Abstract
In regions with high prevalence of chronic hepatitis B virus (HBV) infection and dietary aflatoxin B1 (AFB1) exposure, hepatocellular carcinomas (HCCs) often contain TP53 mutation at codon 249 (R249S). Furthermore, a C-terminal truncated HBx protein expressed from hepatocyte integrated HBV is associated with HCC development. This study evaluates the association between R249S and HBX status in relation to HCC in West African population. HBX (complete or 3′-truncated) and HBS genes were assessed by PCR in cell-free DNA (CFDNA) from plasma of subjects recruited in a hospital-based case–control study (325 controls, 78 cirrhotic patients and 198 HCC cases) conducted in The Gambia. These samples had been previously analyzed for R249S and HBV serological status. Complete HBX sequence was frequently detected in CFDNA of HCC-R249S positive (77%, 43/56) compared with HCC-R249S-negative cases (44%, 22/50). Conversely, the proportion of 3′-truncated HBX gene was significantly higher in HCC-R249S negative than positive cases (34%, 17/50, compared with 12%, 7/56) (χ2 = 12.12; P = 0.002; distribution of R249S negative and positive according to HBX status). Occult HBV infection (detected by PCR) was present in 24% of HCC previously considered as negative by HBV serology. Moreover, HBV mutation analysis revealed that double mutation at nucleotides 1762T/1764A was associated with diagnosis of cirrhosis or HCC {cirrhosis: odds ratio (OR): 9.50 [95% confidence interval (CI) 1.50–60.11]; HCC: OR: 11.29 [95% CI 2.07–61.47]}. These findings suggest that in HCC from The Gambia, complete HBX sequences are often associated with the presence of TP53 R249S mutation.
Introduction
Hepatocellular carcinoma (HCC) is a major cause of cancer mortality and morbidity in many parts of the world. Overall, it is the seventh most common cancer globally and the third leading cause of cancer-related deaths (1). Over 80% of HCC occurs in sub-Saharan Africa and Southern and Eastern Asia. In The Gambia, West Africa, incidence rates are 32.84 per 100 000 person-years in men and 14.9 cases per 100 000 in women (2). The main risk factors are chronic hepatitis B virus (HBV), which is endemic in the Gambian population. The proportion of carriers among non-HBV vaccinated Gambians is 15% and aflatoxin B1 (AFB1)-contaminated foodstuffs are commonly consumed by >95% of the population (3–5). Other risk factors such as HCV and alcohol are found to play a relatively minor role, particularly in cases arising <50 years of age (4,6). AFB1 is a mycotoxin produced by the fungus Aspergillus flavus that contaminates the staple diet in many tropical areas, in particular after storage under hot and humid conditions. After ingestion and metabolism in the liver, metabolites of AFB1 can form a DNA adduct at the third base of codon 249 in the TP53 tumor suppressor gene, inducing a G to T transversion (AGG to AGT, arginine to serine; R249S), a mutation that is detected in >75% of HCC from areas with high AFB1 exposure (7–10).
Chronic HBV infection is the dominant global attributable risk factor for HCC, accounting for 55% of cases worldwide and ≥80% in sub-Saharan Africa and South-East Asia (11). The mechanisms by which HBV causes malignant transformation remain uncertain; however, many studies have highlighted a pathogenic role for HBx, the product of the HBV gene HBX, which encodes a 154 amino acid, 17 KDa factor that enhances viral replication and modulates multiple cellular growth signaling pathways (12). One of the molecular hallmarks of HBV-induced HCC is integration of the viral genome into the host cellular genome detected in 80%–90% of these cancers (13,14). Integration of viral DNA may lead to insertional mutagenesis (15). In addition, integration often results in a 3′-terminal truncation of HBX, generating a shorter form of the HBx protein that is deleted at the C-terminal region by 20–40 amino acids. Integration is also often accompanied by multiple point mutations in the HBX sequence. It has been suggested that both truncation and point mutations may enhance the oncogenic activation processes. In experimental models, the C-truncated HBx proteins were shown to transform immortalized liver cell lines (16) and to interact with the mutant p53 protein p.R249S to alter genetic stability and proliferation of non-transformed hepatocytes (17). Other studies found that the mutant p53 protein p.R249S and HBx formed stable complexes in transformed hepatocytes, further suggesting that the two factors could cooperate in hepatocarcinogenesis occurring in a HBV/AFB1 etiological setting (18).
The genotypes/subgenotypes of HBV show heterogeneity in their global distributions and these varied strains have differences in their biological properties (19–21). It is now evident that the heterogeneity in this distribution of HBV genotypes/subgenotypes may account not only for differences in the prevalence of HBV mutations in the different populations but may also be responsible for variation in the clinical outcomes of HBV infections and the response to antiviral therapy (22,23).
In this study, we have explored circulating cell-free DNA (CFDNA) from Gambian HCC patients to further address the contribution of R249S and HBX to cancer risk. Previous studies have shown that plasma or serum of cancer subjects (in particular HCC patients) as well as healthy subjects contains detectable amounts of CFDNA (range: 1–500 ng/ml). This CFDNA often contains mutations and epigenetic alterations identical to those detected in tumor tissues (24). In a case–control study of HCC in The Gambia, we have used CFDNA to demonstrate the presence of R249S in up to 40% of HCC patients while low levels of mutant DNA were also detected in non-cancer controls, in particular among high-risk HBV chronic carriers (25,26). Although the molecular mechanisms of release and the stability of CFDNA are still unclear, this resource appears to represent a convenient minimally invasive repository of biomarkers for HCC in regions where liver biopsies are not routinely available. Using CFDNA from the plasma of subjects recruited in a hospital-based case–control study conducted in The Gambia and previously analyzed for R249S status and for HBV serological status, we have examined HBV genotype by sequencing the HBS gene (encoding HBsAg) and we have characterized HBX integrity and mutational status.
