The machinery that replicates cellular DNA does so with impressive speed and accuracy, operating at a speed of approximately 1,000 bp/sec with an accuracy of 10−7 (1). What happens when such an exquisitely tuned protein machine encounters a lesion in its DNA template and how mutations result from such DNA lesions are questions that have fascinated scientists for decades. A surprising insight from earlier genetic and physiological studies of Escherichia coli and Saccharomyces cerevisiae was that mutagenesis resulting from lesions caused by UV irradiation and most chemicals requires the participation of specialized cellular functions in each of these model prokaryotic and eukaryotic systems (2–4). In particular, umuD and umuC mutants of E. coli are largely nonmutable by such agents but display only a modest increase in their sensitivity to killing, whereas S. cerevisiae rev1, rev3, and rev7 mutants are similarly nonmutable. For many years, UV and chemical mutagenesis was widely presumed to occur by a mechanism of translesion synthesis, a notion reinforced by analyses of mutational spectra, so that it seemed most likely that the umuD and umuC gene products might be acting by modifying a replication fork so that translesion synthesis could occur on a damaged template (2–4). The first direct evidence that this was indeed true was provided by the late Hatch Echols and his colleagues (5) in a paper published in the Proceedings in 1992, in which they reported that translesion synthesis over a synthetic abasic site did not occur with DNA polymerase III holoenzyme, E. coli’s replicative polymerase, unless they also added UmuD′ (a posttranslationally modified form of the umuD gene product), UmuC, and RecA.
Progress in this fundamental area of biology has been slow for two major reasons. One is the complex regulatory circuit governing umuDC expression and activity, which is summarized below. The second has been the decidedly “non-user-friendly” nature of the UmuC protein. Because of the insolubility of UmuC protein, Echols et al. (5) purified UmuC in its denatured state and obtained the small amounts used in their experiments by renaturation. In writing Echols’ obituary, Sankar Adhya (6) praised Echols’ translesion synthesis experiments as being “virtuosic science” while Echols himself, at a 1992 meeting held in Taos, NM, ruefully referred to UmuC as being the “black diamond slope of DNA biochemistry.” Two teams of expert skiers have now begun to chart trails down this slope. One team, led by Myron Goodman, reported its progress in the previous issue of the Proceedings (7), while the other team, led by Zvi Livneh, published its results essentially contemporaneously (8).
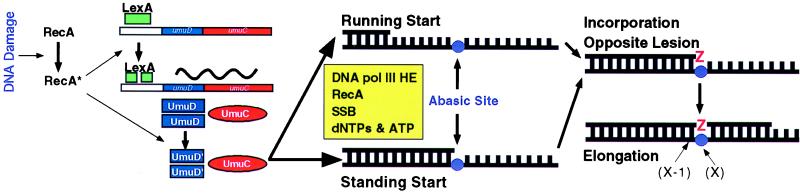
The umuD and umuC genes are subject to both transcriptional and posttranslational regulation as part of E. coli’s SOS response to DNA damage (Fig. 1) (reviewed in ref. 4). The umuDC operon is repressed by the LexA protein and is expressed at higher levels when the LexA protein becomes proteolytically cleaved after interacting with the RecA/single-stranded DNA nucleoprotein filaments (RecA*) that form in cells that have experienced DNA damage. UmuD, the translation product of the umuD gene, is inactive in SOS mutagenesis but undergoes a subsequent proteolytic cleavage that activates it for its role in mutagenesis (9). This cleavage occurs when UmuD interacts with RecA* (10) and removes the N-terminal 24 amino acids. Both LexA cleavage and UmuD cleavage occur by an intriguing RecA*-facilitated autodigestion mechanism (11) in which one of their own lysine residues activates one of their own serine residues that then serves as the nucleophile in the autodigestion reaction. Apart from facilitating LexA and UmuD cleavage, RecA has a “third” role in SOS mutagenesis (4); the RecA1730 derivative is specifically defective in this role (12). DNA polymerase II, a nonessential polymerase encoded by the SOS-regulated polB gene, is clearly not required for UV and chemical mutagenesis (13), but the possibility remains that it could play a role that is physiologically redundant. Although limited genetic evidence suggests that DNA polymerase III participates in SOS mutagenesis in vivo, the relationship of the holoenzyme and its various subunits to this process has remained murky (4).
Figure 1.
(Left) Summary of how the SOS regulatory system operates at both transcriptional and posttranslational levels to control the accumulation of the UmuD′ and UmuC proteins after a cell has experienced DNA damage. (Center and Right) The two classes of DNA substrates carrying a synthetic abasic site that were used by Tang et al. (7) and the events that occur during translesion synthesis.
