Abstract
Factors regulating nucleotide excision repair probably contribute to the heterogenous response of advanced stage lung cancer patients to drugs such as cisplatin. Studies to identify the genes in the nucleotide excision repair pathway most closely associated with resistance to cisplatin have not been conclusive. We hypothesized that Xeroderma pigmentosum complementation group A (XPA), because of its dual role in sensing and recruiting other DNA repair proteins to the damaged template, would be critical in defining sensitivity to cisplatin. Studies were conducted to identify factors regulating transcription of XPA, to assess its role in modulating sensitivity to cisplatin and its expression in primary lung tumors. Hypoxia-inducible factor 1 alpha (HIF1α) subunit was found to bind with strong affinity to a hypoxia response element sequence in the promoter of XPA. Modulating expression of HIF1α by small interfering RNA or cobalt chloride markedly reduced or increased transcription of XPA in lung cancer cell lines, respectively. Protein levels of XPA were strongly correlated with sensitivity to cisplatin (r = 0.88; P < 0.001) in cell lines and sensitivity could be increased by small interfering RNA depletion of XPA. Expression of XPA determined in 54 primary lung tumors was elevated on average 5.2-fold when compared with normal bronchial epithelial cells and correlated with levels of HIF1α (r = 0.58; P < 0.01). Together, these studies identify XPA as a novel target for regulation by HIF1α whose modulation could impact lung cancer therapy.
Introduction
The repair of DNA damage stemming from continuous endogenous cellular byproducts and exogenous exposure to environmental toxicants and carcinogens is critical for preventing cell death and cancer (1). Several DNA damage repair pathways have evolved dependent on the nature of the DNA damage (2). Nucleotide excision repair (NER) is a universal and versatile process that includes two subpathways: global genome NER (GG-NER) and transcription coupled NER (TC-NER) (3). The NER pathway consists of >30 proteins involved in damage recognition, initial incision/excision, gap refill and re-ligation. XPA plays a key role in damage complex conformation in both GG-NER and TC-NER pathway that is responsible for repair of ultraviolet radiation-induced photoproducts and DNA adducts induced by chemical carcinogens. XPA is a zinc finger motif containing protein that possesses high affinity to damaged DNA that in turn is enhanced by forming a complex with replication protein A (4,5). The direct interaction of XPA with the excision repair cross-complementing rodent repair deficiency, complementation group 1 (ERCC1)-xeroderma pigmentosum, complementation group F (XPF) proteins is also critical for localization or loading of an incision complex for DNA repair (6–8). Thus, XPA plays a dual role in NER through acting as a DNA damage sensor and the recruitment of other DNA repair proteins to the damaged template. Although definitive studies have not been conducted to identify the protein(s) rate limiting for NER, mutation of XPA impairs NER capacity and sensitizes the host to sunlight, supporting a critical function for this protein (9).
DNA repair capacity is also a two-edge sword in which efficient repair is prognostic for survival of lung cancer patients after surgical resection and associated with resistance of advanced stage cancer patients to chemotherapeutics, most notably cisplatin that is widely used to treat many types of cancers including sarcomas, lung cancer, lymphomas and germ cell tumors (10). The underlying mechanism for the antitumor activity of platinum-containing drugs is intercalation and covalent cross-link with DNA, which impedes essential physiological processes such as replication and transcription and eventually induces apoptosis (11). NER is primarily responsible for the repair of cisplatin-induced DNA damage (12). Some studies support an association between transcript levels of NER genes and sensitization to cisplatin; however, the major gene-impacting efficacy of this chemotherapeutic has not been clearly substantiated. Transfection of XPA antisense RNA into human lung adenocarcinoma cells reduced DNA repair capacity and sensitized tumor cells to cisplatin (13). Reduced expression of XPA or ERCC1 could also sensitize some prostate cancer cell lines to cisplatin (14). Furthermore, ABCC5, ERCC2, XPA and XRCC1 transcript abundance all correlated with cisplatin chemoresistance in non-small-cell lung cancer cell lines (15). However, in vitro studies failed to identify a correlation between resistance to platinum-based therapy in lung cancer cell lines and messenger RNA or protein levels of ERCC1 (16,17). Furthermore, Saviozzi et al. (18) cast some doubt on the in vivo contribution of the NER pathway as a whole to platinum resistance by demonstrating no significant increase in transcript levels of NER genes in primary non-small-cell lung cancer compared with distant normal tissue.
