Abstract
Background
There is an increasing need for quantitative technologies suitable for molecular detection in a variety of settings for applications including food traceability and monitoring of genetically modified (GM) crops and their products through the food processing chain. Conventional molecular diagnostics utilising real-time polymerase chain reaction (RT-PCR) and fluorescence-based determination of amplification require temperature cycling and relatively complex optics. In contrast, isothermal amplification coupled to a bioluminescent output produced in real-time (BART) occurs at a constant temperature and only requires a simple light detection and integration device.
Results
Loop mediated isothermal amplification (LAMP) shows robustness to sample-derived inhibitors. Here we show the applicability of coupled LAMP and BART reactions (LAMP-BART) for determination of genetically modified (GM) maize target DNA at low levels of contamination (0.1-5.0% GM) using certified reference material, and compare this to RT-PCR. Results show that conventional DNA extraction methods developed for PCR may not be optimal for LAMP-BART quantification. Additionally, we demonstrate that LAMP is more tolerant to plant sample-derived inhibitors, and show this can be exploited to develop rapid extraction techniques suitable for simple field-based qualitative tests for GM status determination. We also assess the effect of total DNA assay load on LAMP-BART quantitation.
Conclusions
LAMP-BART is an effective and sensitive technique for GM detection with significant potential for quantification even at low levels of contamination and in samples derived from crops such as maize with a large genome size. The resilience of LAMP-BART to acidic polysaccharides makes it well suited to rapid sample preparation techniques and hence to both high throughput laboratory settings and to portable GM detection applications. The impact of the plant sample matrix and genome loading within a reaction must be controlled to ensure quantification at low target concentrations.
Background
As the world's agricultural systems endeavour to sustain an expanding population, technologies have become available to increase the yield and viability of cultivated crops including the introduction of novel traits into crops using genetic transformation of foreign DNA to produce GM varieties. However, public resistance to commercialization of genetically modified plants is still widespread in Europe [1,2]. Existing European regulation limits the extent of GM presence in non-GM foodstuffs, and the increasing introduction of GM products into Europe is likely to result in parallel GM and non-GM ("conventional") supply chains. In addition, the more widespread planting of GM crops in Europe will lead to the need for on-farm confirmation of GM status. Together these factors are likely to lead to a substantial increase in the extent and frequency of testing for the presence of DNA of a GM-derived origin.
The European Union has currently defined the proportion of GM that can be present to be no more than 0.9% GM in a non-GM product [3-5]. As a consequence, diagnostic tests must be deployed that can accurately quantify the GM proportion for monitoring [6]. Careful sampling and handling techniques are required to ensure the analysis is statistically relevant and appropriate controls are also needed to compare the presence of a transgene to a suitable reference gene.
Several nucleic acid amplification techniques (NAATs) are available for the detection of GM contamination in plants and food [7,8] of which the polymerase chain reaction (PCR) is by far the most widely used. However PCR requires rapid thermo-cycling to denature the target DNA strands, prior to and during amplification [9,10], which imposes specific equipment requirements. Since the discovery of DNA polymerases with strand displacement activity, novel amplification methods have been developed which operate under isothermal conditions (iNAAT) and propagate the initial target sequence by promoting strand displacement using enzymes or modified oligonucleotides.
Loop-mediated isothermal amplification (LAMP) is a sensitive, rapid and specific nucleic acid amplification technology. It is characterized by the use of 4 different primers, specifically designed to recognize 6 distinct regions on the target DNA template, and proceeds at a constant temperature driven by invasion and strand displacement [11-13]. Amplification and detection of target genes can be completed in a single step at a constant temperature, by incubating DNA template, primers and a strand displacement DNA polymerase. It provides high amplification efficiency, with replication of the original template copy 109-1010 times during a 15-60 min reaction [13]. The primer pairs used in LAMP are given specific designations; LAMP primers that generate hairpin loops, the outer displacement primers, and LOOP primers that accelerate the reaction by amplifying from the hairpin previously created by the LAMP primers [13,14].
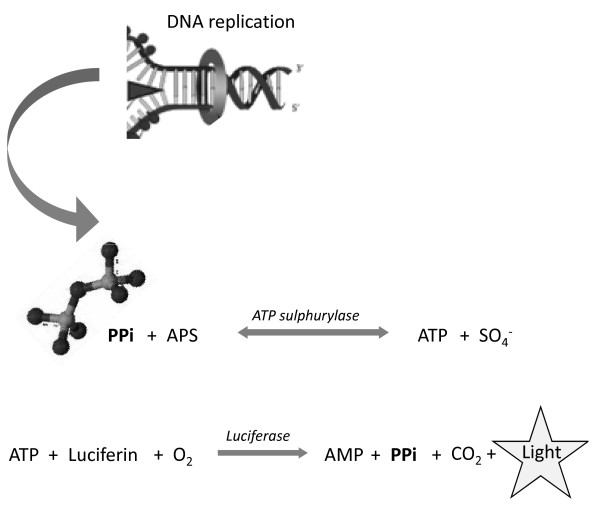
Several methods exist to determine the extent that DNA has been amplified either after or during a given reaction, of which the most frequently used are the incorporation of fluorescent primers into the amplification product or the use of intercalating fluorescent dyes. Other techniques monitor side products of the DNA synthesis responsible for the amplification reaction. For example, turbidity and fluorescence techniques can also used to detect inorganic pyrophosphate liberated during nucleic acid amplification [15,16]. A recently described bioluminescence real time assay [BART] [17-19] allows the quantitative analysis of iNAATs, in real time. The biochemistry of BART is based on the 'Enzymatic Luminometric Inorganic pyrophosphate Detection Assay, or "ELIDA" [20,21] (Figure 1). Unlike previous applications of the ELIDA assay (most notably Pyro-sequencing™), BART allows dynamic changes in pyrophosphate levels to be monitored continuously in real-time over extended periods at 60°C for up to 2 hours. During a BART reaction, the level of light output increases to a peak whose timing under the same assay conditions reflects the initial concentration of the targeted DNA. Hence quantification of BART reactions utilises the time to peak light output and is not dependent on absolute light intensity produced, which greatly simplifies data interpretation and the hardware requirements, as well as making assays robust to turbidity and suspended solids [19].
Figure 1.
Chemistry of the BART bioluminescent coupled assay. PPi liberated during DNA synthesis reacts with adenosine-5'-O-persulfate (APS) in a reaction catalysed by adenosine triphosphate sulfurylase, to form adenosine triphosphate (ATP). A recombinant thermostable firefly luciferase liberates light, CO2, PPi and adenosine monophosphate (AMP) in the presence of the substrates luciferin and oxygen.