Materials and methods
Study participants
This study has been conducted using specimens and data from The Gambia Liver Cancer Study, for which design, ethical approval, recruitment procedures and laboratory testing methods have been previously reported (4). Briefly, The Gambia Liver Cancer Study was a hospital-based case–control study in which incident cases of HCC and cirrhosis were recruited from three tertiary hospitals sites in The Gambia from September 1997 to January 2001. The diagnosis of HCC was based on concordant clinical and ultrasonography findings and on serum levels of α-fetoprotein of ≥100 ng/ml. Cirrhosis was diagnosed using clinical parameters and ultrasonography (27). A minority of the cases was confirmed by histopathology of liver biopsies (4). Hospital controls with no clinical evidence for liver disease were frequency matched by age (within 10 years), gender and study site. All study participants provided informed consent and both Gambian and international institutional review boards approved the study protocol.
HBV serology, DNA extraction and quantitation of R249S mutation in CFDNA
Methods and results for HBV serology, DNA extraction and quantitation of R249S mutation in CFDNA have been reported previously (4,25,26). Briefly, HBV surface antigens (HBsAg) and HBV e antigens (HBeAg) were determined as markers of chronic HBV carriage or viral replication, respectively, using standard laboratory kits (Murex Diagnostics Ltd, Dartford, UK and Sorin Biomedica Diagnostics, Vercelli, Italy). CFDNA was extracted from 200 μl of plasma using QiAmp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's blood and body fluid spin protocol. Purified CFDNA was eluted from the QiAmp Silica column with two volumes of water (2 × 50 μl) (PCR-grade, Sigma, St Louis, MO). Quantitation of extracted DNA was performed by fluorimetry using picogreen (Molecular Probes, Eugene, OR). R249S was detected using two methods, restriction fragment length polymorphisms (RFLP) (25) or short oligonucleotide mass analysis (SOMA). All samples were analyzed by RFLP and the status of 38% was confirmed by SOMA. The latter method allowed the quantitation of R249S against a synthetic, internal standard as described previously (26) and results were considered as ‘R249S-positive’ at a cutoff number of 67 copies of mutant DNA/ml. Overall, RFLP and SOMA results were highly concordant.
Analysis of HBX truncation status
Detection of truncated versus complete integrated HBX was performed by PCR and sequencing based upon a method initially described by Ma et al. (16) and modified as described previously (28). Briefly, CFDNA was used to produce four overlapping amplicons from HBX gene of 139, 192, 334 or 425 bp. The 425 bp amplicon encompasses the entire HBX sequence and the three shorter amplicons correspond to fragments initiated at the 5′-end of HBX and covering progressive lengths of its sequence. Amplification of all four fragments signals the presence of a complete HBX, whereas amplification of one or several shorter fragments signals the presence of 3′-truncated HBX. The same forward primer X1F was used in each reaction (5′-GGGACGTCCTTTGTCTACGT-3′). The four reverse primers were X1R (5′-GGGAGACCGCGTAAAGAGAG-3′), X2R (5′-GTGCAGAGGTGAAGCGAAGT-3′), X3R (5′-CCCAACTCCTCCCAGTCTTT-3′) or X4R (5′-GGCAGAGGTGAAAAAGTTGCA-3′). PCR was carried for 50 cycles after activation with GoTaq Hot Star Polymerase at 94°C for 2 min (0.5 U, Promega). PCR cycles included denaturation at 94°C for 45 s, annealing at 62°C for 45 s and extension at 72°C for 45 s followed by a final extension at 72°C for 10 min. Five microliters of PCR products were purified using standard ExoSap-IT (Usb Corporation, Cleveland, OH) and nucleotide sequences were determined for both strands by automated dideoxy sequencing (sequencer AbiPrism 3100, Applied Biosystems, Foster city, CA) using the same primers as for PCR. Sequences (nucleotide and predicted protein) were analyzed against HBX reference genes from GenBank using MEGA5 software (29). Because of the presence of a gap and of the covalent binding of the viral polymerase at the 5′ end of this gap, this PCR method does not readily amplify the partially double-stranded viral genome. Rather, it amplifies HBX sequences released into CFDNA either as HBV replication intermediates or as integrated viral DNA in the host’s genome. The identity of all PCR products was confirmed by a second PCR and sequencing.
HBV genotyping
HBV genotyping was performed by sequencing HBS after amplification using a heminested PCR as described elsewhere (30). For the first reaction, 2 μl of DNA were used with primers S_HBVpol1 (5′-CCTGCTGGTGGCTCCAGTTCA-3′) and S_HBVporv2 (5′-AAAGCCCAAAAGACCCACAAT-3′); round setting was 95°C (15 min); 40 cycles of 95°C (30 s), 60°C (30 s), 72°C (1 min); followed by extension at 72°C for 7 min. The second step used 2 μl of the first reaction and primers S_HBV123s (5′-TCGAGGATTGGGGACCCTG-3′) and S_HBVporv2; round setting was 95°C (15 min); 45 cycles of 95°C (30 s), 58°C (30 s), 72°C (1 min); followed by extension at 72°C for 7 min. Five microliters of PCR products were purified using standard ExoSap-IT (Usb Corporation) and both strands were directly sequenced by automated dideoxy sequencing (sequencer AbiPrism 3100; Applied Biosystems) using primers S_HBV123s, S_HBVporv2 and S_HBV778r (5′-GAGGTATAAAGGGACTCAAG-3′). HBV genotypes and subtypes were determined as previously described (31) and phylogenetic trees were built using HBV references from GenBank and the software MEGA5 (29).
