-targeting peptide, an MTT assay was performed to assess cell viability in Figure 4G. The cells were cultured with the indicated concentration of CRKL-targeting peptide or dimethyl sulfoxide (DMSO) at 37°C for 72 h, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (Sigma-Aldrich, St. Louis, MO) was then added at a final concentration of 0.25 mg/mL. After incubation at 37°C for 4 h, absorbance was measured at a wavelength of 570 nm using a microplate reader. Cells grown in complete medium with DMSO alone were used as controls. The final concentration of DMSO was set to 0.2%. To assess cell proliferation in Figure 4H, the cells were cultured with CRKL targeting peptide or DMSO at 37°C for 72 h. Cell proliferation was measured by directly counting the cells using a hemocytometer, as described previously [20].
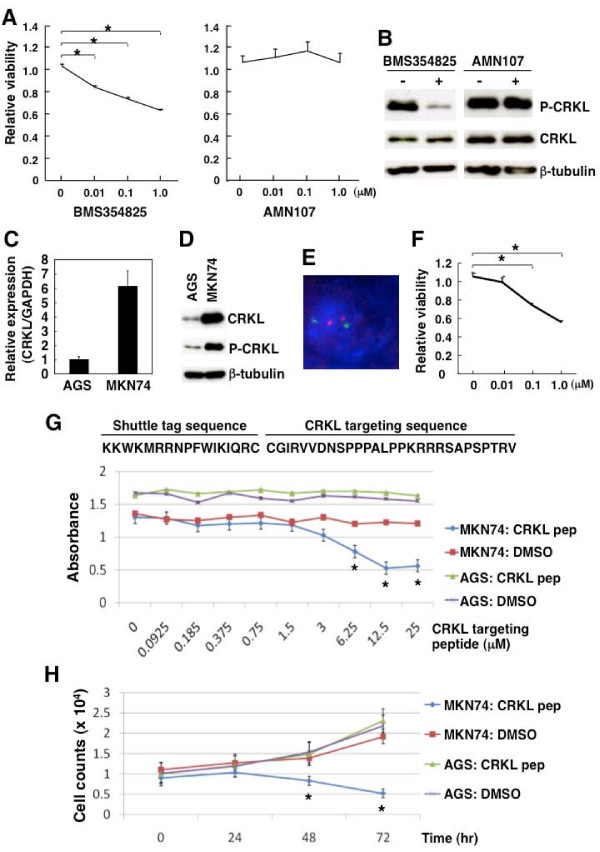
Figure 4.
Responses of the MKN74 gastric cancer cell line withCRKLamplification to treatment with BMS354825 (a dual Src/BCR-ABL kinase inhibitor) and CRKL-targeting peptide. (A) Viability of MKN74 cells treated with BMS354825 but not those treated with AMN107 (a highly selective BCR-ABL kinase inhibitor) is decreased. The cells were seeded in 96-well microplates at a density of 1 × 104 per well; after 24 h, the drug (0.01–1.0 μM) or 0.1% DMSO solution was added. Viability was examined in the MKN74 cells after 72 h of treatment at the indicated concentration using WST-8 reagent. The number of viable cells after treatment with each inhibitor was normalized to the number of viable cells without treatment, and the relative viability is shown in the graph. Values are the mean ± standard error. P values were calculated using the Dunnett’s multiple comparison test, and * indicates a statistically significant decrease. (B) Effective inhibition of CRKL phosphorylation in MKN74 cells treated with BMS354825. Cells were treated with each inhibitor (DMSO only or 0.01 μM of drug) for 90 min, and the expression of CRKL protein was examined using a western blot analysis with anti-phospho CRKL polyclonal antibody (Y207; 1:1,000 dilution) or anti-CRKL monoclonal antibody (Y244; 1:500 dilution). The expression of β-tubulin protein was analyzed as an internal control. (C) Comparison of CRKL mRNA transcripts between AGS and MKN74 cells using real-time QRT-PCR analysis. The amounts of CRKL transcripts normalized to the amount of GAPDH transcripts are shown in the graph. (D) Comparison of expression of CRKL protein between AGS and MKN74 cells using a western blot analysis. The expression of CRKL was examined using the primary antibodies shown in (B). The expression of β-tubulin protein was analyzed as an internal control. (E) Detection of CRKL gene copy number in AGS cells using a FISH analysis. The CRKL signal is red, and the control signal for chromosome 22 is green. Nuclei are stained with DAPI. (F) Viability of AGS cells decreased after BMS354825 treatment. Viability was examined as described in (A). Values are the mean ± standard error. P values were calculated using a t-test, and * indicates a statistically significant decrease. (G) MKN74 cells with CRKL amplification and AGS cells without CRKL amplification were seeded in 96-well microplates at a density of 1 × 104 per well. 24 h after seeding, cells were treated with CRKL-targeting peptide (0.0925–25 μM) or 0.2% DMSO solution at the indicated concentration. The sequence of the CRKL-targeting peptide that was used is shown above the graph. After 72 h of incubation, viability was determined using an MTT assay. The results are presented as the mean ± standard deviation of three independent experiments. P values were calculated using a t-test, and * indicates a statistically significant difference between the cells treated with CRKL-targeting peptide and those treated with DMSO. (H) Cell proliferation of MKN74 and AGS cells treated with CRKL-targeting peptide (6.25 μM) or DMSO as measured by counting cells using a hemocytometer. Cells (1 × 104) were seeded in 24-well plates and treated with CRKL-targeting peptide or DMSO. The cell counting was performed every 24 h for 3 days. Data are shown as the mean ± standard deviation of three independent experiments. P values were calculated using a t-test, and * indicates a statistically significant difference between the cells treated with CRKL-targeting peptide and those treated with DMSO.