The accuracy of molecular diagnostic tests is dependent on appropriate integrity, purity and concentration of the input DNA and therefore on the choice of sample extraction procedure [22-24]. Plant tissues contain a variety of well-known compounds that can be inhibitory to molecular amplifications [25], including acidic polysaccharides, a variety of salts, secondary metabolites and phytochelatins. Most plant genomic DNA extraction technologies are designed to reduce or eliminate these contaminants. Polysaccharides can be removed by exploiting their differential solubilisation in solutions containing detergents, and affinity resins have also been used for the same purpose [26,27]. Hydrophobic cell constituents such as lipids and poly-phenols are routinely excluded from DNA extracts by partitioning with organic solvents, such as chloroform and alcohol. Unfortunately, many of the reagents used to extract and stabilize DNA, such as ethylene diamine tetra-acetic acid (EDTA), phenol, and the ionic detergents, sodium dodecyl sulfate (SDS) and cetyl tri-methyl ammonium bromide (CTAB) also tend to affect NAAT performance [28-31]. Measures to avoid carrying-over these contaminants can make these protocols labour intensive and time consuming to yield DNA of a sufficient quality for PCR.
Several published reports demonstrate that LAMP amplifications tolerate higher levels of certain inhibitors than PCR [32-34]. This suggests that LAMP could have a capacity to amplify polynucleotides from rapidly processed and crude sample matrix derived from plant material [34,35]. Other factors that affect the reliable detection and quantification of low target copy polynucleic acids using this technique are likely to include overall DNA loading within a reaction, which can have a impact upon sensitivity, as it possibly influences non-specific primer interactions [36]. Hence genome size, ploidy and unknown sources of contaminating DNA could affect amplification performance by altering the ratio of target to non-target DNA presence and hence potentially making target quantification and comparisons with reference samples and standards inaccurate. Here we demonstrate the use of LAMP-BART to detect GM events at low copy number levels in samples derived from maize, which has a large genome size and hence a relatively high proportion of non-target DNA. We show that LAMP-BART tolerates crude plant extracts without significant inhibition and examine the characteristics of the sample matrix that impact upon the quantitative nature of this technique and demonstrate its suitability in fieldable systems.
Methods
Plant material
Wild-type (Pure Gold) and transgenic (Mon810) maize were grown in 4 inch pots containing Sinclair Multipurpose compost for 4 weeks in a glasshouse that maintained a temperature of 25°C and a 16-h photoperiod, supplemented when necessary to a photon flux density of 350 μ mol m-2 s-1. Analysis of GM reference samples were performed on blends of lyophilized powdered Bt11 maize and Roundup Ready Soya (European Reference Material; with GM contents verified to be 0, 0.1, 0.5, 1, 2 & 5% w/w).
Conventional Genomic DNA extractions
40 mg of lyophilized or 200 mg fresh tissue were extracted using the Genome Wizard kit (Promega), Nucleon Phytopure kit (GE Healthcare) both according to the manufacturer's instructions, or the CTAB (cetyl trimethyl ammonium bromide) extraction method, which included RNase and proteinase digestions [37]. Once extracted the genomic DNA was resuspended in 100 μl of 100 mM DNAse-free Tris-HCl (pH 8.0) and refrigerated in non-stick plastic micro tubes (Ambion; Life Technologies) until required for analysis.
Rapid genome extraction
50 mg of fresh or lyophilized plant material was ground in 500 ul of genome extraction buffer (700 mM NaCl, 5% Chelex dissolved in 10 mM Tris-HCl buffer; pH 8). The extract was maintained at 100°C for 10 min, mixing regularly. 100 μl of the boiled extract was then desalted using a 0.5 ml Zeba column (Pierce; pre-equilibrated with 3 washes of Tris-EDTA buffer, pH 8), using a syringe to displace the DNA. The final elute contained the partially purified genomic DNA extract suitable for LAMP-BART reactions.
DNA quantity and purity
Genomic DNA was quantified by measuring the sample absorbance between 230 and 300 nm on a NanoDrop spectrophotometer. 1 μl of each DNA sample was analysed to check the quality and quantity of DNA. DNA was also quantified by agarose gel electrophoresis. 10 μl of diluted DNA (10-50 ng) was resolved on 0.8% TAE agarose gels (containing a 10-5 dilution of Gel Red; Biotium) by electrophoresis at 100 v for 60 min, and visualized by UV fluorescence using an Ingenius Gel Documentation System (Syngene). Light densities from the resolved DNA samples were quantified by comparing amplified product UV intensity against standard amounts of titrated λ-DNA resolved in the same way.
Gel analysis of amplified products
After amplification, samples were routinely resolved on 2% TAE agarose gels (containing 10-5 volumes of gel red; Biotium) at 100 volts for 60 min. The resolved amplicon was visualized and photographed over UV light, using an Ingenious Gel Documentation System (Syngene).
Copy Number Estimation
Target gene/transgenic element copy number was estimated by calculation assuming: the length of the maize genome [38]; 2 (diploid) copies of target polynucleotide/extracted genome; the average weight of a base pair (bp) is 650 Daltons; each bp has the same mass; the inverse of the calculated molecular weight is equivalent to the number of moles per gram and that using Avogadro's constant (6.022 × 1023) gives the copies of template/gram sample http://www.uri.edu/research/gsc/resources/cndna.html
LAMP-BART reaction mixture
LAMP-BART reactions were performed in a total volume of 20 μl. A LAMP-BART master mix that contained 1.6 μM of each LAMP primer, 0.8 μM each loop primer and 0.4 μM of each displacement primer, 300 μM each dNTP (Invitrogen), 87 mM trehalose (Sigma), 10 mM DTT (Sigma), 3.5 mM luciferin (Europa Bioproducts Ltd), 250 μM APS (Biolog Institute), Ultraglow Luciferase (Promega; 5.6 μg/ml), ATP sulphurylase (NEB; 375 milliunits/ml), 6.4 U Bst polymerase (NEB), PVP (Sigma; 0.4 mg/ml), 60 mM KCl (Sigma), 2 mM MgSO4 (NEB), diluted in the required amount of Thermopol buffer (NEB). Each reaction was made up to volume by adding the specified amounts of the target DNA or molecular grade water.
BART analysis
All LAMP-BART coupled amplifications were performed on dedicated instruments that simultaneously control temperature and quantify bioluminescence during a given assay. Two variations of the hardware were used; a static thermally controlled machine, equipped with a charged coupled device camera http://www.lumora.co.uk, that has no theoretical limit of sample numbers or configurations; and a portable device (19; photodiode quantification PDQ; http://www.lumora.co.uk), that quantifies light using photo-diodes, which is presently limited to the analysis of 16 samples. All LAMP-BART reactions were performed in suitable nuclease free plastic tubes under molecular grade mineral oil, at 60°C for 90 min.
RT-PCR analysis
Each 25 μl PCR reaction was performed using the JumpStart SYBR Green ready mix (Sigma) supplemented with 5 pmol of respective primers (a dedicated pair for each target; Table 1). Reaction mixtures were denatured for 2 min at 94°C (to disassociate the polymerase from its protective antibody). Each cycle was: 94°C for 30 s, 50°C for 30 s, 72°C for 30 s, for 40 cycles. Amplification and analysis was performed using an ABI Prism 7000 sequence detection system (Applied Biosystems). Results were processed using Applied Biosystems SDS 2.312 software.