Statistical analyses
Odds ratios (ORs), χ2 and P values were calculated using STATA 11.1 (College Station, TX). All ORs were adjusted for age and gender.
Results
Characteristics of study participants
In this study, we analyzed a total of 325 controls, 78 patients with cirrhosis and 198 patients with HCC recruited in the Gambia Liver Cancer Study (4). Characteristics of study participants are presented in Table I. Despite frequency matching within 10 years age strata, HCC patients were significantly older than controls and more frequently male than patients with cirrhosis or than controls. HBV chronic carriage (positivity for HBsAg) was significantly more frequent in HCC and in cirrhosis than in controls. HBsAg was detected in 56% (44/78) and in 54% (106/198) of cirrhosis and HCC, respectively; compared with 13% (43/325) of controls {cirrhosis: OR 13.36 [95% confidence interval (CI) 1.08–165.04]; HCC: OR 12.19 [95% CI 1.29–115.59]}. Among HBsAg-positive subjects, 32% of cirrhosis and 16% of HCC were HBeAg-positive, compared with 5% of controls [OR for HCC versus controls: 0.77 (95% CI 0.14–4.34); for cirrhosis versus controls: 5.66 (95% CI 1.03–31.06)]. When compared with patients with cirrhosis, patients with HCC were less frequently positive for HBeAg [OR: 0.30 (95% CI 0.12–0.76)]. Median viral loads were higher in cirrhosis and HCC (9.5.106 and 3.4.105 copies/ml, respectively) than in controls (2.5.103 copies/ml).
Table I.
Characteristics of the study participants
| Clinical status | Controls, n (%) | Cirrhosis, n (%) | HCC, n (%) |
| Number of subjects | 325 | 78 | 198 |
| Age | |||
| ≤30 | 84 (26) | 20 (25.5) | 23 (12) |
| 31–40 | 62 (19) | 17 (22) | 36 (18) |
| 41–50 | 54 (17) | 21 (27) | 54 (27) |
| ≥51 | 125 (38) | 20 (25.5) | 85 (43) |
| Sex | |||
| Men | 224 (69) | 50 (64) | 160 (81) |
| Women | 101 (31) | 28 (36) | 38 (19) |
| HBsAg | |||
| Positive | 43 (13) | 44 (56) | 106 (54) |
| Negative | 274 (85) | 34 (44) | 88 (44) |
| N/A | 8 (2) | 0 (0) | 4 (2) |
| HBeAga | |||
| Positive | 2 (5) | 14 (32) | 17 (16) |
| Negative | 39 (90) | 27 (61) | 87 (82) |
| N/A | 2 (5) | 3 (7) | 2 (2) |
| Viral loadsa | |||
| > Copies/mlb (n) | 2.5.103 (22) | 9.5.106 (29) | 3.4.105 (76) |
| > <200, n (%) | 21 (49) | 15 (34) | 30 (28) |
N/A: not available.
only for HBsAg-positive subjects.
Median.
Presence of R249S in CFDNA of HCC patients is associated with detection of complete HBX sequence
Integration of HBX in the host cell genome is a hallmark of HCC and this integration often involves truncation of the portion of HBX encoding the C-terminus of the protein. This truncation is thought to somehow activate the oncogenic potential of HBx. Since previous results have shown that the mutant protein p.R249S interacts with HBx protein and since this interaction appears to involve the C-terminus of HBx, we reasoned that integration of the complete HBX sequence may occur more frequently in HCC with R249S mutations than in HCC without such R249S mutation. To test this hypothesis, we have analyzed HBX gene status by PCR amplification of four fragments covering overlapping portions of the gene, from the 5′-end to the complete sequence, in the CFDNA of HBV chronic carriers (HBsAg-positive subjects). This strategy allowed to distinguish between absence of HBX sequence (no PCR amplification), presence of truncated HBX (amplification of 1–3 PCR fragments covering regions of progressive length) and presence of complete HBX (amplification of all four PCR fragments, the longer one containing the region encoding the C-terminus).
Table II shows HBX status (undetectable, 3′-truncated or complete) in HBV carriers in relation with detection of R249S mutation in CFDNA. The detection of R249S in these subjects using two methods, RFLP or SOMA, has been reported previously (25,26). HBX sequences were detected in 14/43 HBV carrier controls (33%), in 32/44 HBV carrier patients with cirrhosis (73%) and in 89/106 HBV carrier patients with HCC (84%). Among each group, complete HBX sequence was detected in 19% of controls, in 57% of patients with cirrhosis and in 61% patients with HCC. However, when subjects in each group were classified according to R249S status, a clear difference emerged between HCC patients and other groups (Table II). In HCC patients, complete HBX sequences were detected in 43/56 (77%) of R249S+ subjects, as compared with 22/50 (44%) in R249S− subjects. In contrast, the number of HCC patients with truncated HBX was significantly higher among R249S− HCC patients (17/50; 34%) than among R249S+ subjects (7/56, 12%) (test for distribution of R249S+ and R249S− subjects according to HBX status among HCC cases: χ 2 = 12.12; P = 0.002). It should be noted, however, that whatever the mutation status, the group of patients with complete HBX is larger than with 3′-truncated HBX (44% versus 34% in R249S-negative patients and 77% versus 12% in R249S-positive patients). Overall, these results suggest that integration of complete HBX sequence is more common in HCC cases with R249S mutation than in cases without this mutation. In the latter cases, 3′-truncation of HBX upon integration appears to be a more common event.