Clinical trials have focused mainly on correlating RNA or protein levels of ERCC1 with response and overall survival to platinum drugs. Overall, ERCC1 protein and messenger RNA expression were highly correlated allowing comparison across studies (19). Interestingly, a meta-analysis revealed a significant improvement in response and overall survival for Asian but not European lung cancer patients with low versus high ERCC1 levels (20). The three largest clinical trials conducted in the USA observed different results when assessing the association between ERRC1 levels and disease response or overall survival. These three results included no association to either end point, associated with disease response but not survival or associated with survival but not disease response (21–23).
We hypothesize that regulation of XPA may be critical in defining sensitivity to cisplatin because of its dual role in sensing and recruiting other DNA repair proteins to the damaged template for NER. The purpose of the current study was to identify factors regulating the transcription of XPA and to assess its role along with ERCC1 in modulating sensitivity to cisplatin in vitro. In addition, expression levels of XPA and ERCC1 were defined in primary tumors relative to non-malignant human bronchial epithelial cell lines (NHBECs) or normal lung tissue.
Materials and methods
Tissue samples and cell lines
Primary lung tumors (n = 54) were obtained from frozen tumor banks at the Lovelace Respiratory Research Institute and Mayo Clinic (24). NHBEC were obtained from cancer-free smokers at the New Mexico Veteran Health Care System. NHBEC were collected through diagnostic bronchoscopy and expanded in short-term tissue culture as described (25). Human bronchial epithelial cell lines (HBECs) (obtained from Drs Shay and Minna, Southwestern Medical Center, Dallas, TX) were immortalized as described from two different people (HBEC1; smoker without lung cancer; HBEC2; smoker with cancer (26)). All subjects gave informed consent according to institutional guidelines and studies were Institutional Review Board approved. Eleven lung cancer-derived cell lines (Calu6, H2009, H2023, H2085, H2228, H23, H358, H441, HCC827, HCC4006, SK-LU-1) were obtained from and authenticated by the American Type Culture Collection.
Gene expression analysis
Total RNA was extracted from cells and tissue using Trizol reagent (Sigma). Total complementary DNA was synthesized from 1 μg of total RNA using High Capacity cDNA RT Kit (Applied Biosystems, Foster City, CA). To avoid PCR products from contaminating DNA, RNA isolation was done in the presence of DNase, and large introns were included in the reverse transcription–PCR amplification product. TaqMan assays [XPA (Hs00166045_ml), ERCC1 (Hs01012159_ml) and hypoxia-inducible factor 1 alpha (HIF1α) (Hs00936371_ml)] from Applied Biosystems were used. Real-time PCR was performed with the ABI PRISM 7900HT (Applied Biosystems). All experiments were normalized to β-actin and performed in triplicate.
Western blotting
Approximately 15 μg of total protein was electrophoresed on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred to polyvinylidene difluoride membranes 2 h on the ice. The membranes were incubated with 5% milk and hybridized with antibodies against human XPA (1:1000; New England Biolabs), HIF1α (1:200; Santa Cruz Biotechnology) or β-actin (1:1000; Sigma) overnight at 4°C. The membranes were washed 6 × 5 min with 1× TBST (Tris-buffered saline plus 0.05% Tween-20) at room temperature and incubated with secondary anti-mouse IgG-horseradish peroxidase (1:2000; Santa Cruz Biotechnology) for 1 h at room temperature. The membranes were washed 6 × 5 min with 1× TBST at room temperature and then were visualized by enhanced chemiluminescence reagent according to the manufacturer’s instructions (Thermo Scientific). The intensity of the individual bands was quantified by densitometry (Bio-Rad) and normalized to the corresponding input control (β-actin) bands.
Cytotoxicity assay
The MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) assay was used to determine cell viability as an indicator for the relative sensitivity of the cells to cisplatin. Cells growing in the logarithmic phase were seeded in 96-well plates (5 × 103 per well), allowed to attach overnight and then were treated with varying doses (0, 1, 2, 5, 10, 20, 30, 40 or 60 μM) of cisplatin (Sigma) for 72 h. Twenty-five microliters of MTT (5 mg/ml; Sigma) was added to each well and after 2 h, color formation was quantified by a spectrophotometric plate reader (VersaMax; Molecular Devices) at 570 nm wavelength after solubilizing in 200 μl of dimethyl sulfoxide.
Transfections
Cells were seeded at 50% confluency. After 24 h, cells were transfected using Lipofectamine 2000 (Invitrogen) with the following small interfering RNA sequence: XPA 5′-ACACAAGCUUAUAACCAAAtt-3′; HIF1α 5′-CCGAAUUGAUGGGAUAUGAtt-3′. Knockdown efficiency was determined by western blotting at designated time points.