QRT-PCR
Total RNA was extracted using Isogen (Nippongene, Tokyo, Japan) or an RNeasy Plus mini kit (Qiagen, Valencia, CA) and converted to cDNA using the SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Real-time QRT-PCR was performed using the cDNA and Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA) on a StepOne Real-Time PCR system (Applied Biosystems). The following PCR primers were used: 5'-CAA CCT GCC TAC AGC AGA AGA TAA-3' and 5'-CGG CAT CAT TCC CAG GAA-3' for the CRKL transcript, and 5'-GGT GGT CTC CTC TGA CTT CAA CA-3' and 5'-GTT GCT GTA GCC AAA TTC GTT GT-3' for the transcript of a housekeeping gene, GAPDH. The relative amounts of CRKL transcript were normalized to those of the GAPDH transcript.
Western blot analysis
Cells were lysed, and the protein concentration was quantified using a BCA protein assay kit (Pierce, Rockford, IL). The proteins were electrophoresed and transferred to a PVDF membrane (GE Healthcare Bio Science, Piscataway, NJ). After blocking with non-fat milk or Blocking One-P (Nakalai Tesque, Kyoto, Japan), the membrane was incubated with rabbit anti-CRKL monoclonal antibody (Y244; Abcam), rabbit anti-phospho CRKL polyclonal antibody (Y207; Cell Signaling, Beverly, MA), or mouse anti-β-tubulin (2-28-33, Sigma-Aldrich). The immunoreactive proteins were visualized using horseradish peroxidase-coupled secondary antibody and enhanced chemiluminescence detection reagents (GE Healthcare Bio Science) [21].
Small interfering RNA (siRNA) knockdown
A stealth siRNA duplex oligonucleotide (Invitrogen) was used for siRNA knockdown. The following CRKL sequence was used: 5'-UCG UGA AAG UCA CAA GGA UGA AUA U-3'. A low GC Duplex #2 (Invitrogen) was used as a negative control. MKN74 cells were reverse-transfected with the siRNA oligonucleotides (20 nM) using HiPerFect Transfection Reagent (Qiagen), according to the manufacturer’s instructions.
BMS354825 and AMN107 treatment
BMS354825, a dual Src/BCR-ABL kinase inhibitor, was kindly provided by Bristol-Myers Squibb (New York, NY), and AMN107, a highly selective BCR-ABL kinase inhibitor, was kindly provided by Novartis Pharmaceuticals (Basel, Switzerland) [22-25]. Stock solutions (10 mM) of BMS354825 and AMN107 were prepared in DMSO. The cells were incubated with BMS354825 or AMN107 at a final concentration of 0.01 to 1.0 μM for 72 h. The final concentration of DMSO was set to 0.1%.
Preparation of CRKL targeting peptide
In this study, we used the peptides, which has been reported to be disrupted complexes between BCR-ABL and CRKL depend on the SH3 domain of CRKL in CML cells [26]. Peptides used in the experiments are followed: CRKL-targeting peptide; KKW KMR RNP FWI KIQ RC – CGI RVV DNS PPP ALP PKR RRS APS PTR V, control peptide; KKW KMR RNP FWI KIQ RC – CGI RVV DNS PPG ALG PLL RRS APS PTR V. The KKW KMR RNP FWI KIQ RC was the shuttle tag sequence performing a receptor-independent cell entry. The chimeric peptide was synthesized and purified by using reverse-phase high performance liquid chromatography (HPLC) (Toray Research Center, Otsu, Japan). Peptide stocks were prepared in DMSO and stored in aliquots at -80°C.