Table II.
Number and percentage of subjects according to HBX and R249S in HBsAg-positive subjects
| Status | HBX status, n (%) | Total | |||
| R249S | Complete | 3′-truncated | Undetectable | ||
| Control | − | 8 (21) | 5 (13) | 25 (66) | 38 |
| + | 0 (0) | 1 (20) | 4 (80) | 5 | |
| Cirrhosis | − | 16 (52) | 5 (16) | 10 (32) | 31 |
| + | 9 (69) | 2 (15.5) | 2 (15.5) | 13 | |
| HCC | − | 22 (44) | 17 (34) | 11 (22) | 50 |
| + | 43 (77) | 7 (12) | 6 (11) | 56 | |
Occult HBV infection based on HBX detection
Although the analysis described above was focused on those subjects identified as HBV carriers solely on the basis of HBsAg status, we investigated whether HBX DNA could be detected among HBsAg-negative subjects as a possible marker of occult HBV infection. Of a total of 396 HBsAg-negative subjects, 34 (9%) were found to harbor HBX sequences in CFDNA (Table III). These 34 subjects included 8/274 controls (3%), 5/34 patients with cirrhosis (15%) and 21/88 (24%) in patients with HCC.
Table III.
Number and percentage of subjects according to HBX and HBC in HBsAg-negative subjects
| Status | HBX, n (%) | Total | ||
| HBC a, n (%) | ||||
| Complete | 3′-truncated | Undetectable | ||
| Control | 1 (0.4) | 7 (3) | 266 (96.6) | 274 |
| 1 (100) | 5 (71) | N/A | ||
| Cirrhosis | 0 (0) | 5 (15) | 29 (85) | 34 |
| N/A | 4 (80) | N/A | ||
| HCC | 3 (3) | 18 (20) | 67 (77) | 88 |
| 3 (100) | 16 (89) | N/A | ||
N/A: not applicable.
Positive for anti-HBc antibody or HBC gene, percentages were calculated on HBX-positive subjects.
To further assess the significance of HBX sequence detection in HBsAg-negative subjects, we performed a PCR analysis to detect the presence of the HBC gene and correlated it with positivity for anti-HBc antibodies (Table III). Of the 34 HBX-positive subjects, 33 could be analyzed for HBC, among which 29 (85%) were found to be positive for HBC gene or anti-HBc antibodies, thus showing a remarkable correlation between HBX and HBC and anti-HBc antibodies positivity. Overall, these results suggest that detection of HBX sequence is a marker for occult infection and there is a trend toward more frequent occult infection in chronic liver disease and in HCC.
Predominance of HBV genotype E
In previous studies, genotype E was found to be the most common HBV genotype in subjects from The Gambia as well as other parts of West Africa (20,30). In the present study, the HBS gene could be amplified from CFDNA by PCR in 66 of 170 HBX-positive subjects (39%) and sequenced to determine subtypes and establish a phylogenetic tree. Genotype E was detected in 75.5% of these subjects, including 2/4 controls, 15/19 cirrhosis and 33/43 HCC, with a majority of ayw4 subtype (88%) (Supplementary Table 1, available at Carcinogenesis Online). Twenty-three percent of subjects carried genotype A with 93% of subtype ayw1 and only one subject (1.5%; HCC patient (28)) carried genotype D subtype ayw2. HBS sequences of genotype E were heterogeneously distributed among the existing phylogenetic tree of this genotype, with no relation to disease status (Supplementary Figure 2A, available at Carcinogenesis Online). In contrast, all 15 HBS genotype A sequences were distributed into a single group corresponding to subgenotype A3 (Supplementary Figure 2B, available at Carcinogenesis Online). There was no difference in the distribution of genotypes and subgenotypes according to either presence of R249S or clinical status.
HBX sequences and mutations
Since HBS sequences were not available for 104 subjects, we used HBX sequences to identify HBV genotypes. HBX sequences from the 66 subjects genotyped based on HBS sequences were used to establish a phylogenetic tree (Supplementary Figure 3, available at Carcinogenesis Online). This tree separates HBX sequences associated with genotypes A, D or E. By identifying their closest neighbor HBX sequence, it thus became possible to assign HBV genotypes in 76 of the 104 subjects for whom HBS sequences were not available. This analysis further established the presence of genotype E, A and D in 66%, 16.5% and 1% of the subjects, respectively. The remaining 16.5% of HBX sequences could not be clearly classified.