Electrophoretic mobilitiy shift assay
LightShift Chemiluminescent EMSA Kit (Thermo Scientific) was used. Whole cell extracts were prepared from H358 and H2228. The protein concentrations were determined using the BCA assay (Thermo Scientific). A typical double-stranded consensus oligonucleotide for HIF1α binding (5′-GAGGTCTTCTACGTGCCAGGTTATT-3′) and a scrambled DNA sequence (5′-GAGGTCTTCTTTTCCCCAGGTTATT-3′) with 5′ biotin labeled were purchased from Integrated DNA Technology (San Diego, CA). Protein extract was incubated with DNA probes in binding buffer for 20 min at room temperature in a final volume of 20 μl. DNA–protein complexes were separated on native 6% polyacrylamide gel electrophoresis gels and transferred by electrophoresis to polyvinylidene difluoride membrane. Shift band was detected by chemiluminescence according to the manufacturer’s instruction.
Chromatin immunoprecipitation assay
Antibody specific to HIF1α from Santa Cruz Biotechnology was used to capture protein–DNA complexes. Anti-RNA polymerase ‖ antibody and normal mouse IgG was used as positive and negative control, respectively. Results were generated by reverse transcriptase quantitative PCR carried out in triplicate, using Power SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences and PCR conditions are available on request. Results were quantified using a 2−ΔΔCt method (27).
Data analysis
Spearman rank order was used to determine the correlation between transcript or protein levels of XPA and cisplatin IC50, and gene expression levels between XPA and HIF1α. Fold enrichment for HIF1α at the XPA promoter was compared with normal mouse IgG using a T-test.
Results
HIF1α regulates the expression of XPA in lung cancer cell lines
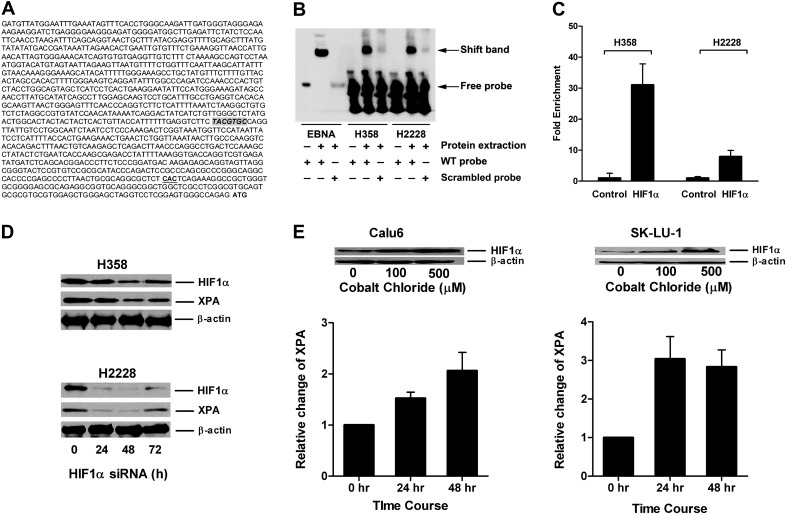
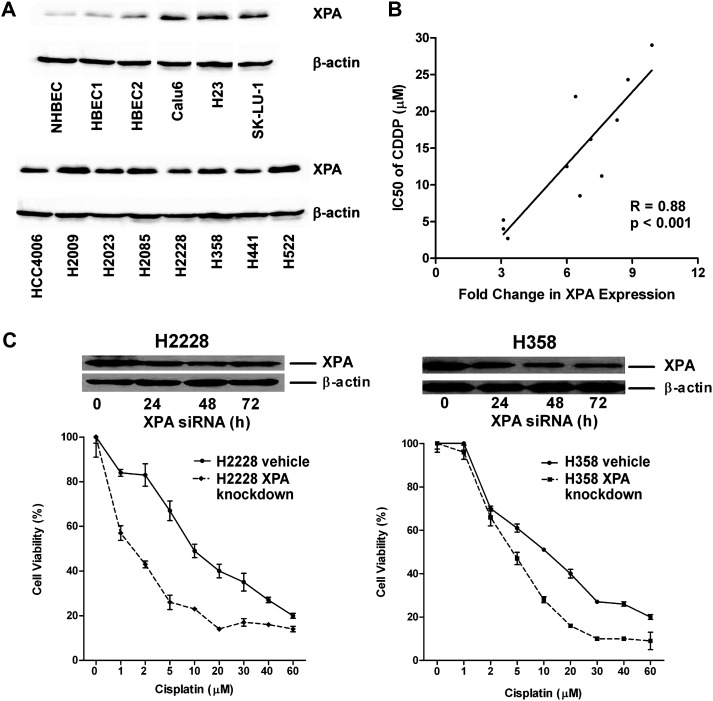
Transcription factor-binding sites within the XPA promoter were identified using MatInspector software that locates binding sites and assigns quality ratings to the resulting matches within a sequence (28). A hypoxia response element (HRE) sequence TACGTGC with perfect homology to that described for the HIF1α protein was present 477 bp upstream from the XPA transcriptional start site (Figure 1A). Prediction scores ≥95% for binding of GATA1, MZF1, CdxA and Nkx-2 proteins were also observed. We focused on HRE because it is the identical sequence present in the promoters of the HIF1α target genes vascular endothelial growth factor and carbonic anhydrase 9 (29,30). This response element was not present in the promoter region of other NER genes, including ERCC1 (data not shown). Expression and protein levels of XPA were determined in lung cancer cell lines to facilitate studies to assess binding of HIF1α to this HRE and the effect on transcription of XPA. RNA transcript (expression) and protein