Goodman and his colleagues (14) earlier had reported the purification of a soluble UmuD′2C complex. Tang et al. (7) describe their characterization of the influence of this complex on translesion synthesis. The DNA template they used in their experiments was a linearized 7.2-kb M13 single-stranded molecule that carries a single synthetic abasic site 50 nucleotides from its 5′ end (Fig. 1). The synthetic abasic site mimics the lesion created by the loss of a purine or pyrimidine base from DNA, an event that can occur spontaneously or be catalyzed by a DNA glycosylase. Two different oligodeoxyribonucleotide primers were used in their studies (Fig. 1). In their “running start” experiments, the 3′-hydroxyl of the primer was located 46 bp upstream of the lesion so that the polymerase would have been replicating normal DNA before encountering the lesion. In their “standing start” experiments, the 3′-hydroxyl of the primer was located just before the lesion, thereby mimicking the situation that would occur if a replication fork stalled at a lesion.
In their initial experiments, which recapitulated the findings of Echols and his colleagues (5), Tang et al. (7) tested the effect of various combinations of DNA polymerase III holoenzyme, UmuD′2C, RecA, and SSB on translesion synthesis by using a running start substrate. In their experiments, DNA polymerase III was added as two components: (i) the β processivity clamp and the γ-complex clamp loader and (ii) the core, which includes α (the polymerization subunit) and ɛ (the proofreading subunit). In the absence of RecA and UmuD′2C, DNA synthesis initiated at the primer terminated just before the lesion (position X-1). Although RecA stimulated replication on the normal DNA leading up to the lesion, it did not permit bypass so that synthesis still terminated at X-1. Only when the UmuD′2C complex was added (at a ratio of 200:1 relative to DNA polymerase III core) was translesion synthesis observed. An intense pause still was observed at X-1, suggesting that incorporation opposite the lesion was rate limiting or that the 3′ → 5′ exonuclease of the ɛ subunit was constantly removing any nucleotide incorporated opposite the lesion. Strikingly, although a limited number of chains were extended to the end of the template, DNA synthesis was essentially distributive past the lesion.
Tang et al. (7) obtained a most unexpected result when they omitted the DNA polymerase III core and observed a pattern of bypass that was extremely similar to that obtained in its presence except for the subtle difference that an additional band terminating at the X position was also present. They also observed that their UmuD′2C preparation permitted translesion synthesis without the addition of a known DNA polymerase on a standing start substrate as well, but found that the addition of increasing amounts of DNA polymerase III core, polymerase III α subunit, or DNA polymerase II, resulted in increasing amounts of full-length and intermediate products. Thus their UmuD′2C preparation, which they estimate is >95% pure, contains a weak DNA polymerase activity that is stimulated by RecA. To date Tang et al. have shown that this is not caused by pol I contamination but they have not yet determined whether the polymerase activity could be caused by contamination by pol III or pol II, or whether the activity is intrinsic to the UmuD′2C complex. They are cautious with respect to this point, appropriately so given that the UmuD′2C complex is present in their reactions at a 200-fold higher concentration (200 nM) than that they used for DNA polymerase III core (1 nM). Evidence that this polymerase activity in their UmuD′2C preparation is relevant to the in vivo situation is provided by their observation that bypass synthesis in the absence of added polymerase no longer occurred when RecA+ was substituted by RecA718, which is defective in the third role of RecA in SOS mutagenesis.
Rather than directly analyzing the identity of the nucleotides incorporated during these translesion synthesis experiments, Tang et al. (7) measured incorporation opposite the lesion and immediately afterward in standing start experiments in which they provided only one deoxyribonucleoside triphosphate at a time. Under these conditions, there was essentially no stable incorporation catalyzed by DNA polymerase III core (1 nM) opposite the abasic site whereas low levels of incorporation were observed with pol II (0.2 nM), consistent with their previous report (15). In striking contrast, in reactions carried out by using the UmuD′2C preparation (200 nM) but no additional polymerase, up to several nucleotides could be added onto the primer when the investigators provided dATP, dGTP, dCTP, or dTTP. The issue of whether one nucleotide was favored when all four were present has not yet been addressed. Moreover, incorporations of any of the four deoxyribonucleotides and also the ribonucleotide A were observed when the standing start substrate had a normal T instead of the synthetic abasic site. Combining these observations with those discussed above, Tang et al. conclude that the most noteworthy property of UmuD′2C lies in its ability to stimulate both nucleotide misincorporation and mismatch extension at aberrant and normal template sites. Additional work will be required to determine whether the formation of the new phosphodiester bonds in their experiments is catalyzed by an intrinsic activity of the UmuD′2C complex, by a contaminant in their UmuD′2C preparation, or by a combination of both. Although not explicitly discussed by Tang et al., it would seem possible that the limited “misincorporations” they observe in the vicinity of the lesion are the result of untemplated additions by a specialized nucleotidyl transferase activity related to that of S. cerevisiae Rev1 (16), with which UmuC shares homology (17), and that the subsequent chain extensions were carried out by a minor amount of DNA polymerase present in the UmuD′2C preparation.