Table 1.
Details of the primers used in the LAMP-BART and RT-PCR amplifications
| Primer Type | Orientation | Target (5' base) | Primer Sequence (5'-3') |
|---|---|---|---|
| Displacement | sense | ADH1 (7) | CTTTGGATCGATTGGTTTC |
| Displacement | antisense | ADH1 (287) | CCCAAAATTACTCAACG |
| LAMP | sense | ADH1 (116) | GGTGATCAAGTGCAAAGGTCTTTTCATAAACCAAGATTAGTCAGATCAAG |
| LAMP | antisense | ADH1 (94) | CCCCTCCGCAAATCTTCGAACAGTTTTGTAACTGGTGAAGGACTGAG |
| LOOP | sense | ADH1 (68) | CGCCTTGTTTCTCCTCTGTC |
| LOOP | antisense | ADH1 (136) | CCAAATCATCCACTCCGAGAC |
| Displacement | sense | CaMV-35 S-p (7214) | AGGAAGGGTCTTGCG |
| Displacement | antisense | CaMV-35 S-p (7404) | ATAAAGGAAAGGCCATCG |
| LAMP | sense | CaMV-35 S-p (7317) | GTCTTCAAAGCAAGTGGTTTTGGATAGTGGGATTGTGCG |
| LAMP | antisense | CaMV-35 S-p (7296) | TTCCACGATGCTCCTCGTTTTCCTCTGCCGACAGTGG |
| LOOP | sense | CaMV-35 S-p (7274) | TCCACTGACGTAAGGG |
| LOOP | antisense | CaMV-35 S-p (7350) | GGGGTCCATCTTTGGG |
| Displacement | sense | NOS-t (1850) | CGCGATAATTTATCCTAGTTTG |
| Displacement | antisense | NOS-t (2053) | CGTTCAAACATTTGGCAAT |
| LAMP | sense | NOS-t (1962) | GCATGACGTTATTTATGAGATGGGTTTTCGCTATATTTTGTTTTCTATCGCG |
| LAMP | antisense | NOS-t (1947) | CATGCTTAACGTAATTCAACAGTTTTTGAATCCTGTTGCCGGTC |
| LOOP | sense | NOS-t (2007) | GATTAGAGTCCCGCAATTATAC |
| LOOP | antisense | NOS-t (1925) | AAATTATATGATAATCATCGCAA |
| PCR | sense | ADH1 (1297) | AATTTTGGGGAAAGCTTCGT |
| PCR | antisense | ADH1 (1369) | TTCACCACGATTGCAGGATA |
| PCR | sense | CaMV-35 S-p (7133) | GATTCCATTGCCCAGCTATC |
| PCR | antisense | CaMV-35 S-p (7215) | CAACGATGGCCTTTCCTTTA |
| PCR | sense | NOS-t (1854) | TCGTTCAAACATTTGGCAAT |
| PCR | antisense | NOS-t (1885) | AAGACCGGCAACAGGATTC |
Underscored bases of the LAMP primers are additional foreign nucleotides, introduced to link different homology segments (CAMV-35 S-p LAMP primers contain four linker bases, while the other LAMP primers only contain 3 linker bases)
Primer design and synthesis
Previously published LAMP primers [39], (Table 1) were used to target the cauliflower mosaic virus 35 S promoter (CaMV 35 S-p; GenBank: X79465), and the Agrobacterium tumefaciens nopaline synthetase gene terminator (NOS-t; GenBank: V00087; 41), while the LAMP primers used to target Zea mays alcohol dehydrogenase reference gene (ADH1; GenBank: NM_001111939) were designed according to http://loopamp.eiken.co.jp/e/lamp/primer.html. The same three genes were also amplified by PCR (see Table 1); these primers were designed using Primer 3 http://frodo.wi.mit.edu/primer3/. All the primers were synthesisized by Eurofins MWG Operon as desalted, unmodified deoxribonucleotide oligonucleotides.
Regression analysis
Regression analysis was performed on experimental data sets where amplification procedures were assessed against various titrations of maize genomic DNA extracted using the 3 commercial extraction techniques and Lumora's simplified technique described above. The variation, linearity and efficiencies of the amplifications were calculated according to the mathematical algorithms stipulated by Pfaffl [40].
Comparative inhibition
Bt11 maize genomic DNA samples (5% GM; comprising 105 and 5 × 103 copies of wt and GM genomic copies respectively) prepared using Promega's Genome Wizard Kit were subjected to RT-PCR and LAMP amplifications as described earlier in the presence or absence of the following concentration of inhibitors (introduced into each assay at the following concentrations): SDS (sodium dodecyl sulphate; (0.005%; 0.01%; w/v); CTAB (cetyl trimethyl amonium bromide; 0.005%; 0.01%; w/v); NaCl (sodium chloride; 25 mM; 50 mM); Xylan (0.1%; 0.25%; w/v); Starch (0.1%; 0.25%; w/v); Humic Acid (0.01 ng; 0.1 ng); CaCl2 (1 μM; 100 μM).
Effect of carrier DNA on low copy amplification
The maize reference materials (0.1%, 0.5%, 1%, 2% and 5% GM), were extracted using Promega's Genome Wizard kit according to the manufacturer's instructions. The final air-dried pellets were hydrated in TE buffer and stored at 4°C. Extracted maize genomic DNA was quantified using the Nano Drop and electrophoresis methods described above. DNA from each extracted reference sample equivalent to 300, 200, 100 or 50 copies of 35 S promoter were assayed using the respective LAMP-BART assay to determine whether the genomic load has an impact upon the kinetics, reproducibility or sensitivity of this amplification method.