Statistical analysis
The statistical analysis was performed using an unpaired t-test, chi-square test, or Dunnett’s test. JMP version 7.0.1 software (SAS Institute, Cary, NC) was used for the analyses. P values less than 0.05 were considered statistically significant.
Results
Identification of CRKL amplification in gastric cancer
To search for highly amplified genes in gastric adenocarcinoma, we adopted a genome-wide high-resolution SNP microarray approach in three cell lines of differentiated gastric adenocarcinoma: MKN7, MKN28, and MKN74. Genotype calls were obtained at more than 95% of the 262,264 SNP sites on the array, meaning that the SNP microarray analysis had been performed properly. The SNP microarray data were then used to determine the chromosomal copy number using the CNAG program (Figures 1A and 1B). Five highly amplified regions with a copy number of more than 6 (9p13, 17q12-q21, 19q12, 19q13, and 22q11) were identified, as shown in Table 1. These regions contained various kinds of genes, a total of 22 genes (Table 1). Among them, we decided to focus on the CRKL gene at chromosome 22q11.21, the product of which is an SH2 and SH3 domain-containing adaptor protein that shares homology with the CRK oncoprotein, because CRKL is a known substrate of BCR-ABL kinase in Philadelphia chromosome-positive leukemia [27,28] and its role in gastric cancer has not been previously analyzed. To confirm that CRKL gene amplification was detectable in the MKN74 cell line, we performed a FISH analysis using a probe specific for CRKL. As expected, an extreme increase in the CRKL copy number was detected in the MKN74 cells using a FISH analysis (Figure 1C). When the level of CRKL mRNA expression was examined in MKN74 cells using a real-time QRT-PCR analysis, the level was much higher than that in non-cancerous gastric tissue (Figure 1D). Moreover, a western blot analysis showed that the level of CRKL protein expression was higher in MKN74 cells than in non-cancerous gastric tissue (Figure 1E). These results suggested that the CRKL gene is highly amplified and that CRKL is overexpressed in a subset of gastric cancer cell lines.
Table 1.
Detection of chromosomal regions with a high copy number (more than 6) in the gastric cancer cell lines MKN7, MKN28, and MKN74 using a genome-wide SNP microarray analysis
| Chromosomal regionsa | Genes with a high copy number in the region |
|---|---|
| 9p13 |
PAX5 |
| 17q12-q21 |
FBXL20, MED1, PERLD1, ERBB2, IKZF3, ZPBP2 |
| 19q12 |
CCNE1 |
| 19q13 |
CD22 |
| 22q11 | DGCR8, USP41, ZNF74, SCARF2, KLHL22, MED15, PI4KA, SERPIND1, SNAP29, CRKL, THAP7, P2RX6, LOC729526 |
a If more than four consecutive SNP probes with a copy number of more than six were detected in either of the three cell lines, the chromosomal region was regarded as being a “highly amplified region” and was listed in this table.
Ability of CRKL to control gastric cell proliferation
To explore the functional significance of CRKL amplification in gastric cancer, we attempted to examine the effect of overexpressed CRKL on gastric cell proliferation. For this purpose, we prepared MKN74 cells with distinct CRKL expression levels using the siRNA knockdown of CRKL expression. CRKL-specific siRNA transfection effectively decreased the level of CRKL protein expression in MKN74 cells by approximately 70% of the levels observed in negative control siRNA-transfected cells (Figure 2A). A cell proliferation assay showed that the number of CRKL siRNA-transfected MKN74 cells was significantly lower at 3 and 4 days after transfection than the number of negative control siRNA-transfected cells (Figure 2B), meaning that CRKL has the ability to upregulate cell proliferation.