levels of XPA was determined in 11 cell lines and compared with that seen for NHBEC, HBEC1 and HBEC2 to assess differences between nonmalignant and malignant cells. Expression of XPA did not differ significantly between normal lung cell lines and was elevated 2.6- to 6.7-fold in cancer lines (Table I). XPA protein levels were elevated 3- to 10-fold when compared with NHBEC and HBECs. Protein levels on average were 1.6-fold higher (range, 0- to 3.7-fold) than transcript levels (Table I; Figure 2A).
Fig. 1.
HIF1α regulated expression of XPA in lung cancer cell lines. (A) Location of HRE-binding site TACGTGC in XPA promoter relative to the transcriptional start site (CAC) and the translational start site (ATG) is shown. (B) The binding of HIF1α to HRE is demonstrated by EMSA assay. Whole cell extracts from H358 and H2228 were incubated with HRE-containing probe and then subjected to EMSA. Shift band is shown compared with the positive control (Epstein-Barr Nuclear Antigen, EBNA) in the presence of protein extract and consensus oligonucleotide for HIF1α binding (wild-type probe) compared with the scrambled probe. (C) Binding of HIF1α protein to the XPA promoter. Antibody to HIF1α was used to assess binding of this protein to the XPA promoter using EZ ChIP™. The fold enrichment of HIF1α in XPA promoter in H358 and H2228 compared with normal mouse IgG is shown. (D) Inhibition of HIF1α by small interfering RNA (siRNA) reduces expression of XPA in lung cancer cell lines. Expression levels determined by western blotting of HIF1α and XPA following HIF1α transient knockdown in H358 and H2228 are shown at indicated time points posttransfection. (E) Cobalt chloride treatment increases expression of HIF1α and XPA in lung cancer cell lines. Calu6 and SK-LU-1 cells were treated up to 48 h with 0, 100 or 500 μM cobalt chloride. Dose response increase in expression of HIF1α is shown along with increased expression of XPA seen with the 500 μM dose.
Table I.
RNA and protein levels of XPA and corresponding IC50 for cisplatin in lung cancer cell lines
| Cell line | XPA level (fold elevation) | IC50 for cisplatin (μM)c | |
| RNAa | Proteinb | ||
| Calu6 | 3.1 ± 0.4 | 3.1 | 4.0 ± 0.6 |
| H23 | 3.2 ± 0.2 | 3.1 | 5.2 ± 0.7 |
| SK-LU-1 | 3.3 ± 0.3 | 3.3 | 2.7 ± 0.4 |
| HCC4006 | 2.6 ± 0.4 | 6.1 | 22.0 ± 5.5 |
| H2009 | 2.7 ± 0.2 | 9.9 | 29.0 ± 0.6 |
| H2023 | 6.7 ± 0.9 | 8.8 | 24.3 ± 5.2 |
| H2085 | 3.5 ± 0.9 | 7.1 | 16.2 ± 4.0 |
| H2228 | 1.9 ± 0.1 | 6.6 | 8.5 ± 2.8 |
| H358 | 5.8 ± 0.7 | 7.6 | 11.2 ± 1.3 |
| H441 | 4.4 ± 0.9 | 6.0 | 12.5 ± 0.5 |
| H552 | 5.3 ± 0.7 | 8.3 | 18.8 ± 1.6 |
Values for RNA levels are mean ± standard deviation from three separate RT-qPCR experiments that were normalized to β-actin and expressed as fold elevation compared with average expression seen for NHBEC, HBEC1 and HBEC2.