Results & Discussion
DNA extraction procedure affects quality estimations
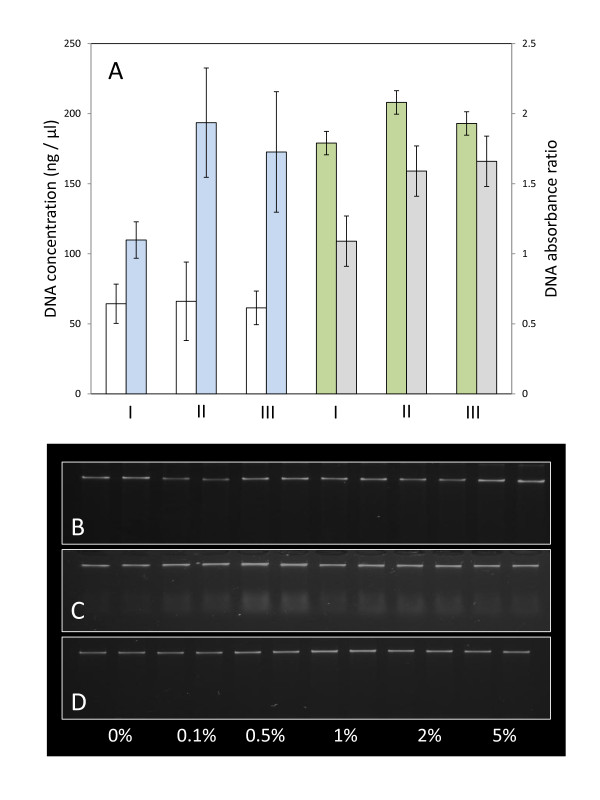
The quality of maize genomic DNA samples, extracted from Bt11 seed reference material were compared after extraction, using three different and commonly used plant DNA extraction methods (CTAB; Nucleon Phytopure™; Promega Genome Wizard™). When DNA was extracted using the Nucleon Phytopure kit, low molecular weight contamination was observed on gels after resolution of extracted samples by electrophoresis. Gel analysis of comparative DNA samples, extracted using the other methods, revealed little or no contamination (Figure 2B, C &2D). All three extraction methods yielded DNA apparently suitable for both PCR and LAMP-BART analysis, as defined by absorbance ratio (Figure 2A). We noted that DNA quantification was dramatically influenced by the choice of extraction and quantification technique. Gel images of the Nucleon Phytopure kit show fluorescence below the high molecular weight band, and may be consistent with RNA contamination (Figure 2C). RNA contamination may affect quantification, which is markedly higher (2 fold) when extracts were assessed using the spectrophotometric method (NanoDrop), compared to those made by gel density estimations. However, this discrepancy in calculated DNA yield was less obvious in samples of DNA extracted using the Promega Genome Wizard kit. Given potential differences in sample purity and composition, we suggest that the gel density method is likely to be a more reliable indication of comparative genomic DNA yield than spectrometry.
Figure 2.
Estimation of genomic DNA quantity and quality. Bar graph represents average estimated DNA concentrations from at least 50 extracted samples (A; ng/μl), as defined by gel density analysis (white bars), or spectrophotometric determinations using a NanoDrop spectrophotometer (blue bars). Results were obtained for DNA samples extracted using Promega's Genome Wizard kit (I), the Nucleon Phytopue Kit (II) or the CTAB-homebrew method (III). Collated absorbance ratios obtained from the same samples at 260: 280 nm (green bars), or 260: 230 nm (grey bars) are shown. DNA quantity and integrity was assessed by resolving DNA samples (extracted from Bt11 maize reference material; 0%, - 5%) on 0.8% TAE agarose gels by electrophoresis. Representative samples extracted using Promega's Genome Wizard kit (B), the Nucleon Phytopue Kit (C) or the CTAB-homebrew method (D).
Detection of maize GM Bt11 event using LAMP-BART and effect of DNA extraction procedures
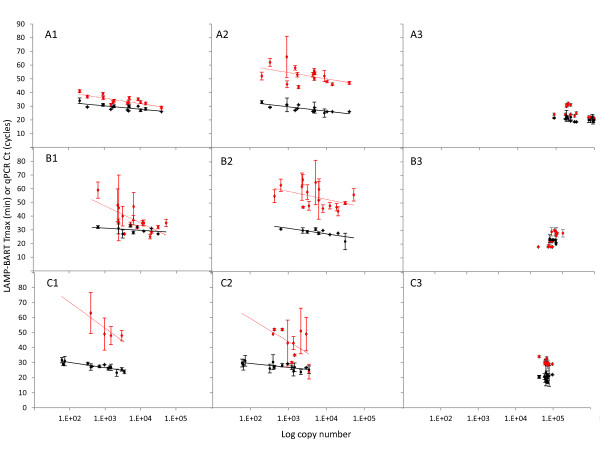
A comparison of LAMP-BART and PCR techniques was made using a titration series of maize Bt11 reference genome samples containing certified proportions of the Bt11 genome in a background of non-transgenic maize (0, 0.1, 0.5, 1.0, 2.0 & 5.0%), extracted from maize meal powder using three commercial genomic DNA extraction procedures. In each case, the total DNA load per assay remained constant, while GM copy number varied (50 to 103 copies/reaction) due to the different GM proportions in the original samples, from which the DNA was extracted. Both the GM LAMP-BART and RT-PCR techniques developed for this investigation were sufficiently sensitive to amplify reproducibly the cauliflower mosaic virus (CaMV) 35 S gene promoter (35 S-p) and nopaline synthase terminator (NOS-t) sequences, present in the transgene of the GM component of the reference samples, containing only 0.1% Bt11 maize powder. Both techniques amplified the endogenous ADH1 reference gene reproducibly for a given genome copy number (Figure 3).
Figure 3.
Influence of the DNA extraction method on RT-PCR and LAMP-BART efficiency. LAMP-BART (red points) and RT-PCR (black points) standard curves made by serial dilutions of DNA isolated using Promega's Genome Wizard kit (A), the Nucleon Phytopure Kit (B) or the CTAB-homebrew method (C; diluted 10 fold to reduce inhibitory nature of this preparation). DNA isolated using each extraction technique was amplified with the CaMV 35 S-p (1), NOS-t (2) or ADH1 (3) primer sets. Template concentration in LAMP-BART comparisons are plotted as a function of time to light peak (Tmax), while RT-PCR comparisons are based on cycle number (Ct). Each RT-PCR cycle is approximately equivalent to 2 min.
Each set of GM-LAMP-BART and RT-PCR assays was shown to be linear with respect to GM copy number, with comparable velocities (each Ct value presented represents 2 minutes). The linear regression analysis, demonstrated major differences in observed velocity, sensitivity and reproducibility for each LAMP-BART assay, which was largely extraction dependent (Figure 3). Assays exhibited greater variability and a reduced sensitivity when DNA was extracted using either the CTAB or Phytopure chemistries (Figure 3B and 3C). DNA extracted with the Promega Genome Wizard procedure had little impact upon either amplification technology (Figure 3, Table 2), and greater sensitivity and more reproducible results were achieved.
Table 2.