Overexpression of CRKL protein in gastric cancer
Next, we investigated the expression status of CRKL protein in primary gastric cancer using an immunohistochemical analysis with anti-CRKL monoclonal antibody (Y243). CRKL was mainly observed in the cytoplasm, consistent with previous reports [29]. When we compared the level of CRKL expression between non-cancerous gastric foveolar epithelium (n = 41) and gastric cancer (n = 360), the level of CRKL expression in gastric cancer (mean ± standard deviation = 0.42 ± 0.63) was significantly higher than that in non-cancerous tissue (0.20 ± 0.26) (P = 0.032) (Figures 3A3E). When an expression level of 1.00, which corresponds to a value 5-fold of the mean expression level in non-cancerous gastric foveolar epithelium, was used as a cutoff value for the expression status in gastric cancer (i.e., low expression group, 0–0.99; high expression group, 1.00–3.00), 88 (24.4%) of the 360 primary gastric cancers were included in the high expression group (Figure 3F). To examine whether CRKL overexpression is associated with CRKL amplification in gastric cancer, we performed a FISH analysis for the CRKL gene in the 360 primary gastric cancers and compared the prevalence of CRKL amplification between the low expression group and the high expression group. As expected, the percentage of gastric cancer cells with CRKL amplification was significantly higher in the high expression group (9.1%; 8/88 cases) than in the low expression group (2.2%; 6/272 cases) (P = 0.028, chi-square test). This result suggests that CRKL amplification contributes to CRKL overexpression in primary gastric cancer. We further investigated whether the levels of CRKL expression is associated with clinicopathological features in primary gastric cancer patients, the high CRKL expression was observed significantly more often in male and differentiated-type gastric cancer (Table 2). These results suggested that CRKL protein is overexpressed partly due to CRKL amplification in a subset of primary gastric cancers and is associated with the gender and histopathology.
Table 2.
Association between CRKL expression and clinicopathological factors in 360 patients with primary gastric cancer
| CRKL expression level | ||||
|---|---|---|---|---|
| Factor |
Patient |
Low (n = 272) |
High (n = 88) |
P |
| Age |
|
|
|
|
| Year, mean ± SDa |
62.0 ± 11.2 |
61.7 ± 11.7 |
62.9 ± 11.4 |
0.3936b |
| (range) |
(29–86) |
(29–86) |
(31–85) |
|
| Gender |
|
|
|
|
| Male |
255 |
182 (66.9%) |
73 (83.0%) |
0.0028c |
| Female |
105 |
90 (33.1%) |
15 (17.0%) |
|
| Histological type |
|
|
|
|
| Differentiated |
172 |
118 (43.4%) |
54 (61.4%) |
0.0033c |
| Undifferentiated |
188 |
154 (56.6%) |
34 (38.6%) |
|
| pT stage |
|
|
|
|
| pT1 |
143 |
103 (37.9%) |
40 (45.5%) |
0.2082c |
| pT2-pT4 | 217 | 169 (62.1%) | 48 (54.5%) | |
a SD, standard deviation. bt-test. c Chi-square test.
Decrease in the viability of CRKL-expressing MKN74 cells treated with BMS354825
Finally, we tested the possibility of using CRKL as a therapeutic target in MKN74 cells with CRKL amplification. Since Philadelphia chromosome-positive leukemia expressing BCR-ABL is responsive to BMS354825 (a dual Src/BCR-ABL kinase inhibitor) and AMN107 (a highly selective BCR-ABL kinase inhibitor) [22,24], we checked the response of MKN74 cells to both inhibitors. Cell viability was significantly decreased in BMS354825-treated (0.01–1.0 μM) MKN74 cells, compared with cells treated with the solvent only, while it was not significantly decreased in AMN107-treated cells (Figure 4A). When the status of CRKL phosphorylation was examined in the MKN74 cells using western blot analysis with an anti-phospho CRKL antibody, CRKL phosphorylation was found to be inhibited more effectively by BMS354825 than by AMN107 (Figure 4B). These results suggested that BMS354825 has the potential to suppress the viability of MKN74 cells expressing CRKL, likely via the inhibition of CRKL phosphorylation.
To further characterize the role of CRKL in the BMS354825-induced suppression of MKN74 cell viability, we examined the effect of BMS354825 on gastric cancer cells without CRKL amplification. Since the AGS gastric cancer cell line had lower CRKL mRNA and CRKL protein expression levels than MKN74 cells (Figures 4C and D) and had a normal CRKL genomic copy number (Figure 4E), these cells were treated with BMS354825. Unexpectedly, the viability of the BMS354825-treated (0.1–1.0 μM) AGS cells decreased significantly (Figure 4F). Moreover, although the IC50 value (inhibitory concentration producing a 50% response) for BMS354825 was slightly higher in AGS cells than in MKN74 cells, the values were not much different between AGS and MKN74 cells (data not shown). These results suggest that BMS354825 has the potential to suppress the viability of AGS cells, likely via a CRKL-independent pathway.