Values are fold elevation in protein levels relative to average levels for NHBEC, HBEC1 and HBEC2 after normalization to β-actin.
Values are the mean ± standard deviation for three separate determination of IC50.
Fig. 2.
XPA protein levels in lung cancer cell lines are associated with sensitivity to cisplatin. (A) Western blot depicting protein levels of XPA in 11 lung cancer cell lines. (B) XPA expression and sensitivity to cisplatin (IC50) are highly correlated in lung cancer cell lines (r = 0.88; P < 0.001). (C) XPA knockdown sensitizes H2228 and H358 to cisplatin. Transient knockdown was confirmed by western blotting up to 72 h posttransfection. The viability of cells treated with different doses of cisplatin was measured by MTT assay. Each point represents the mean for three wells (mean ± standard error of the mean).
The binding of a HIF1α protein complex to this HRE consensus DNA sequence was examined by electrophoretic mobilitiy shift assay. A mobility shift band was detected with the HRE probe but not by the scrambled probe incubated with extract from H358 and H2228, two cell lines that express high levels of XPA protein (Table I; Figure 1B). Chromatin immunoprecipitation was used to confirm the binding of HIF1α to the XPA promoter. A 30- and 10-fold enrichment compared with normal mouse IgG in binding of HIF1α to the promoter sequence encompassing the HRE was seen in H358 and H2228, respectively (Figure 1C). Further evidence for a critical role by HIF1α in regulating the transcription of XPA was provided through small interfering RNA studies. Protein levels of XPA were reduced 50% following transient knock down of HIF1α in H358 and H2228 (Figure 1D). Finally, we assessed whether increasing levels of HIF1α in cell lines Calu6 and SK-LU-1 that express low levels of XPA could increase the expression of this gene. Hypoxia was mimicked by treatment of cell lines with cobalt chloride (100 or 500 μM) for up to 48 h. Protein levels of HIF1α were markedly increased by the high dose of cobalt chloride concomitant with a 1.5- to 3-fold increase in expression of XPA (Figure 1E). The H358 cell line that has high levels of XPA protein concomitant with enrichment of the HIF1α protein at the XPA promoter was also treated with cobalt chloride. As expected, this treatment did not increase protein levels of HIF1α or expression of XPA (Supplementary Figure S1 is available at Carcinogenesis Online).
XPA protein level is strongly correlated with cisplatin resistance in lung cancer cell lines
The strength of the association between expression of XPA and ERCC1 to cisplatin resistance in lung cancer cell lines was defined. Cisplatin sensitivity determined by the MTT assay revealed that 3 of 11 cell lines were highly sensitive (IC50 < 7.5 μM), whereas HCC4006, H2023 and H2009 were most resistant with IC50 > 20 μM (Table I). Protein levels of XPA were strongly correlated with sensitivity to cisplatin (r = 0.88; P < 0.001; Figure 2B) in cell lines. In contrast, no correlation was evident when comparing levels of XPA transcript to cisplatin sensitivity (r = 0.2449; data not shown). Transient knockdown of XPA in cell lines H2228 and H358 that express high levels of this gene sensitized them to cisplatin, with IC50 decreasing 6- and 2-fold, respectively (Figure 2C). In marked contrast to XPA, levels of expression of ERCC1 were only modestly elevated in lung cancer cell lines (no increase to 2.1-fold) and were correlated with protein levels. There was no correlation between ERCC1 expression and sensitivity to cisplatin (r = 0.19; data not shown).