Regression analysis was performed on PCR and LAMP-BART data sets obtained using various DNA extractions performed on a titration series of Bt11 maize reference tissue (0.1 - 5%; nd - not determined)
| Target | Amplification | Extraction | Slope | Lowest detectable | Amplification |
|---|---|---|---|---|---|
| Sequence | Technique | Technique | (R2) | copy No (0.1% GM) | Efficiency E (+/- SD) |
| CaMV-35 S-p | PCR | Promega Wizard | 0.522 | 205 | 2.64 (0.46) |
| CaMV-35 S-p | PCR | Nucleon Phytopure | 0.131 | 438 | 2.32 (0.08) |
| CaMV-35 S-p | PCR | CTAB (10-1) | 0.495 | 70 | 2.47 (0.82) |
| CaMV-35 S-p | PCR | Lumora's Simplified | nd | nd | nd |
| NOS-t | PCR | Promega Wizard | 0.657 | 205 | 2.16 (0.41) |
| NOS-t | PCR | Nucleon Phytopure | 0.471 | 438 | 2.13 (0.18) |
| NOS-t | PCR | CTAB (10-1) | 0.775 | 70 | 1.91 (0.19) |
| NOS-t | PCR | Lumora's Simplified | nd | nd | nd |
| CaMV-35 S-p | LAMP-BART | Promega Wizard | 0.565 | 205 | 1.55 (0.22) |
| CaMV-35 S-p | LAMP-BART | Nucleon Phytopure | 0.471 | 641 | 1.24 (0.13) |
| CaMV-35 S-p | LAMP-BART | CTAB (10-1) | 0.247 | 350 | 1.1 (0.03) |
| CaMV-35 S-p | LAMP-BART | Lumora's Simplified | 0.403 | 40 | 1.21 (0.07) |
| NOS-t | LAMP-BART | Promega Wizard | 0.215 | 205 | 1.70 (0.73) |
| NOS-t | LAMP-BART | Nucleon Phytopure | 0.196 | 438 | 1.31 (0.33) |
| NOS-t | LAMP-BART | CTAB (10-1) | 0.740 | 350 | 1.15 (nd) |
| NOS-t | LAMP-BART | Lumora's Simplified | 0.119 | 40 | 1.25 (0.07) |
In all sets of analysis undertaken by RT-PCR, CaMV 35 S-p was amplified more efficiently than NOS-t, whereas efficiencies of amplification obtained using LAMP-BART were similar regardless of target sequence. However RT-PCR had a lower threshold of detection than LAMP-BART more reproducible data was achieved using the RT-PCR technique regardless of the extraction adopted in this experiment (Figure 3).
These results indicate that the DNA extracted using the CTAB and Phytopure methods are higher in contaminants or sample impurities incompatible with the LAMP-BART chemistry, perhaps reflecting their original development to service PCR. LAMP-BART and RT-PCR amplifications may therefore be affected by different inhibitors, either derived from the sample or the extraction procedure. The observed differences in the quantification data discussed previously (Figures 2 and 3) would lend weight this hypothesis. We therefore assessed the response of LAMP-BART to classical PCR inhibitors.
Inhibitors of RT-PCR and LAMP-BART
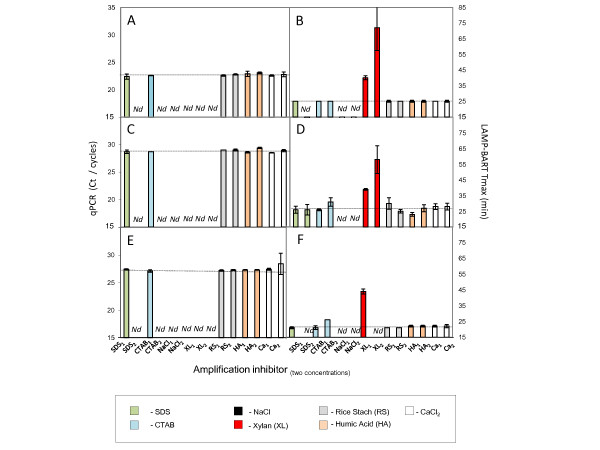
The effect of a range of inhibitors known to affect Taq polymerase [25,27] was tested on LAMP-BART to assess whether they affect the Bst polymerase used in LAMP or the LAMP-BART reaction couple. Comparative assessment of RT-PCR and LAMP-BART kinetics was therefore carried out in the presence of known PCR inhibitors. Promega Wizard extracted Bt11 maize genome was used during this investigation to standardize the GM target in each assay (5% maize reference genome; 105 copies ADH or 103 copies of the GM targets per assay). Moreover, the previous experiments confirmed that the DNA extracted using this technique was likely to contain fewer impurities and consequently, the lowest innate influence over either amplification methods (Figure 3, Table 2).
The monovalent salt (NaCl) abolished both types of amplification at 25 mM, regardless of the target and template concentration (Figure 4). This salt is likely to be perturbing these amplifications via Cl-, as this anion is known to compete for the active site of polymerase enzymes more effectively than phosphate, glutamate and acetate [41]. Clearly both polymerase enzymes are susceptible to this type of inhibition. The RT-PCR reaction was also strongly inhibited by the higher concentration of SDS and CTAB used (0.01%), and 0.1% of the acidic polysaccharide xylan. These inhibitors had little or no effect upon the kinetics of the equivalent LAMP-BART reactions (Figure 4B, D, F), although these detergents did reduce the output light intensity of the BART reporter, and hence were probably affecting one or more of the enzymes used to generate the light signal rather than LAMP amplification (19; data not shown). Neither starch, humic acid nor CaCl2 affected the kinetics of either amplification technique (Figure 4), at the concentrations used. It is also notable that RT-PCR reactions were either apparently unaffected or completely inhibited by specific inhibitors, and these effects are common to the different primer pairs tested. In contrast, LAMP-BART showed differences between the responses of different primer sets and also displayed increased reaction times in the presence of certain inhibitors particularly xylan. This points to the need for appropriate controls for sample inhibition in quantitative assays.
Figure 4.
Comparison of inhibitors of PCR and LAMP-BART. Bt11 maize genomic DNA samples (5% GM) were subjected to RT-PCR amplification using ADH1 primers (A); CaMV 35 S-p primers (C) or NOS-t primers (E). A comparative set of analysis was performed using LAMP-BART, conducted in the presence of ADH1 primers (B); CaMV 35 S-p primers (D) or NOS-t primers (F). The extracted sample contained either 105 copies of wild type genome or 5 × 103 copies of transgenic genome. The analysis was performed in the presence and absence of final inhibitor concentration defined: sodium dodecyl sulphate; (SDS; 0.005% [1]; 0.01% [2] – GREEN BARS); cetyl trimethylamonium bromide (CTAB; 0.005% [1]; 0.01% [2] – BLUE BARS); sodium chloride (NaCl; 25mM [1]; 50mM [2]- BLACK BARS); xylan (XL; 0.1% [1]; 0.25% [2] – RED BARS); rice starch (RS; 0.1% [1]; 0.25% [2] – GREY BARS); humic acid (HA; 0.01ng [1]; 0.1ng [2] – BROWN BARS); calcium chloride (CaCl2; (1µM [1]; 100µM [2] – WHITE BARS). The broken line (----------), represents the average Ct or Tmax value for each set of amplifications performed in the absence of inhibitor. In some instances amplification or reporting of this activity was completely inhibited (not determined; Nd).
These results indicate that Taq polymerase is more prone to inhibition by plant acidic polysacharrides than Bst polymerase. It is therefore likely that more rapid solutions for DNA extraction may be appropriate for isothermal amplifications from matrixes that utilize these displacement polymerases as will be demonstrated later in this manuscript.