Decrease in the viability/proliferation of CRKL-expressing MKN74 cells treated with a CRKL-targeting peptide
We then planned to use a more specific inhibitor of CRKL and examined the response of MKN74 and AGS cells to a CRKL-targeting peptide [26]. Cell viability decreased significantly in MKN74 cells treated with the CRKL-targeting peptide (6.25–25 μM), compared with DMSO (solvent)-treated cells, but a similar decrease was not found in AGS gastric cancer cells without CRKL amplification (Figure 4G). When cell proliferation was compared after treatment with 6.25 μM of the CRKL-targeting peptide, the cell proliferation was significantly suppressed in MKN74 cells treated with the peptide, compared with DMSO-treated MKN74 cells, but no inhibition of cell proliferation was seen in the AGS cells (Figure 4H). Control peptide had no effect on the gastric cancer cell proliferation. These results suggested that the CRKL-targeting peptide has the potential to suppress the viability/proliferation of gastric cells exhibiting CRKL amplification, but not of gastric cells that do not exhibit CRKL amplification.
Discussion
Through a genome-wide SNP microarray analysis performed in this study, the CRKL gene was identified as a highly amplified gene in gastric cancer. An increase in the copy number was confirmed in MKN74 gastric cancer cells with CRKL amplification using a FISH analysis, and a high CRKL expression level was also observed in these cells. The ability of CRKL to upregulate cell proliferation was shown in MKN74 cells by comparing the cell proliferation rate between CRKL siRNA-transfected cells and negative control siRNA-transfected cells. CRKL protein was overexpressed in 24.4% of the primary gastric cancers, and its level in the gastric cancer was associated with the gender and histopathology. CRKL amplification was more frequently found in primary gastric cancers with high CRKL protein expression levels than in those with low CRKL expression levels. Finally, we showed that MKN74 cells with CRKL amplification were responsive to the kinase inhibitor BMS354825, likely via the inhibition of CRKL phosphorylation, and a CRKL-targeting peptide. Our current findings suggest that CRKL has an important role in the development of a subset of gastric cancers and has the potential to be a molecular therapy target for gastric cancer.
CRKL is an adaptor cell signaling protein that contains an SH2 domain and two tandem SH3 domains, both of which mediate protein-protein interactions [27,28,30]. CRKL is well known as a surrogate substrate of BCR-ABL kinase in chronic myeloid leukemia and acute lymphoblastic leukemia [11,27,28], and intensive studies of CRKL in Philadelphia chromosome-positive leukemia have been performed. However, only one paper by Kim et al.[31] has reported the CRKL status in gastric cancer. They revealed that the expression of CRKL mRNA in a cancer cell line was stimulated by proteins released by Helicobacter pylori, although the underlying mechanism was not resolved and the CRKL genomic copy number was not analyzed. Our genome-wide SNP microarray analysis successfully revealed, for the first time, that the CRKL gene is highly amplified in a subset of gastric cancers. We also showed that the CRKL protein can upregulate cell proliferation using the RNA-interference-mediated knockdown of CRKL in a gastric cancer cell line with CRKL amplification. Thus, CRKL overexpression arising from genomic amplification likely contributes to the aggressiveness of gastric cancer.
Recent progress in the development of molecular cancer therapy has revealed new molecular-targeting drugs, such as EGFR-targeting drug ZD1839 (Iressa) and HER2-targeting anti-HER2 monoclonal antibody trastuzumab (Herceptin), to be potent therapies for specific cancers [32-34]. In this study, BMS354825, a dual inhibitor for Src and BCR-ABL kinases, but not AMN107, a BCR-ABL specific inhibitor, showed an inhibitory effect on the survival of MKN74 cells with CRKL amplification. A decrease in CRKL phosphorylation through the inhibition of a currently unknown Src kinase seems to be one of the main mechanisms of BMS354825-mediated cytotoxicity in MKN74 cells. BMS354825 is currently being studied clinically in colorectal cancer, prostate cancer, breast cancer, lung cancer, and Philadelphia chromosome-positive leukemia [22,23,35]. Our results suggest that the CRKL protein may be a target of BMS354825-mediated therapy for a subset of gastric cancers. In our analyses, BMS354825 suppressed the viability of AGS cells without CRKL amplification as well as the viability of MKN74 cells with CRKL amplification, suggesting that a CRKL-independent pathway, which has been previously implicated [36], may also be involved in the BMS354825-mediated cytotoxicity seen in gastric cancers. We also presented the usefulness of a CRKL-targeting peptide for suppressing the proliferation of MKN74 cells with CRKL amplification. Our results should contribute to the establishment of CRKL-targeting therapy for a subset of gastric cancers in the future.
In the present study, a genome-wide, high-resolution SNP microarray analysis was successfully performed and five highly amplified chromosome regions containing 22 genes were identified in gastric cancers, as listed in Table 1. Although the ERBB2 gene, a well-known oncogene that is often amplified in gastric cancer [4], was included in this list, the roles of the most of the genes in the Table have not been studied in gastric cancer. Further investigation of these roles is needed in the future.