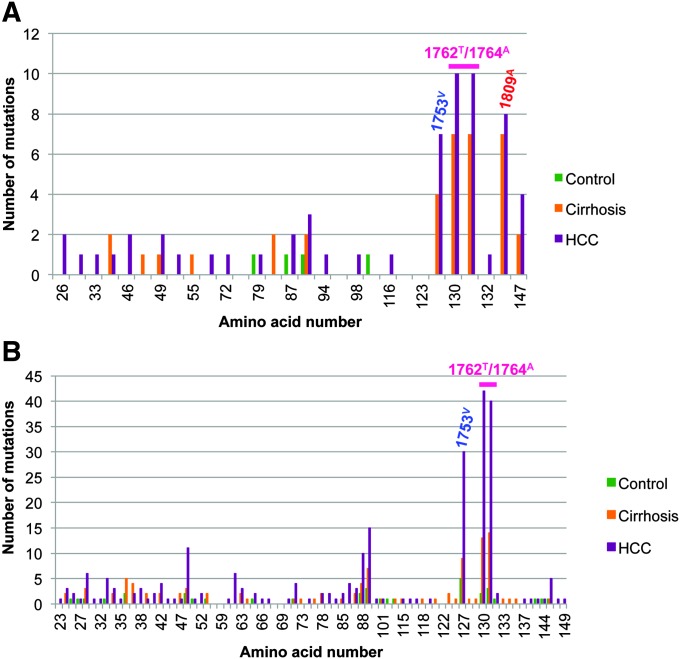
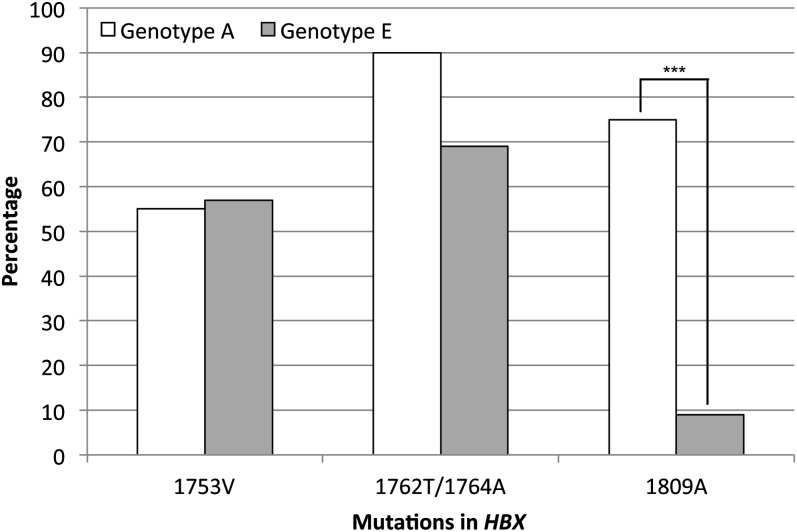
Further analysis of HBX sequences was performed to identify recurrent point mutations (Figure 1). Three mutation sites were detected in genotypes A and E, including 1753V, the double mutation 1762T/1764A and 1809A. However, the relative frequency of these mutations varied among genotypes (Figure 2). Although 1753V and 1762T/1764A were equally found in both genotypes, 1809A was more common in genotype A than E (75% versus 9%), suggesting a possible genotype-specific effect. An association was observed between the HBV 1762T/1764A double mutations and clinical status. Carriers with this double mutation had an elevated risk of cirrhosis or HCC when compared with carriers without this mutation [cirrhosis: OR: 9.50 (95% CI: 1.50–60.11); HCC: OR: 11.29 (95% CI: 2.07–61.47)]. No significant association was detected between mutations and R249S positivity.
Fig. 1.
Number of variations and mutations along HBX gene. The two mutation points 1753V and 1762T/1764A are commonly found in genotype A (A) and E (B), whereas the mutation 1809A is genotype A specific.
Fig. 2.
Frequency of the three specific HBX mutations 1753V, 1762T/1764A and 1809A in genotypes A and E. Mutations 1753V and 1762T/1764A are present in >50% and 70% of subjects with no difference between genotypes. On the contrary, a higher proportion of 1809A mutations was observed in genotype A compared with genotype E (***P < 0.001).
Discussion
In this study, we have used circulating cell-free DNA (CFDNA) from the plasma as a surrogate material to analyze the associations between the status of complete sequences of HBX and the presence of the TP53 mutation R249S in individuals from The Gambia, a country with high incidence of HCC in West Africa. Previous studies have shown that CFDNA in patients with HCC or chronic liver disease was a good source of DNA originating from the liver (24).
Our interpretation is based on the assumption that HBX sequences detected in CFDNA are representative of the integrated status of HBX in hepatocytes. Plasma DNA has proven to be a convenient source to access and analyze liver CFDNA. In previous studies, we have shown that there was an excellent overall concordance between presence of TP53 R249S mutation in the plasma and in HCC tissues of patients from The Gambia (32). This excellent performance of CFDNA as a surrogate for liver DNA provides a convenient approach for studying the molecular pathology of chronic liver disease and HCC in low-resource countries where liver biopsies are not routinely available. Moreover, the use of plasma DNA is well adapted to case–control study designs in which blood samples can be equally obtained from patients and controls. To detect HBX and assess its truncation status in CFDNA, we have adapted a PCR-based method previously described by Ma et al. (16), which generates products of different lengths, the longest one corresponding to full-length HBX sequence. The structure of HBV genome is such that the viral DNA is partially double-stranded, not covalently closed and contains the site of covalent binding of the RNA polymerase in a region overlapping with HBX. This structure precludes the amplification of viral genomes using the PCR approach we have used. Thus, this method preferentially, if not exclusively, detects double-stranded HBX sequences released from liver cells in the bloodstream either in the form of fragmented HBV replication intermediates or in the form of fragments of cellular genomic DNA containing integrated viral sequences. In agreement with this interpretation, we did not observe a higher proportion of complete HBX sequences in patients with the highest viral loads (cirrhosis), which should have been expected if viral genomes were detected.
We have analyzed HBX gene status in the CFDNA of subjects with HCC, cirrhosis or controls (601 samples in total). To our knowledge, this is the first systematic study on HBX genetic status in subjects from West Africa, one of the areas of the world with the highest population carriage of HBV. We found that complete HBX is highly associated with HCC-R249S+ compared with R249S− (P = 0.002), whereas 3′-truncated HBX appears to be more frequent in R249S− HCC, defining an inverse relationship between 3′-truncated HBX and presence of R249S mutation. A study performed in a Chinese population using the same approach has identified 3′-truncated HBX in >70% of HCC cases although it did not examine R249S mutation status (16). Thus, our results suggest that entire HBX is often retained in HCC from this West African population, in particular when the tumor also contains R249S. This observation suggests that HCC in this population may develop according to different mechanism, perhaps corresponding to different mechanism of interplay between the major etiological risk factors, chronic HBV infection and exposure to AFB1. We propose that the association between complete HBX and R249S may reflect a particular mechanism of cooperation between the HBx protein and the mutant p.R249S protein.