The effect of total DNA concentration on LAMP-BART
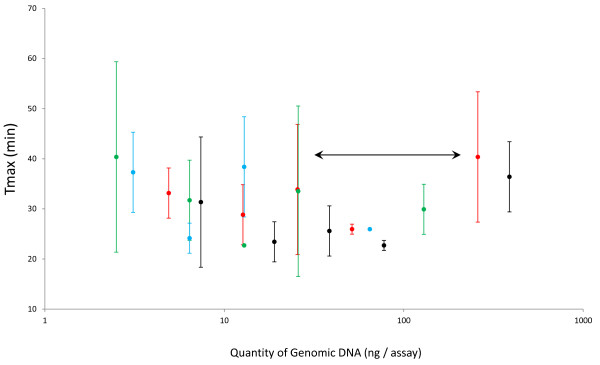
A further contributor to the sensitivity of molecular amplifications is the total quantity of DNA in the reaction, as this is thought to affect the retention of polynucleotides to plastic ware, and sequester primers and/or polymerase, and can thereby reduce mis-amplification events [36,42]. We therefore investigated the reproducibility of LAMP-BART assays across a wide range of total DNA concentration from 5-750 ng per assay containing a constant amount of the target sequence. Results were determined for 50, 100, 200 and 300 copies of target. The data (Figure 5) show that reproducibility of the CaMV 35 S-p LAMP-BART significantly deteriorates when the total amount of genome is below 50 ng or above 500 ng total assay DNA, regardless of the target copy number; the converse is apparent when the target polynucleotides are assayed between these copy numbers (Figure 5), where a much higher degree of reproducibility is achieved.
Figure 5.
Effect of varying total DNA concentration on CaMV 35 S-p LAMP-BART assay kinetics and reproducibility. Graph of average Tmax values obtained for a constant target copy number present in a range of total maize genomic DNA concentrations, for each of 4 target copy numbers: 300 copies/assay (black circle symbol); 200 copies/assay (red circle symbol); 100 copies/assay (green circle symbol) and 50 copies/assay (blue circle symbol). Errors represent standard deviation of the mean (three replicates). The least variability in Tmax data was observed between the concentrations of genome marked with an arrow (50-500 ng).
LAMP-BART quantification relies on dependable assay kinetics and accurate estimations of the ensuing time-to-maximum light output (Tmax; 19). These data show that the total DNA load within a given reaction must therefore be taken into consideration. Similar considerations apply when preparing standards for reference curves [43]. In the case of PCR, reports in the literature are sparse, but a few demonstrate the potential for carrier DNA to positively impact upon the sensitivity and specificity of low copy PCR [36,43]. However, this is not observed when a hot start method is adopted [42]. It is thought that the carrier DNA affects the dominance of side reactions that occur before thermal cycling commences, while reactions are being formulated on the bench. If the polymerase is active during this phase in the procedure, then it can potentially propagate primer oligomerization and mis-priming events. Carrier DNA is thought to quench these side reactions at limited copy numbers, by sequestering the DNA polymerase and primers [36,44]. It is clear that the effect of carrier DNA is not limited to PCR, and is likely to be more significant in isothermal reactions, such as LAMP, where multiple primers are used to drive the amplification process.
Other reports describe how large amounts of non-target DNA can become limiting at the threshold of PCR detection, affecting both sensitivity and analytical kinetics [44], as we observed here (Figure 5). It is likely that higher genome loads and DNA concentrations compete for the DNA polymerase and primers intended for the target nucleotide. Together, this data defines a requirement to keep the carrier DNA within a given window for quantitative analysis. Furthermore, both biological samples and standards should be compared at equivalent concentrations of DNA.
Rapid fieldable DNA extraction procedure
A rapid and simple extraction technique was devised that capitalized on the increased tolerance of LAMP to acidic polysaccharide inhibition. Maize leaf discs (wt or Mon810) were extracted in sodium chloride (cell lysate) and CHELEX, a resin with a high affinity for divalent cations, thereby reducing problematic tertiary DNA structures that are known to become exacerbated in the presence of both Mg2+ and Ca2+, and limiting DNA degradation after cell lysis by inhibiting DNase activity, as this enzyme requires Mg2+ [45-49]. The resulting extracts were used successfully in LAMP-BART assays (Figure 6). Interestingly, the same samples were completely inhibitory to RT-PCR, known to be more prone to inhibition by acidic polysaccharides, which is a contaminant likely to be present in these plant extracts.
Figure 6.
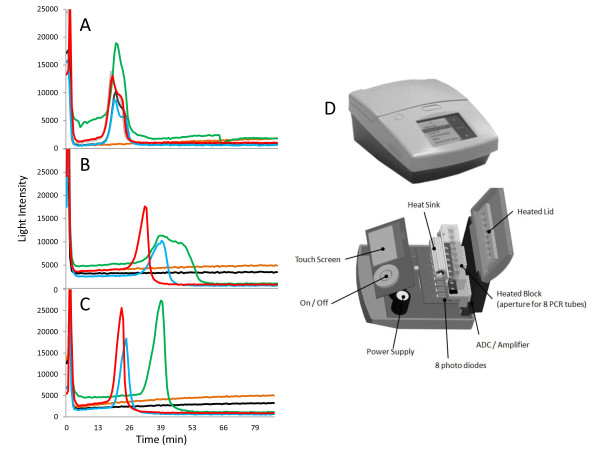
Qualitative determinations of GM maize presence using fieldable extraction and amplification. LAMP-BART light intensities recorded over time in amplifications of ADH1 (A), CaMV 35 S-p (B) and NOS-t (C), performed on DNA extracted from maize reference samples, containing either containing either 0% (brown line), 0.1% (green line), 1.0% (blue line) or 5.0% (red line) genetically modified maize or a MGW control (black line). The analysis was performed in a field environment using a portable instrument (PDQ; Lumora Ltd, Ely, UK); the exploded view illustrates the simplicity of this device (D).
This simple genome extraction method allowed LAMP-BART amplification to be performed on 0.1% Bt11 maize reference sample (Figure 6A, B &6C). The amplification of both CaMV 35 S-p and NOS-t were shown to be linear with respect to target copy (data not shown) and achieved similar efficiencies and thresholds of detection compared to the same assays performed on DNA extracts obtained using commercial DNA extraction kits. Moreover, this extraction technique is suited to qualitative field testing, as it is rapid and only requires simple hardware (Figure 6D).
Conclusions
Here we show the use of the recently described bioluminescent coupling of loop mediated amplification (LAMP) to the real-time bioluminescent reporting of amplification (BART) for the detection of low levels of genomic GM maize DNA, equivalent to contamination of 0.1% or 50 copies of GM target per 20 μl assay. The optimum level of total DNA in such LAMP-BART assays was determined to be in the range 75 ng per reaction (4 ng/μl). The sensitivity and reproducibility of reactions where GM target is limiting can be improved if the carrier DNA is supplemented to 80 ng/assay.