Conclusion
We conclude that CRKL protein is overexpressed in a subset of gastric cancers and is associated with CRKL amplification in gastric cancer. Furthermore, we conclude that CRKL protein has the ability to regulate gastric cell proliferation and has the potential to serve as a molecular therapy target for gastric cancer.
Abbreviations
DAPI: 4',6-diamidino-2-phenylindole; DMSO: Dimethyl sulfoxide; FISH: Fluorescence in situ hybridization; QRT-PCR: Quantitative reverse-transcription-polymerase chain reaction; SNP: Single nucleotide polymorphism; siRNA: Small interfering RNA; TMA: Tissue microarray; SH2/SH3: Src homology 2 and 3.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HN performed the experiments and wrote the paper draft. KS and SN interpreted the data and revised the paper. HT, HI, MS, KN, MG, SN, and HY performed a part of the experiments. MM and HK provided tissue samples. SN performed a part of the experiments and was involved in the experimental design. HS conceived the research, designed the experiment, and revised the paper. All authors have read and approved the manuscript.
Contributor Information
Hiroko Natsume, Email: n-hiroko@hama-med.ac.jp.
Kazuya Shinmura, Email: kzshinmu@akiha.hama-med.ac.jp.
Hong Tao, Email: taohong@hama-med.ac.jp.
Hisaki Igarashi, Email: hisaki@hama-med.ac.jp.
Masaya Suzuki, Email: masaya0325@gmail.com.
Kiyoko Nagura, Email: ellasekisi@yahoo.co.jp.
Masanori Goto, Email: goto-m@hama-med.ac.jp.
Hidetaka Yamada, Email: h-yamada@hama-med.ac.jp.
Matsuyoshi Maeda, Email: rbpjb381@yahoo.co.jp.
Hiroyuki Konno, Email: kon_6416@hama-med.ac.jp.
Satoki Nakamura, Email: satonaka@hama-med.ac.jp.
Haruhiko Sugimura, Email: hsugimur@hama-med.ac.jp.
Acknowledgements
We greatly appreciate the critical reading of the manuscript by Prof.Shinya Tanaka at Hokkaido University. This work was supported by grants from the Ministry of Health, Labour and Welfare (21-1), the Japan Society for the Promotion of Science (21790383, 22590356, and 22790378), the Ministry of Education, Culture, Sports, Science and Technology (20014007 and 221 S0001), and the Smoking Research Foundation.
References
- Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenberger P, Gretschel S. Gastric cancer. Lancet. 2003;362:305–315. doi: 10.1016/S0140-6736(03)13975-X. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Kohno T, Takahashi M, Sasaki A, Ochiai A, Guilford P, Hunter A, Reeve AE, Sugimura H, Yamaguchi N, Yokota J. Familial gastric cancer: clinicopathological characteristics, RER phenotype and germline p53 and E-cadherin mutations. Carcinogenesis. 1999;20:1127–1131. doi: 10.1093/carcin/20.6.1127. [DOI] [PubMed] [Google Scholar]
- Tahara E. Genetic pathways of two types of gastric cancer. IARC Sci Publ. 2004;157:327–349. [PubMed] [Google Scholar]
- Yang S, Jeung HC, Jeong HJ, Choi YH, Kim JE, Jung JJ, Rha SY, Yang WI, Chung HC. Identification of genes with correlated patterns of variations in DNA copy number and gene expression level in gastric cancer. Genomics. 2007;89:451–459. doi: 10.1016/j.ygeno.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, Matsuno Y, Saito D, Sugimura H, Tanioka F, Kato S, Matsukura N, Matsuda N, Nakamura T, Hyodo I, Nishina T, Yasui W, Hirose H, Hayashi M, Toshiro E, Ohnami S, Sekine A, Sato Y, Totsuka H, Ando M, Takemura R, Takahashi Y, Ohdaira M, Aoki K, Honmyo I, Chiku S, Aoyagi K, Sasaki H, Ohnami S, Yanagihara K, Yoon KA, Kook MC, Lee YS, Park SR, Kim CG, Choi IJ, Yoshida T, Nakamura Y, Hirohashi S. Study Group of Millennium Genome Project for Cancer. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730–740. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- Calcagno DQ, Leal MF, Assumpcao PP, Smith MA, Burbano RR. MYC and gastric adenocarcinoma carcinogenesis. World J Gastroenterol. 2008;14:5962–5968. doi: 10.3748/wjg.14.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin VY, Jin H, Ng EK, Cheng AS, Chong WW, Wong CY, Leung WK, Sung JJ, Chu KM. NF-κB targets miR-16 and miR-21 in gastric cancer: involvement of prostaglandin E receptors. Carcinogenesis. 2011;32:240–245. doi: 10.1093/carcin/bgq240. [DOI] [PubMed] [Google Scholar]