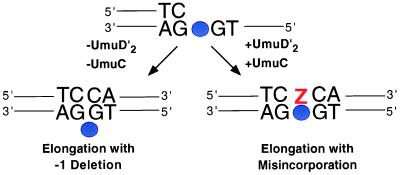
Reuven et al. (8), taking a different approach to the difficulties of working with UmuC, purified and characterized a fusion protein (MBP-UmuC) consisting of maltose-binding protein joined to the N terminus of UmuC. The DNA substrate they used was a gapped plasmid carrying a single abasic site. The abasic site was located within the gap, 10 nucleotides from the 3′-hydroxyl end and thus is of the running start class. In their initial experiments, which recapitulated the observations of Echols and his colleagues (5), they found that the addition of MBP-UmuC (230 nM) and UmuD′ (2.5 μM) to a mixture of DNA polymerase III holoenzyme (1 nM), RecA, and SSB resulted in robust translesion synthesis with up to 70% of the substrates elongated more than 40 nucleotides past the lesion in a short reaction. In contrast to the Tang et al. experiments (7), DNA synthesis past the lesion was quite processive. Some RecA- and SSB-stimulated bypass was observed in the absence of UmuD′ and MBP-UmuC but, by analyzing the translesion synthesis products, Reuven et al. were able to show that the lower amounts of bypass synthesis that occurred in the absence of the Umu protein had resulted from some type of slippage mechanism that most commonly generates a 1-nt deletion (Fig. 2). In contrast, when the Umu proteins were present, an additional product of the expected size was present that had evidently resulted from the insertion of a nucleotide, most commonly an A, opposite the abasic site (Fig. 2). Based on these observations, Reuven et al. make the interesting proposal that UmuD′ and UmuC function to suppress small deletions, a highly disruptive type of mutation within a coding region, and to promote base substitution, a more mild type of mutation within the same context. In vivo experiments provided additional support for their hypothesis.
Figure 2.
Illustration of the two classes of extended translesion synthesis products observed by Reuven et al. (8). In the absence of UmuD′2 and UmuC, the translesion synthesis product most commonly carried a 1-nt deletion that apparently arose by a slippage mechanism. In the presence of UmuD′2 and UmuC, the predominant translesion synthesis product was of the expected length and apparently rose from the introduction of a nucleotide, most often an A, opposite the abasic site.
With respect to the unexpected observation of Tang et al. (7), a significant finding of Reuven et al. (8) was that neither class of extended translesion synthesis product was obtained when DNA polymerase III holoenzyme was omitted from the reaction. As discussed above, an important possibility is that the UmuD′2C preparation, purified by a different procedure, has a contaminating polymerase. Alternatively, in the case of the fusion protein, the MBP moiety might interfere with a polymerase function of UmuC but this seems unlikely given the robust translesion synthesis it promotes. Other factors in experimental design could account for observed differences and need to be explored. These include: (i) the sequence context of the lesion, the Tang et al. substrate could not undergo the facile 1-nt slippage of the Reuven et al. substrate, (ii) the ratio of UmuD′/UmuC, ca. 11:1 in one case [close to the ratio observed in vivo (18)] and 2:1 in the other (the ratio in the purified complex), (iii) the presence of β and the γ complex in the polymerase minus reactions of Tang et al., and (iv) the proportion of the DNA substrate that is single stranded.
These two complementary papers represent a highly significant start toward understanding the mechanisms of translesion synthesis and its relationship to cell survival and mutagenesis. Like most good papers, they raise more questions than they answer. In his recent essay on protein machines, Bruce Alberts (19) pointed out that “Any real clear understanding of the function of a protein machine will require not only its resting structure in atomic detail but also a knowledge of the kinetics and energetics of each of its reaction intermediates.” Learning how the Umu proteins interact with this most fundamental of protein machines, the replisome (20), to permit the replication of damaged DNA templates is sure to be fascinating. Prospects for progress are bright. For example, the monomer structure of UmuD′ is now known from crystallographic analyses (21), the correct homodimer interface of UmuD′2 has been established by NMR studies (22), and monocysteine-directed crosslinking studies have begun to offer insights into the interactions of the UmuD and UmuD′ proteins in solution (23, 24).