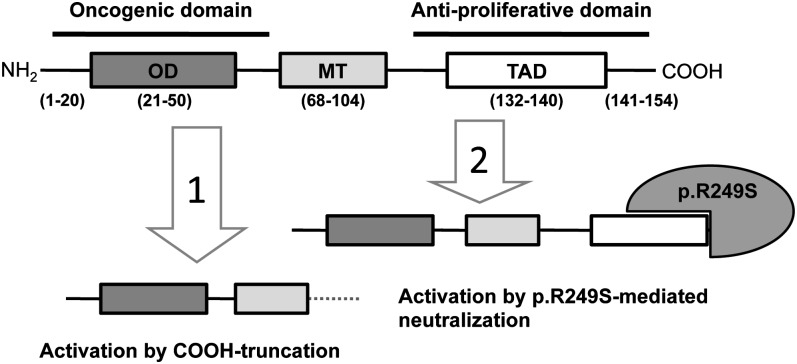
Previous studies have shown that the p.R249S mutant protein forms stable complexes with full-length HBx protein (18). It has also been reported that the C-terminus of HBx contains a domain with antiproliferative proapoptotic effects (33). Thus, we propose that interaction between p.R249S and HBx somehow favors the retention and integration of a full-length HBX, perhaps because the mutant protein neutralizes a suppressive activity located in the C-terminus of HBx, thus making its truncation dispensable during hepatocarcinogenesis (Figure 3). This interaction may contribute to explain the strong association between p.R249S and chronic HBV infection in causing HCC. Indeed, presence of p.R249S may bypass the need for generating a truncated HBX product and therefore accelerates hepatocarcinogenesis. Thus, presence of p.R249S may facilitate the maintenance of complete HBX gene until late stages of HCC progression. This hypothesis remains to be further assessed in patients with HCC from other areas such as parts of China where aflatoxin-induced R249S mutations are also common in HCC.
Fig. 3.
Two hypothetic mechanisms for HBx protein activation in HCC. The protein contains three domains from N- to C-terminal region: an oligomerization domain (OD; aa 21-50); a mitochondria association domain (MT; aa 68-104), which is essential for cell death but not for transactivation function; and a transactivation domain (TAD; aa 132-140). Left: activation by C-truncation (due to integration of 3′-truncated HBX sequence in host genome). Right: activation by neutralization (due to binding of p. R249S to the antiproliferative C-terminal domain of HBx protein encoded by a complete integrated HBX gene).
Another important lesson from our study is the identification of a high prevalence of occult HBV infection in Gambian patients. In our study, 15% and 24% of HBsAg-negative subjects with cirrhosis and HCC, respectively, were positive for HBX in CFDNA, suggesting that these patients may harbor viral sequences despite the absence of HBsAg expression. In recent years, occult HBV infection has emerged as an important and challenging form of persistent HBV infection. The term ‘occult infection’ is commonly used to characterize a heterogeneous group of patients who are HBsAg-negative and either seronegative for all HBV markers or positive for anti-HBc and/or anti-HBs. Many of these patients are positive for HBV DNA detected by PCR in the liver, the serum or both. Some of these patients have underlying liver disease, suggestive of ongoing hepatocellular injury from persistent HBV infection. A meta-analysis by Shi et al. has concluded that occult HBV contributes to the development of HCC. It may serve as cofactor in the development of HCV-related HCC and may also play a direct role in promoting non-B and non-C HCC growth (34). However, further studies are needed to clarify these observations. In any case, our results in Gambian subjects are consistent with the idea that occult infection may play an important part of the burden of HBV-related liver diseases in this population.
The genetic diversity of HBV genotypes is not well documented in The Gambia. In this study, sequencing of S gene confirmed genotype E in 76% of the subjects analyzed, with genotype A in 23% and genotype D in 1.5%, irrespective of disease status. This genotype distribution was essentially confirmed by sequencing HBX. Previous studies have reported over 90% genotype E in asymptomatic HBV carriers in The Gambia and in other West African countries (35). Using the same method as in the present paper, Villar et al. (30) have identified 90% of genotype E in carriers from three villages in rural Gambia. The higher proportion of genotype A in the present study suggests that genotype A might be more widespread than previously suspected, in particular in urban or peri-urban residential areas of the coastal region, where the majority of the participants of this case–control study were recruited.
Mutation analysis of HBX identified several common mutation sites in the 3′-end of HBX. The well-studied mutations 1753V and the double mutation 1762T/1764A were present in >50% and 70% of subjects, respectively, with no difference between genotypes. Among HBV carriers, presence of the double mutation 1762T/1764A was associated with higher risk of cirrhosis or HCC [cirrhosis: OR: 9.50 (95% CI: 1.50–60.11); HCC: OR: 11.29 (95% CI: 2.07–61.47)], consistent with an earlier pilot study in The Gambia (36) and with results in patients from other parts of the world (36–40). The mechanisms by which these mutations enhance the risk of chronic liver disease are still a matter of debate. They may contribute to increase the rate of HBV replication and they may also result into an HBx protein with specific amino acid substitutions enhancing its oncogenic properties (17,41). In contrast with these mutations, the mutation 1809A was more frequently associated with genotype A3 (75%) than genotype E (9%), raising the possibility that this mutation may occur in a genotype-specific manner. Previous studies have shown that mutation at positions 1809–1812 were highly associated with genotype A1 (42,43). These substitutions do not appear to be the result of adaptive changes under immune pressure, since they are also found in HBV isolates obtained from children and in acute hepatitis patients (44,45).