- Mita H, Toyota M, Aoki F, Akashi H, Maruyama R, Sasaki Y, Suzuki H, Idogawa M, Kashima L, Yanagihara K, Fujita M, Hosokawa M, Kusano M, Sabau SV, Tatsumi H, Imai K, Shinomura Y, Tokino T. A novel method, digital genome scanning detects KRAS gene amplification in gastric cancers: involvement of overexpressed wild-type KRAS in downstream signaling and cancer cell growth. BMC Cancer. 2009;9:198. doi: 10.1186/1471-2407-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- Birge RB, Kalodimos C, Inagaki F, Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. 2009;7:13. doi: 10.1186/1478-811X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, Koeffler HP, Ogawa S. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotidepolymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–126. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Nanya Y, Yamamoto G. Genome-wide copy number analysis on GeneChip platform using copy number analyzer for affymetrix GeneChip 2.0 software. Methods Mol Biol. 2007;396:185–206. doi: 10.1007/978-1-59745-515-2_13. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Goto M, Suzuki M, Tao H, Yamada H, Igarashi H, Matsuura S, Maeda M, Konno H, Matsuda T, Sugimura H. Reduced expression of MUTYH with suppressive activity against mutations caused by 8-hydroxyguanine is a novel predictor of a poor prognosis in human gastric cancer. J Pathol. 2011;225:414–423. doi: 10.1002/path.2953. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Iwaizumi M, Igarashi H, Nagura K, Yamada H, Suzuki M, Fukasawa K, Sugimura H. Induction of centrosome amplification and chromosome instability inp53-deficient lung cancer cells exposed to benzo[a]pyrene diol epoxide (B[a]PDE) J Pathol. 2008;216:365–374. doi: 10.1002/path.2422. [DOI] [PubMed] [Google Scholar]
- Sugimura H, Mori H, Nagura K, Kiyose S, Tao H, Isozaki M, Igarashi H, Shinmura K, Hasegawa A, Kitayama Y, Tanioka F. Fluorescence in situ hybridization analysis with a tissue microarray: ‘FISH and chips’ analysis of pathology archives. Pathol Int. 2010;60:543–550. doi: 10.1111/j.1440-1827.2010.02561.x. [DOI] [PubMed] [Google Scholar]
- Sugimura H. Detection of chromosome changes in pathology archives: an application of microwave-assisted fluorescence in situ hybridization to human carcinogenesis studies. Carcinogenesis. 2008;29:681–687. doi: 10.1093/carcin/bgn046. [DOI] [PubMed] [Google Scholar]
- Iwaizumi M, Shinmura K, Mori H, Yamada H, Suzuki M, Kitayama Y, Igarashi H, Nakamura T, Suzuki H, Watanabe Y, Hishida A, Ikuma M, Sugimura H. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut. 2009;58:249–260. doi: 10.1136/gut.2008.149468. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Nagura K, Igarashi H, Tao H, Midorikawa Y, Kitayama Y, Sugimura H. Copy number estimation algorithms and fluorescence in situ hybridization to describe copy number alterations in human tumors. Pathol Int. 2009;59:218–228. doi: 10.1111/j.1440-1827.2009.02354.x. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Hirano I, Okinaka K, Takemura T, Yokota D, Ono T, Shigeno K, Shibata K, Fujisawa S, Ohnishi K. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis. 2010;31:2012–2021. doi: 10.1093/carcin/bgq185. [DOI] [PubMed] [Google Scholar]
- Goto M, Shinmura K, Nakabeppu Y, Tao H, Yamada H, Tsuneyoshi T, Sugimura H. Adenine DNA glycosylase activity of 14 human MutY homolog (MUTYH) variant proteins found in patients with colorectal polyposis and cancer. Hum Mutat. 2010;31:E1861–E1874. doi: 10.1002/humu.21363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti G, Castagnetti F, Gugliotta G, Palandri F, Martinelli G, Baccarani M. Dasatinib and nilotinib in imatinib-resistant Philadelphia-positive chronic myelogenous leukemia: a ‘head-to-head comparison’. Leuk Lymphoma. 2010;51:583–591. doi: 10.3109/10428191003637282. [DOI] [PubMed] [Google Scholar]