The homology between UmuC and Rev1 (17) suggests that the mechanisms of translesion synthesis in prokaryotes and eukaryotes, including humans (25), may be mechanistically related. For example, a plausible, if speculative, model for the action of the Umu proteins is that they interact with components of a DNA polymerase III holoenzyme blocked at a lesion in a way that permits the incorporation of a nucleotide across from the lesion by two different mechanisms. One is to relax the stringency of the DNA polymerase III α subunit for a normal DNA template; depending on the nature of the lesion this mode could potentially be quite accurate (26). The second mechanism is that a Umu-encoded nucleotidyl-transferase activity, similar to the Rev1 activity (16) but with a preference for A addition, adds an untemplated nucleotide to the end of the growing chain, which then becomes repositioned opposite the lesion. The Umu proteins then play additional roles by allowing DNA polymerase III to extend the primer terminus that is opposite the lesion, possibly by suppressing proofreading and altering interactions with the processivity elements. Which of the two modes of incorporation opposite the lesion predominates at a given site would be determined by the nature of the lesion and the sequence context, factors known to influence the mutagenic potential of a translesion synthesis event (26, 27). A related mechanistic strategy with Rev1 (16) replacing the Umu proteins could then explain how translesion synthesis can cause mutagenesis in eukaryotes. DNA polymerase ξ (a complex of the Rev3 and Rev7 proteins) (28) could be involved either in elongation of the imperfect primer terminus opposite the lesion and/or in a Rev1-modulated misinsertion opposite the lesion.
Acknowledgments
I thank Bradley T. Smith for preparing the figures.
Footnotes
The companion to this commentary begins on page 9755 in issue 17 of volume 95 of this issue.
References
- 1.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 2.Witkin E M. Bacteriol Rev. 1976;40:869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker G C. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 5.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman M F, Echols H. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adhya S. Cell. 1993;73:833–834. doi: 10.1016/0092-8674(93)90263-p. [DOI] [PubMed] [Google Scholar]
- 7.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O’Donnell M, Goodman M F. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reuven, N. B., Tomer, G. & Livneh, Z. (1998) Mol. Cell, in press. [DOI] [PubMed]
- 9.Nohmi T, Battista J R, Dodson L A, Walker G C. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Little J W. J Bacteriol. 1993;175:4943–4950. doi: 10.1128/jb.175.16.4943-4950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutreix M, Moreau P L, Bailone A, Galibert F, Battista J R, Walker G C, Devoret R. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kow Y W, Fuandez G, Hays S, Bonner C, Goodman M F, Wallace S S. J Bacteriol. 1993;175:561–564. doi: 10.1128/jb.175.2.561-564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruck I, Woodgate R, McEntee K, Goodman M F. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 15.Bonner C A, Randall S K, Rayssiguier C, Radman M, Eritja R, Kaplan B E, McEntee K, Goodman M. J Biol Chem. 1988;263:18946–18952. [PubMed] [Google Scholar]
- 16.Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 17.Larimer F, Perry J, Hardigree A. J Bacteriol. 1989;171:230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodgate R, Ennis D G. Mol Gen Genet. 1991;229:10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- 19.Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 20.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 21.Peat T S, Frank E G, McDonald J P, Levine A S, Woodgate R, Hendrickson W A. Nature (London) 1996;380:727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- 22.Ferentz A E, Opperman T, Walker G C, Wagner G. Nat Struct Biol. 1997;4:979–983. doi: 10.1038/nsb1297-979. [DOI] [PubMed] [Google Scholar]
- 23.Lee M H, Walker G C. J Bacteriol. 1996;178:7284–7294. doi: 10.1128/jb.178.24.7285-7294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzzo A, Lee M H, Oda K, Walker G C. J Bacteriol. 1996;178:7295–7303. doi: 10.1128/jb.178.24.7295-7303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbs P E, McGregor W G, Maher V M, Nisson P, Lawrence C W. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence C W, Banerjee S K, Borden A, LeClerc J E. Mol Gen Genet. 1990;222:166–168. doi: 10.1007/BF00283040. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence C W, Borden A, Banerjee S K, LeClerc J E. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]