This investigation also highlights the impact that the choice of plant DNA extraction and quantification technique has on RT-PCR and LAMP-BART. The latter is less well suited to some conventional plant DNA extraction procedures, but is less affected by classical PCR amplification inhibitors, particularly acidic polysaccharides. A consequence of this robust nature was LAMP-BART's ability to amplify target DNA from rapidly extracted crude genome samples which were refractory to RT-PCR, and we show the application to a field-based qualitative test for GM maize. Together these data illustrate the potential for rapid testing of GM samples using LAMP-BART and highlights the importance of extraction technique, DNA quality and yield, genome and inhibitor loading on the quantitative nature of molecular tests.
Competing interests
LT and JM are the inventors of Bioluminescent Assay in Real-Time (BART). They filed a patent and co-founded Lumora Ltd, a company that has full rights for the use of the BART patent. JM LT and OG declare financial interests in Lumora Ltd.
Authors' contributions
The following authors were all employees at Lumora Ltd: GK OG CP CJM RJ NA CM and LT. GK PH NB OG CP CJM MR RJ NA CM conceived and designed the experiments detailed in this manuscript, while the experimental work was performed by GK PH NB MR OG, and the data analyzed by GK PH NB OG LT JAHM. The reagents, materials, and analytical tools were provided by GK PH NB OG NA LT JAHM and the manuscript was written by GK OG NA LT JAHM; all other authors have read and approved the final version of this manuscript.
Contributor Information
Guy Kiddle, Email: g.kiddle@Lumora.co.uk.
Patrick Hardinge, Email: hardingep@cardiff.ac.uk.
Neil Buttigieg, Email: buttigiegnd@Cardiff.ac.uk.
Olga Gandelman, Email: O.gandelman@lumora.co.uk.
Clint Pereira, Email: c.pereira@lumora.co.uk.
Cathal J McElgunn, Email: c.mcelgunn@lumora.co.uk.
Manuela Rizzoli, Email: manuriz@inwind.it.
Rebecca Jackson, Email: r.harrall@lumora.co.uk.
Nigel Appleton, Email: n.appleton@lumora.co.uk.
Cathy Moore, Email: catamour17@hotmail.com.
Laurence C Tisi, Email: l.tisi@lumora.co.uk.
James AH Murray, Email: murrayja1@cardiff.ac.uk.
Acknowledgements
We are extremely grateful for the Mon810 maize seed and primers designed and donated by David Lee (National institute of Botany, UK), for the purposes of this work. Thanks also to Sue Howroyd and the other support staff at the Institute of Biotechnology (Cambridge University, UK) for carefully maintaining the maize plants used in these experiments. We thank Marc Bax (Panchromos Ltd) and Staff at Grant Instruments and Syngene, for the development and production of the dedicated hardware required for this investigation.
This work was funded by Lumora Ltd with the financial support of the Co-Extra project: GM and non-GM supply chains, their CO-Existence and TRAcebility (Contract FOOD-2005-CT-007158), funded by the European Commission, through the Sixth Framework Programme under the Food Quality and Safety Priority http://www.coextra.eu, Lumora Ltd, Cambridge and Cardiff Universities. PH and NB were recipients of a Biotechnology and Biological Sciences Research Council (BBSRC) Collaborative Award in Science and Engineering (CASE) PhD studentship.
References
- Heller R. GM Nation? The findings of the public debate. London, HMSO: Department of Trade and Industry; 2003. doi: http://www.aebc.gov.uk/aebc/reports/gm_nation_report_final.pdf. [Google Scholar]
- King D. GM Science Review: Second Report, Prepared by the GM Science Review Panel under the chairmanship of Sir David King for the UK Government. 2004.
- Regulation (EC) No 1830/2003 of the European Parliament and of the Council of 22 September 2003 concerning the traceability and labelling of genetically modified organisms and the traceability of food and feed products produced from genetically modified organisms and amending Directive 2001/18/EC
- Miraglia M, Berdal KG, Brera C, Corbisier P, Holst-Jensen A, Kok EJ, Marvin HJ, Schimmel H, Rentsch J, Van Rie J, Zagon J. Detection and traceability of genetically modified organisms in the food production. Chain Food and Chemical Toxicology. 2004;42(7):1157–1180. doi: 10.1016/j.fct.2004.02.018. [DOI] [PubMed] [Google Scholar]
- Ramessar K, Capell T, Twyman RM, Quemada H, Christou P. "Trace and traceability- a call for regulatory harmony". Nat Biotech. 2008;26(9):975–978. doi: 10.1038/nbt0908-975. doi: http://fbae.org/2009/FBAE/website/images/PDF%20files/biosafety/GMO%20Tracking.pdf. [DOI] [PubMed] [Google Scholar]
- Dong W, Yang L, Shen K, Kim B, Kleter GA, Marvin HJP, Guo R, Liang W, Zhang D. GMDD: a database of GMO detection methods. BMC Bioinformatics. 2008;9(260):1471–2105. doi: 10.1186/1471-2105-9-260. doi: http://www.biomedcentral.com/content/pdf/1471-2105-9-260.pdf. p. 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisset D, Stebih D, Cankar K, Zel J, Gruden K. Alternative DNA amplification methods to PCR and their application in GMO detection: a review. European Food Research and Technology. 2008;227(5):1287–1297. doi: 10.1007/s00217-008-0850-x. doi: http://www.springerlink.com/content/cw7381p6520117x2/fulltext.pdf?page=1. [DOI] [Google Scholar]
- Marmiroli N, Maestri E, Gullì M, Malcevschi A, Peano C, Bordoni R, De Bellis G. Methods for detection of GMOs in food and feed. Anal Bioanal Chem. 2008;392(3):369–384. doi: 10.1007/s00216-008-2303-6. doi: http://www.springerlink.com/content/p0r6tp36823l14t1/fulltext.pdf?page=1. [DOI] [PubMed] [Google Scholar]
- McPherson MJ, Moller SG. PCR the basics. Taylor & Francis. 2006.
- Demidov VV, Broude NE. DNA Amplification Technologies. Current Technologies and Applications Horizon Bioscience. 2004.
- Fukuta S, Mizukami Y, Ishida A, Ueda J, Hasegawa M, Hayashi I, Hashimoto M, Kanbe M. Real-time loop-mediated isothermal amplification for the CaMV-35 S promoter as a screening method for genetically modified organisms. European Food Research & Technology. 2004;218(5):496–500. doi: 10.1007/s00217-003-0862-5. doi: http://www.springerlink.com/content/d1xb009pun3k0yjy/fulltext.pdf?page=1. [DOI] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16(3):223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Whitehead TP, Kricka LJ, Carter TJ, Thorpe GH. Analytical luminescence: its potential in the clinical laboratory. Clin Chem. 1979;25(9):1531–1546. [PubMed] [Google Scholar]
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. http://www.gene-quantification.de/higuchi-1993.pdf. 1993;11(9):1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- Gandelman OA, Church VL, Moore CA, Carne C, Jalal H, Murray JAH, Tisi LC. BART-bioluminescent alternative to real-time PCR. Proceedings of the 14th International Symposium on BL & CL Ed. Szalay A.A. et al. 2006. pp. 95–98.