In conclusion, this analysis of HBX status in a case–control study of HCC in The Gambia has shown that there is an association between the presence of complete HBX sequences and the presence of the aflatoxin-induced mutation R249S. The significance of this association for diagnosis, patient prognosis or tumor cell activity needs to be addressed in further studies. Furthermore, this study indicates that occult HBV infection might be present in up to 24% of HCC cases, suggesting that the role of HBV chronicity as attributable risk of HCC in the Gambian population is even greater than previously suggested (46). It will be important to examine how the implementation of neonatal HBV vaccine, which has started in 1986 in the Gambian population, will affect these molecular parameters of HBV infection.
Supplementary material
Supplementary Table 1 and Figures 1–3 can be found at http://carcin.oxfordjournals.org/.
Funding
This research was supported in part by National Institutes of Health (NIH) grant (P01 ES006052), in part by the National Cancer Institute, NIH, Department of Health and Human Services grant (N02 CP40521), in part by Association pour la Recherche sur le Cancer (ARC-5044) and in part by the American Cancer Society (MRSG-07-284-01-CCE to G.D.K.).
Supplementary Material
Acknowledgments
G.D.K. has provided consulting to GSK and to Merck regarding hepatitis vaccination.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AFB1
aflatoxin B1
- CFDNA
cell-free DNA
- CI
confidence interval
- HBsAg
HBV surface antigens
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- OR
odds ratio
- RFLP
restriction fragment length polymorphism
- SOMA
short oligonucleotide mass analysis
References
- 1.Ferlay J, et al. GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10. Lyon: International Agency for Research on Cancer; 2008. [Google Scholar]
- 2.Sighoko D, et al. Increase in female liver cancer in the Gambia, West Africa: evidence from 19 years of population-based cancer registration (1988–2006) PLoS One. 2011;6:e18415. doi: 10.1371/journal.pone.0018415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wild CP, et al. Dietary intake of aflatoxins and the level of albumin-bound aflatoxin in peripheral blood in The Gambia, West Africa. Cancer Epidemiol. Biomarkers Prev. 1992;1:229–234. [PubMed] [Google Scholar]
- 4.Kirk GD, et al. The Gambia Liver Cancer Study: infection with hepatitis B and C and the risk of hepatocellular carcinoma in West Africa. Hepatology. 2004;39:211–219. doi: 10.1002/hep.20027. [DOI] [PubMed] [Google Scholar]
- 5.Kirk GD, et al. Molecular epidemiology of human liver cancer: insights into etiology, pathogenesis and prevention from The Gambia, West Africa. Carcinogenesis. 2006;27:2070–2082. doi: 10.1093/carcin/bgl060. [DOI] [PubMed] [Google Scholar]
- 6.Badvie S. Hepatocellular carcinoma. Postgrad. Med. J. 2000;76:4–11. doi: 10.1136/pmj.76.891.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressac B, et al. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 8.Hsu IC, et al. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 9.Shen HM, et al. Mutations of the p53 tumor suppressor gene and ras oncogenes in aflatoxin hepatocarcinogenesis. Mutat. Res. 1996;366:23–44. doi: 10.1016/s0165-1110(96)90005-6. [DOI] [PubMed] [Google Scholar]
- 10.Gouas D, et al. The aflatoxin-induced TP53 mutation at codon 249 (R249S): biomarker of exposure, early detection and target for therapy. Cancer Lett. 2009;286:29–37. doi: 10.1016/j.canlet.2009.02.057. [DOI] [PubMed] [Google Scholar]
- 11.Kew MC. Epidemiology of chronic hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol. Biol. (Paris) 2010;58:273–277. doi: 10.1016/j.patbio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Benhenda S, et al. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv. Cancer Res. 2009;103:75–109. doi: 10.1016/S0065-230X(09)03004-8. [DOI] [PubMed] [Google Scholar]
- 13.Paterlini P, et al. Selective accumulation of the X transcript of hepatitis B virus in patients negative for hepatitis B surface antigen with hepatocellular carcinoma. Hepatology. 1995;21:313–321. [PubMed] [Google Scholar]
- 14.Tu H, et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803–7810. [PubMed] [Google Scholar]
- 15.Feitelson MA, et al. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Ma NF, et al. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin. Cancer Res. 2008;14:5061–5068. doi: 10.1158/1078-0432.CCR-07-5082. [DOI] [PubMed] [Google Scholar]
- 17.Jiang W, et al. Cooperation of tumor-derived HBx mutants and p53-249(ser) mutant in regulating cell proliferation, anchorage-independent growth and aneuploidy in a telomerase-immortalized normal human hepatocyte-derived cell line. Int. J. Cancer. 2010;127:1011–1020. doi: 10.1002/ijc.25118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gouas DA, et al. Effects of the TP53 pR249S mutant on proliferation and clonogenic properties in human hepatocellular carcinoma cell lines: interaction with hepatitis B virus X protein. Carcinogenesis. 2010;31:1475–1482. doi: 10.1093/carcin/bgq118. [DOI] [PubMed] [Google Scholar]
- 19.Miyakawa Y, et al. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329–338. doi: 10.1159/000074988. [DOI] [PubMed] [Google Scholar]