XPA expression is elevated in primary lung tumors and correlates with expression of HIF1α
The level of XPA expression was evaluated in 54 tumors and distant normal lung tissues (DNLTs) obtained from similar numbers of patients with adenocarcinoma (smokers and never-smokers) and squamous cell carcinoma. Exposure and clinical covariates are summarized in Supplementary Table 1, available at Carcinogenesis Online. XPA expression in DNLT was on average 18-fold higher than in NHBECs and varied from 3.3- to 53-fold (data not shown). The elevated expression and heterogeneity of XPA levels among DNLT may be due to different basal levels of expression of this gene within the multiple cell types comprising the lung parenchyma, a finding also seen in our expression studies with 6-O-endosulfatase and O 6-methylguanine-DNA methyltransferase (31,32). Therefore, because tumors studied for expression of XPA and ERCC1 were selected based on tumor cell content >70%, we compared expression levels of these genes with both DNLT and NHBECs. The NHBEC also represents a pure cell population that can give rise to lung tumors and thus may in fact be a better source for comparison of expression to tumors. Expression levels of XPA and ERCC1 determined in nine NHBEC lines, HBEC1 and HBEC2 showed a variance of 7 and 5%, respectively. Only modest (1.1- to 1.3-fold) to no elevated expression of XPA was seen when compared with DNLT, a finding similar to that shown by Saviozzi et al. (18). In marked contrast, expression levels of XPA were elevated on average 5.2-fold (range, 1.2–22.5) when compared with NHBECs and to a similar extent in adenocarcinoma from smokers and never-smokers and squamous cell carcinoma with or without adjustment for age, gender and pack years. (Figure 3A). Expression of XPA was increased >5-fold in 20 of 54 tumors. Limited tissue precluded the ability to assess protein levels in tumor tissue. Similar to results in cell lines, expression of ERCC1 was not increased significantly when compared with either NHBECs or DNLT (Figure 3A; data not shown).
Fig. 3.
Expression of XPA is increased in primary lung tumors and correlates with levels of HIF1α in tumors. (A) Box plots depict the mean and standard deviation of fold increase in expression of XPA and ERCC1 compared with the average expression seen in nine NHBEC lines, HBEC1 and HBEC2 (expression level set to 1) in adenocarcinoma from smokers (Adc S) and never-smokers (Adc NS) and smokers with squamous cell carcinoma (SCC S). XPA expression is highly correlated to HIF1α in lung cancer cell lines (B) and primary tumors (C). Note that the scale for the x-axis differs between the three graphs.
RT-qPCR was used to assess the correlation between expression of HIF1α and XPA in lung cancer cell lines (n = 11) and primary lung tumors (n = 54). Expression was highly and moderately correlated in cell lines (r = 0.90, P < 0.001) and primary tumors (r = 0.58, P < 0.01; Figure 3B and C), respectively.
Discussion
These studies identify HIF1α as a key protein in regulating transcription of XPA in lung cancer cell lines and primary tumors. Levels of XPA expression but not ERCC1 were tightly correlated with response to cisplatin, supporting the interrogation of this protein in lung tissue biopsies from patients to help guide treatment strategies. Furthermore, inhibiting HIF1α could sensitize tumor cells to cisplatin and improve therapeutic response.
There is a clear need to better predict response to common chemotherapeutic options for treatment of non-small-cell lung cancer. Support for ERCC1 levels as a biomarker for sensitivity to cisplatin in clinical trials has been inconsistent in predicting response or overall survival (21–23). This outcome may stem from the fact that expression of this gene in our study showed very little variation across lung cancer cell lines or primary tumors when compared with either DNLT or NHBEC and was not predictive of sensitivity of cell lines to cisplatin. In contrast, expression and protein levels of XPA varied significantly across cell lines and tumors with a highly significant correlation between protein levels and drug sensitivity. Moreover, protein levels of XPA were often increased disproportionately to RNA, suggesting an important role for posttranscriptional and translational regulation that could be mediated in part by microRNAs (33).
The major role identified for HIF1α in regulating transcription of XPA could support testing new strategies for therapy. The expansion of tumor cell growth relies on nutrient supply and oxygen limitation can control neovascularization, glucose metabolism, survival and tumor spread (34). However, hypoxia is common in solid tumors and is associated with malignant progression, therapy resistance, metastasis and poor prognosis (35,36). Tumor cells adapt to low oxygen by inducing angiogenesis, increasing glucose consumption and switching to glycolysis. This response is regulated by two transcription factors, HIF1 and HIF2 (37,38). HIF1α is critical for hypoxic induction of VEGF that plays a key role in tumor angiogenesis (31). Previous studies have shown in HeLa and Hep3B cells that HIF1α and β messenger RNAs are constitutively expressed, whereas only protein levels of HIF1β are detected under normoxic conditions (39). However, under hypoxic conditions, HIF1α protein is stabilized and activated through heterodimerizing with HIF1β (40). Chemotherapy resistance related to hypoxia is usually attributed to its limited oxygen availability, but our findings support another potential possibility, that cisplatin resistance in lung cancer is also conferred by HIF1α regulated expression of XPA. Even under normoxic conditions, levels of HIF1α protein were high in some cancer cell lines and modulation of protein levels through small interfering RNA markedly decreased protein levels of XPA. Moreover, in cell lines with low endogenous levels of HIF1α protein, simulating hypoxia through treatment with cobalt chloride concurrently induced HIF1α and XPA proteins. This tight regulation of XPA suggests a second mechanism and scenario by which clinically targeting HIF1α could improve efficacy of chemotherapeutics. This strategy is supported by a recent study in which noscapine, an inhbitor of HIF1α, sensitized ovarian cancer cells to cisplatin (41). In addition, KC7F2 a novel small molecule that inhibits translation of HIF1α was effective in inducing cytotoxicity of glioma, breast and prostate cancer cells under hypoxic conditions (42). Finally, a retrospective study evaluated the clinical significance of HIF1α and ERCC1 in patients with small-cell lung cancer treated with front-line platinum-based chemotherapy (43). High expression of HIF1α determined by immunohistochemistry was associated with a significant reduction in survival of advanced stage lung cancer patients (hazard ratio = 3.0, P = 0.004). Thus, our studies identify XPA as a novel target for regulation by HIF1α whose modulation should impact lung cancer therapy.