- Borriello A, Caldarelli I, Bencivenga D, Cucciolla V, Oliva A, Usala E, Danise P, Ronzoni L, Perrotta S, Della Ragione F. p57Kip2 is a downstream effector of BCR-ABL kinase inhibitors in chronic myelogenous leukemia cells. Carcinogenesis. 2011;32:10–18. doi: 10.1093/carcin/bgq211. [DOI] [PubMed] [Google Scholar]
- Weisberg E, Manley PW, Breitenstein W, Brüggen J, Cowan-Jacob SW, Ray A, Huntly B, Fabbro D, Fendrich G, Hall-Meyers E, Kung AL, Mestan J, Daley GQ, Callahan L, Catley L, Cavazza C, Azam M, Neuberg D, Wright RD, Gilliland DG, Griffin JD. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Nakamura S, Okinaka K, Hirano I, Ono T, Shigeno K, Shinjo K, Ohnishi K. HOXA10 expression induced by Abl kinase inhibitors enhanced apoptosis through PI3K pathway in CML cells. Leuk Res. 2008;32:962–971. doi: 10.1016/j.leukres.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kardinal C, Konkol B, Schulz A, Posern G, Lin H, Adermann K, Eulitz M, Estrov Z, Talpaz M, Arlinghaus RB, Feller SM. Cell-penetrating SH3 domain blocker peptides inhibit proliferation of primary blast cells from CML patients. FASEB J. 2000;14:1529–1538. doi: 10.1096/fj.14.11.1529. [DOI] [PubMed] [Google Scholar]
- Sattler M, Salgia R. Role of the adapter protein CRKL in signal transduction of normal hematopoietic and BCR/ABL-transformed cells. Leukemia. 1998;12:637–644. doi: 10.1038/sj.leu.2401010. [DOI] [PubMed] [Google Scholar]
- ten Hoeve J, Arlinghaus RB, Guo JQ, Heisterkamp N, Groffen J. Tyrosine phosphorylation of CRKL in Philadelphia + leukemia. Blood. 1994;84:1731–1736. [PubMed] [Google Scholar]
- Nakamura T, Komiya M, Sone K, Hirose E, Gotoh N, Morii H, Ohta Y, Mori N. Grit, a GTPase-activating protein for the Rho family, regulates neurite extension through association with the TrkA receptor and N-Shc and CrkL/Crk adapter molecules. Mol Cell Biol. 2002;22:8721–8734. doi: 10.1128/MCB.22.24.8721-8734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hoeve J, Morris C, Heisterkamp N, Groffen J. Isolation and chromosomal localization of CRKL, a human crk-like gene. Oncogene. 1993;8:2469–2474. [PubMed] [Google Scholar]
- Kim N, Park WY, Kim JM, Park JH, Kim JS, Jung HC, Song IS. Gene expression of AGS cells stimulated with released proteins by Helicobacter pylori. J Gastroenterol Hepatol. 2008;23:643–651. doi: 10.1111/j.1440-1746.2007.05241.x. [DOI] [PubMed] [Google Scholar]
- Ko JC, Ciou SC, Cheng CM, Wang LH, Hong JH, Jheng MY, Ling ST, Lin YW. Involvement of Rad51 in cytotoxicity induced by epidermal growth factor receptor inhibitor (gefitinib, IressaR) and chemotherapeutic agents in human lung cancer cells. Carcinogenesis. 2008;29:1448–1458. doi: 10.1093/carcin/bgn130. [DOI] [PubMed] [Google Scholar]
- Holden J, Garrett Z, Stevens A. NICE guidance on trastuzumab for the treatment of HER2-positive metastatic gastric cancer. Lancet Oncol. 2011;12:16–17. doi: 10.1016/S1470-2045(10)70276-X. [DOI] [PubMed] [Google Scholar]
- Norell H, Poschke I, Charo J, Wei WZ, Erskine C, Piechocki MP, Knutson KL, Bergh J, Lidbrink E, Kiessling R. Vaccination with a plasmid DNA encoding HER-2/neu together with low doses of GM-CSF and IL-2 in patients with metastatic breast carcinoma: a pilot clinical trial. J Transl Med. 2010;8:53. doi: 10.1186/1479-5876-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo J, Logothetis C. Dasatinib: a potent SRC inhibitor in clinical development for the treatment of solid tumors. Cancer Treat Rev. 2010;36:492–500. doi: 10.1016/j.ctrv.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian HM, Cortes J, La Rosée P, Hochhaus A. Optimizing therapy for patients with chronic myelogenous leukemia in chronic phase. Cancer. 2010;116:1419–1430. doi: 10.1002/cncr.24928. [DOI] [PubMed] [Google Scholar]