- 20.Norder H, et al. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 21.Kramvis A, et al. Hepatitis B virus genotypes. Vaccine. 2005;23:2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 22.Kramvis A, et al. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J. Viral Hepat. 2005;12:456–464. doi: 10.1111/j.1365-2893.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu CJ, et al. Therapeutic implications of hepatitis B virus genotypes. Liver Int. 2005;25:1097–1107. doi: 10.1111/j.1478-3231.2005.01177.x. [DOI] [PubMed] [Google Scholar]
- 24.Gormally E, et al. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat. Res. 2007;635:105–117. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Kirk GD, et al. 249(ser) TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene. 2005;24:5858–5867. doi: 10.1038/sj.onc.1208732. [DOI] [PubMed] [Google Scholar]
- 26.Lleonart ME, et al. Quantitative analysis of plasma TP53 249Ser-mutated DNA by electrospray ionization mass spectrometry. Cancer Epidemiol. Biomarkers Prev. 2005;14:2956–2962. doi: 10.1158/1055-9965.EPI-05-0612. [DOI] [PubMed] [Google Scholar]
- 27.Kuniholm MH, et al. Aflatoxin exposure and viral hepatitis in the etiology of liver cirrhosis in the Gambia, West Africa. Environ. Health Perspect. 2008;116:1553–1557. doi: 10.1289/ehp.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abedi-Ardekani B, et al. TP53 mutations and HBX status analysis in hepatocellular carcinomas from Iran: evidence for lack of association between HBV genotype D and TP53 R249S mutations. Hepat. Res. Treat. 2011;2011:475965. doi: 10.1155/2011/475965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villar S, et al. Seasonal variation in TP53 R249S-mutated serum DNA with aflatoxin exposure and hepatitis B virus infection. Environ. Health Perspect. 2011;119:1635–1640. doi: 10.1289/ehp.1103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kidd-Ljunggren K, et al. Genetic variability in hepatitis B viruses. J. Gen. Virol. 2002;83:1267–1280. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 32.Szymanska K, et al. Ser-249TP53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the Gambia, West Africa. Int. J. Cancer. 2004;110:374–379. doi: 10.1002/ijc.20103. [DOI] [PubMed] [Google Scholar]
- 33.Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2011;26(suppl. 1):144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, et al. Association between occult hepatitis B infection and the risk of hepatocellular carcinoma: a meta-analysis. Liver Int. 2011;32:231–240. doi: 10.1111/j.1478-3231.2011.02481.x. [DOI] [PubMed] [Google Scholar]
- 35.Forbi JC, et al. Epidemic history and evolutionary dynamics of hepatitis B virus infection in two remote communities in rural Nigeria. PLoS One. 2010;5:e11615. doi: 10.1371/journal.pone.0011615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendy ME, et al. Application of a novel, rapid, and sensitive oligonucleotide ligation assay for detection of cancer-predicting mutations in the precore and basal core promoter of hepatitis B virus. J. Clin. Microbiol. 2008;46:2723–2730. doi: 10.1128/JCM.01622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan JM, et al. Prospective evaluation of hepatitis B 1762(T)/1764(A) mutations on hepatocellular carcinoma development in Shanghai, China. Cancer Epidemiol. Biomarkers Prev. 2009;18:590–594. doi: 10.1158/1055-9965.EPI-08-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan HL. JGH Foundation emerging leadership lecture. Significance of hepatitis B virus genotypes and mutations in the development of hepatocellular carcinoma in Asia. J. Gastroenterol. Hepatol. 2011;26:8–12. doi: 10.1111/j.1440-1746.2010.06514.x. [DOI] [PubMed] [Google Scholar]
- 39.Fang ZL, et al. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am. J. Gastroenterol. 2008;103:2254–2262. doi: 10.1111/j.1572-0241.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuang SY, et al. Hepatitis B 1762T/1764A mutations, hepatitis C infection, and codon 249 p53 mutations in hepatocellular carcinomas from Thailand. Cancer Epidemiol. Biomarkers Prev. 2005;14:380–384. doi: 10.1158/1055-9965.EPI-04-0380. [DOI] [PubMed] [Google Scholar]
- 41.Iyer S, et al. Interaction of mutant hepatitis B X protein with p53 tumor suppressor protein affects both transcription and cell survival. Mol. Carcinog. 2011;50:972–980. doi: 10.1002/mc.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimbi GC, et al. Distinctive sequence characteristics of subgenotype A1 isolates of hepatitis B virus from South Africa. J. Gen. Virol. 2004;85:1211–1220. doi: 10.1099/vir.0.19749-0. [DOI] [PubMed] [Google Scholar]
- 43.Sugauchi F, et al. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype A. J. Gen. Virol. 2004;85:811–820. doi: 10.1099/vir.0.79811-0. [DOI] [PubMed] [Google Scholar]
- 44.Ahn SH, et al. Sequence variation upstream of precore translation initiation codon reduces hepatitis B virus e antigen production. Gastroenterology. 2003;125:1370–1378. doi: 10.1016/j.gastro.2003.07.016. [DOI] [PubMed] [Google Scholar]
- 45.Chang MH. Hepatitis B virus mutation in children. Indian J. Pediatr. 2006;73:803–807. doi: 10.1007/BF02790390. [DOI] [PubMed] [Google Scholar]
- 46.Viviani S, et al. 20 years into the Gambia Hepatitis Intervention Study: assessment of initial hypotheses and prospects for evaluation of protective effectiveness against liver cancer. Cancer Epidemiol. Biomarkers Prev. 2008;17:3216–3223. doi: 10.1158/1055-9965.EPI-08-0303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.