Supplementary material
Supplementary Table 1 and Figure S1 can be found at http://carcin.oxfordjournals.org/.
Funding
National Institutes of Health (R01 ES015262 to S.A.B).
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- DNLT
distant normal lung tissue
- ECRR1
excision repair cross-complementing rodent repair deficiency, complementation group 1
- HBEC
human bronchial epithelial cell line
- HIF1α
hypoxia-inducible factor 1 alpha
- HRE
hypoxia response element
- MTT
(3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide
- NER
nucleotide excision repair
- NHBEC
non-malignant human bronchial epithelial cell line
- XPA
Xeroderma pigmentosum complementation group A
- XPF
xeroderma pigmentosum, complementation group F
References
- 1.Jackson SP, et al. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helleday T, et al. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 3.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 4.Morita EH, et al. Implications of the zinc-finger motif found in the DNA-binding domain of the human XPA protein. Genes Cells. 1996;1:437–442. doi: 10.1046/j.1365-2443.1996.d01-252.x. [DOI] [PubMed] [Google Scholar]
- 5.Krasikova YS, et al. Interaction of nucleotide excision repair factors XPC-HR23B, XPA, and RPA with damaged DNA. Biochemistry (Mosc.) 2008;73:886–896. doi: 10.1134/s0006297908080063. [DOI] [PubMed] [Google Scholar]
- 6.Tsodikov OV, et al. Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J. 2007;14:4768–4776. doi: 10.1038/sj.emboj.7601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nocentini S, et al. DNA damage recognition by XPA protein promotes efficient recruitment of transcription factor II H. J. Biol. Chem. 1997;272:22991–22994. doi: 10.1074/jbc.272.37.22991. [DOI] [PubMed] [Google Scholar]
- 8.Li L, et al. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl Acad. Sci. USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka K, et al. UV-induced skin carcinogenesis in xeroderma pigmentosum group A (XPA) gene-knockout mice with nucleotide excision repair-deficiency. Mutat. Res. 2001;477:31–40. doi: 10.1016/s0027-5107(01)00093-8. [DOI] [PubMed] [Google Scholar]
- 10.Sleijfer DT, et al. Cisplatin: a review of clinical applications and renal toxicity. Pharm. Weekbl. Sci. 1985;7:237–244. doi: 10.1007/BF01959196. [DOI] [PubMed] [Google Scholar]
- 11.Johnes J, et al. Gene-specific formation and repair of cisplatin intrastrand adducts and interstrand cross-links in Chinese hamster ovary. J. Biol. Chem. 1991;266:7107–7107. [PubMed] [Google Scholar]
- 12.Sancar A. Excision repair in mammalian cells. J. Biol. Chem. 1995;270:15915–15918. doi: 10.1074/jbc.270.27.15915. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, et al. Sensitization to the cytotoxicity of cisplatin by transfection with nucleotide excision repair gene xeroderma pigmentosun group A antisense RNA in human lung adenocarcinoma cells. Clin. Cancer Res. 2003;9:5874–5879. [PubMed] [Google Scholar]
- 14.Cummings M, et al. XPA versus ERCC1 as chemosensitising agents to cisplatin and mitomycin C in prostate cancer cells: role of ERCC1 in homologous recombination repair. Biochem. Pharmacol. 2006;72:166–175. doi: 10.1016/j.bcp.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Weaver DA, et al. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol. Cancer. 2005;4:18. doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhimizu J, et al. mRNA expression of RRM1, ERCC1 and ERCC2 is not associated with chemosensitivity to cisplatin, carboplatin and gemcitabine in human lung cancer cell lines. Respirology. 2008;13:510–517. doi: 10.1111/j.1440-1843.2008.01302.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, et al. Resistance to platinum-based chemotherapy in lung cancer cell lines. Cancer Chemother. Pharmacol. 2010;66:1103–1111. doi: 10.1007/s00280-010-1268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saviozzi S, et al. Non-small cell lung cancer exhibits transcript overexpression of genes associated with homologous recombination and DNA replication pathways. Cancer Res. 2009;69:3390–3396. doi: 10.1158/0008-5472.CAN-08-2981. [DOI] [PubMed] [Google Scholar]