- Tisi LC, Murray JA. H. (WO/2004/062338) Method for determining the amount of template nucleic acid present in a sample. BART; Patent no EP20050767491. http://www.wipo.int/pctdb/en/wo.jsp?WO=2004062338&IA=WO2004062338&DISPLAY=DESC
- Gandelman OA, Church VL, Moore CA, Kiddle G, Carne CA, Parmar S, Jalal H, Tisi LC, Murray JA. Novel bioluminescent quantitative detection of nucleic acid amplification in real-time. PLoS One. 2010;5(11):e14155. doi: 10.1371/journal.pone.0014155. doi: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0014155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamohamed S, Nyrén P. Real-time detection and quantification of adenosine triphosphate sulfurylase activity by a bioluminometric approach. Anal Biochem. 1999;271(1):81–85. doi: 10.1006/abio.1999.4130. [DOI] [PubMed] [Google Scholar]
- Eriksson J, Karamohamed S, Nyrén P. Method for Real-Time Detection of Inorganic Pyrophosphatase. Activity Analytical biochemistry. 2001;293:67–70. doi: 10.1006/abio.2001.5106. [DOI] [PubMed] [Google Scholar]
- Vroh Bi I, Harvengt L, Chandelier A, Mergeai G, Du Jardin P. Improved RAPD amplification of recalcitrant plant DNA by the use of activated charcoal during DNA extraction. Plant Breed. 1996;115:205–206. doi: 10.1111/j.1439-0523.1996.tb00905.x. [DOI] [Google Scholar]
- Zimmermann A. A sensitive detection method for genetically modified maisGard™ corn using a nested PCRsystem Lebensm.-Wiss. U-Technol. 1998;31:664–667. [Google Scholar]
- Cankar K, Stebih D, Dreo T, Zel J, Gruden K. Critical points of DNA quantification by real-time PCR--effects of DNA extraction method and sample matrix on quantification of genetically modified organisms. BMC Biotechnol. 2006;6:37. doi: 10.1186/1472-6750-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeke T, Adams RP. The effects of plant polysaccharides and buffer additives on PCR. Biotechniques. 1992;12(3):332–334. [PubMed] [Google Scholar]
- Puchooa D, Shahnoo S, Khoyratty S. Genomic DNA extraction from Victoria amazonica. Plant Molecular Biology Reporter. 2004;22(2):195–196. doi: 10.1007/BF02772727. [DOI] [Google Scholar]
- Peist R, Honsel D, Twieling G, Löffert D. PCR inhibitors in plant DNA preparations. 2001. http://www1.qiagen.com/literature/qiagennews/0301/1017317_QN301_ITJPDIS-7-9.pdf
- Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62(3):1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. doi: http://aem.asm.org/cgi/reprint/62/3/1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology. 1998;5(2):69–76. doi: 10.1007/BF00020088. doi: http://www.springerlink.com/content/m184517145445166/fulltext.pdf?page=1. [DOI] [PubMed] [Google Scholar]
- Spoth B, Strauss E. Screening for genetically modified organisms in food using Promega's wizard resin. 1999.
- Melzak KA, Sherwood CS, Turner RFB, Haynes CA. Driving forces for DNA adsorption to silica in perchlorate solutions. J Colloid Interface Sci. 1996;181:635–644. doi: 10.1006/jcis.1996.0421. [DOI] [Google Scholar]
- Curtis KA, Rudolph DL, Owen SM. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP) J Virol Methods. 2008;151(2):264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Kawana T, Fukushima E, Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. Journal of Biochemical and Biophysical Methods. 2007;70(3):499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, Perkins MD, Schrenzel J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol. 2011;62(1):41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- Lee D, Mura M, Allnutt T, Powell W, Greenland A. Isothermal Amplification of Genetically Modified DNA Sequences Directly from Plant Tissues Lowers the Barriers to High-Throughput and Field-Based Genotyping. Journal of Agricultural and Food Chemistry. 2009;57(20):9400–9402. doi: 10.1021/jf902455j. [DOI] [PubMed] [Google Scholar]
- Chou Q, Russell M, Birch DE, Raymond J, Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992;20(7):1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11. [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Ann Bot. 2005;95(1):127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, La Mura M, Allnutt TR, Powell W. Detection of genetically modified organisms (GMOs) using isothermal amplification of target DNA sequences. BMC Biotechnol. 2009;9:7. doi: 10.1186/1472-6750-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR Nucleic. Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. doi: http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=55695&blobtype=pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep MA, McHenry CS. Glutamate overcomes the salt inhibition of DNA polymerase III holoenzyme. J Biol Chem. 1989;264(19):11294–11301. [PubMed] [Google Scholar]
- Ellison SL, English CA, Burns MJ, Keer JT. Routes to improving the reliability of low level DNA analysis using real-time PCR. BMC Biotechnol. 2006;6:33. doi: 10.1186/1472-6750-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt F, Barbati C, Paoletti C, Querci M, Kay S, De Beuckeleer M, Van den Eede G. Real-time polymerase chain reaction-based approach for quantification of the pat gene in the T25 Zea mays event. J AOAC Int. 2004;87(6):1342–1355. [PubMed] [Google Scholar]
- Garner TW. Genome size and microsatellites: the effect of nuclear size on amplification potential. Genome. 2002;45(1):212–215. doi: 10.1139/g01-113. [DOI] [PubMed] [Google Scholar]
- Kwon HB, Su HS, Jong SL, Sung RM, Suk MK, Jang RL, Dongsu C, Won JJ. Rapid and simple method for DNA extraction from plant and algal species suitable for PCR amplification using a chelating resin Chelex 100. Plant Biotechnology Reports. pp. 49–52.
- Dobi A, van Agoston D. Submillimolar levels of calcium regulates DNA structure at the dinucleotide repeat (TG/AC)n. Proc Natl Acad Sci USA. 1998;95(11):5981–5986. doi: 10.1073/pnas.95.11.5981. doi: http://www.pnas.org/content/95/11/5981.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H, Dahlberg JE. Topology and formation of triple-stranded H-DNA. Science. 1989;243(4898):1571–1576. doi: 10.1126/science.2648571. doi: http://www.sciencemag.org/cgi/content/aBstract/243/4898/1571. Review. [DOI] [PubMed] [Google Scholar]
- Kohwi Y, Kohwi-Shigematsu T. Altered gene expression correlates with DNA structure. Genes Dev. 1991;5(12B):2547–2554. doi: 10.1101/gad.5.12b.2547. doi: http://genesdev.cshlp.org/content/5/12b/2547.full.pdf. [DOI] [PubMed] [Google Scholar]
- Roberts RW, Crothers DM. Specificity and stringency in DNA triplex formation. Proc Natl Acad Sci USA. 1991;88(21):9397–9401. doi: 10.1073/pnas.88.21.9397. doi: http://www.pnas.org/content/88/21/9397.full.pdf+html. [DOI] [PMC free article] [PubMed] [Google Scholar]