- 19.Friboulet L, et al. Molecular characteristics of ERCC1-negative versus ERCC1-positive tumors in resected NSCLC. Clin. Cancer Res. 2011;17:5562–5572. doi: 10.1158/1078-0432.CCR-11-0790. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, et al. The platinum-based treatments for advanced non-small cell lung cancer, is low/negative ERCC1 expression better than high/positive ERCC1 expression? A meta-analysis. Lung Cancer. 2010;70:63–70. doi: 10.1016/j.lungcan.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Lord RV, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin. Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 22.Rosell R, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin. Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds C, et al. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J. Clin. Oncol. 2009;27:5808–5815. doi: 10.1200/JCO.2009.21.9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tessema M, et al. Concomitant promoter Methylation of multiple genes in lung adenocarcinomas from current, former and never smokers. Carcinogenesis. 2009;30:1132–1138. doi: 10.1093/carcin/bgp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belinsky SA, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 26.Ramirez RD, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, et al. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Quandt K, et al. MatInd and matInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura H, et al. Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood. 2000;95:189–197. [PubMed] [Google Scholar]
- 30.Kopacek J, et al. MAPK pathway contributes to density- and hypoxia-induced expression of the tumor-associated carbonic anhydrase IX. Biochim. Biophys. Acta. 2005;1729:41–49. doi: 10.1016/j.bbaexp.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Tessema M, et al. SULF2 methylation is prognostic for lung cancer survival and increases sensitivity to topotecan via induction of ISG15. Oncogene, 2011 doi: 10.1038/onc.2011.577. doi:10.1038/onc.2011.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leng S, et al. The A/G allele of Rs16906252 predicts for MGMT methylation and is selectively silenced in premalignant lesions from smokers and in lung adenocarcinoma. Clin. Cancer Res. 2011;17:2014–2023. doi: 10.1158/1078-0432.CCR-10-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janga SC, et al. MicroRNAs as post-transcriptional machines and their interplay with cellular networks. Adv. Exp. Med. Biol. 2011;722:59–74. doi: 10.1007/978-1-4614-0332-6_4. [DOI] [PubMed] [Google Scholar]
- 34.Pouyssegur J, et al. Hypoxia signaling in cancer and approaches to enforcer tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 35.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 36.Pugh CW, et al. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat. Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 37.Nakayama K. Cellular signal transduction of the hypoxia response. J. Biochem. 2009;146:757–765. doi: 10.1093/jb/mvp167. [DOI] [PubMed] [Google Scholar]
- 38.Rey S, et al. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodeling. Cardiovasc. Res. 2010;86:236–242. doi: 10.1093/cvr/cvq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang LE, et al. Activation of hypoxia-inducible transcription factor depends primarily upon redoxsensitive stabilizaton of its α subunit. J. Biol. Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 40.Wang FS, et al. Ras inductiono f superoxide activates ERK-dependent angiogenic transcription factor HIF-1alpha and VEGF-A expression in shock wave-stimulated osteoblasts. J. Biol. Chem. 2004;279:10331–10337. doi: 10.1074/jbc.M308013200. [DOI] [PubMed] [Google Scholar]
- 41.Su W, et al. Noscapine sensitizes chemoresistant ovarian cancer cells to cisplatin through inhibition of HIF-1α. Cancer Lett. 2011;305:94–99. doi: 10.1016/j.canlet.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 42.Narita T, et al. Identification of a novel small molecule HIF-1alpha translation inhibitor. Clin. Cancer Res. 2009;25:6128–6136. doi: 10.1158/1078-0432.CCR-08-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee G-W, et al. Hypoxia-inducible factor-1α and excision repair cross-complementing 1 in patients with small cell lung cancer who received front-line platinum-based chemotherapy. A retrospective study. J. Thorac. Oncol. 2012;7:528–534. doi: 10.1097/JTO.0b013e3182417830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.