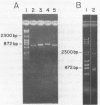
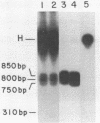
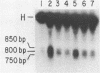
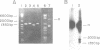Abstract
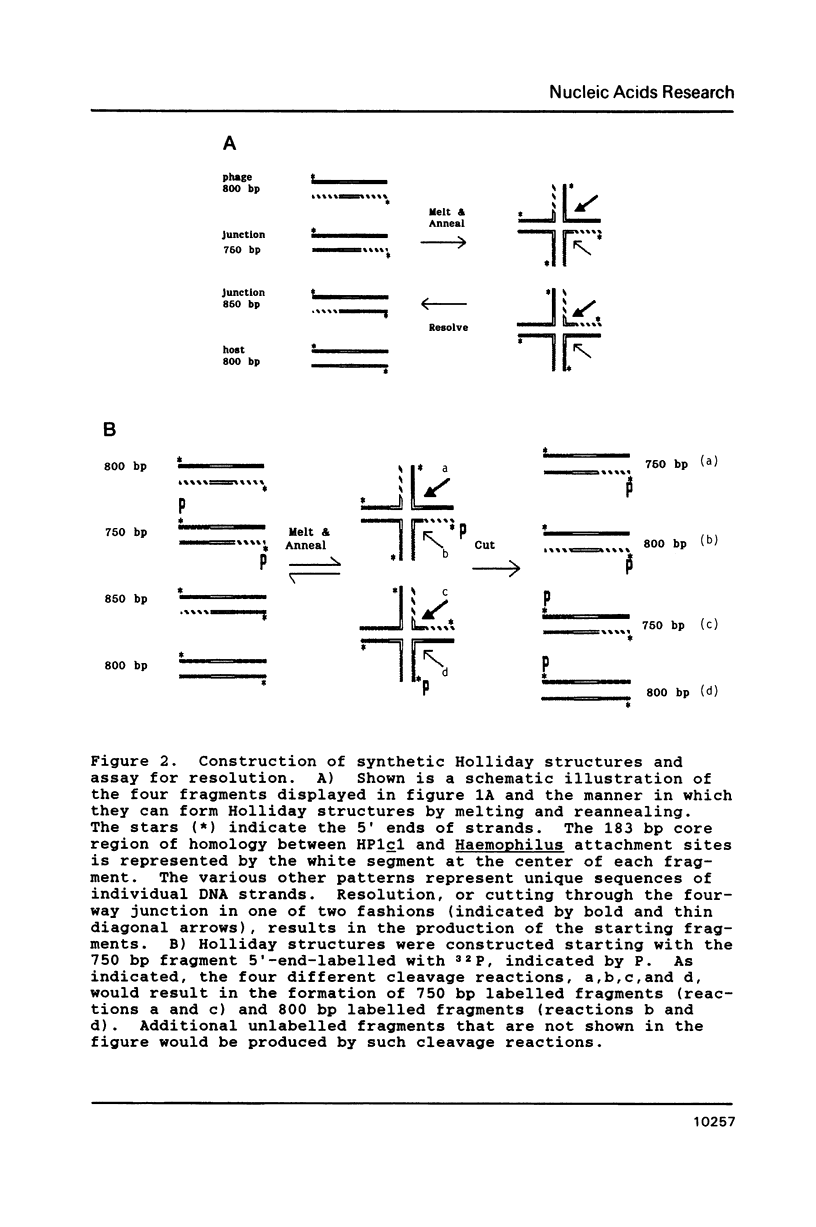
Virtually all models for recombination between homologous DNA sequences invoke a branched intermediate known as a Holliday structure. The terminal steps of recombination are postulated to involve a specific cleavage through the four-way junction of a Holliday structure, in a process known as resolution. We have constructed a synthetic Holliday structure in which the position of the junction of the DNA duplexes can branch migrate through approximately 185 bp. Using this structure, we have found that a component of a cytoplasmic extract of Hela cells is capable of cleaving the central junction of the substrate in a manner consistent with resolution. The activity requires a divalent cation but does not require an exogenous energy source. This is the first reported resolution activity from a mammalian source.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell L., Byers B. Occurrence of crossed strand-exchange forms in yeast DNA during meiosis. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3445–3449. doi: 10.1073/pnas.76.7.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M. E. Interaction of a protein from rat liver nuclei with cruciform DNA. EMBO J. 1988 Mar;7(3):843–849. doi: 10.1002/j.1460-2075.1988.tb02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. L., Yang L., Rowe T. C., Halligan B. D., Tewey K. M., Liu L. F. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984 Nov 10;259(21):13560–13566. [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Enzymes of general recombination. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- Elborough K. M., West S. C. Specific binding of cruciform DNA structures by a protein from human extracts. Nucleic Acids Res. 1988 May 11;16(9):3603–3616. doi: 10.1093/nar/16.9.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. H., Kolodner R. Construction of a synthetic Holliday junction analog and characterization of its interaction with a Saccharomyces cerevisiae endonuclease that cleaves Holliday junctions. J Biol Chem. 1987 Jul 5;262(19):9160–9165. [PubMed] [Google Scholar]
- Evans D. H., Kolodner R. Effect of DNA structure and nucleotide sequence on Holliday junction resolution by a Saccharomyces cerevisiae endonuclease. J Mol Biol. 1988 May 5;201(1):69–80. doi: 10.1016/0022-2836(88)90439-1. [DOI] [PubMed] [Google Scholar]
- Ganea D., Moore P., Chekuri L., Kucherlapati R. Characterization of an ATP-dependent DNA strand transferase from human cells. Mol Cell Biol. 1987 Sep;7(9):3124–3130. doi: 10.1128/mcb.7.9.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick G., Spear B. B., Veomett G. Intracellular localization of mouse DNA polymerase-alpha. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1136–1139. doi: 10.1073/pnas.73.4.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y., Tabata S., Bouchard R. A., Piñon R., Stern H. General recombination mechanisms in extracts of meiotic cells. Chromosoma. 1985;93(2):140–151. doi: 10.1007/BF00293161. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Meyn M. S., Camerini-Otero R. D. Partial purification and characterization of a recombinase from human cells. Cell. 1986 Mar 28;44(6):885–894. doi: 10.1016/0092-8674(86)90011-5. [DOI] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E., Kroeger P., Holliday R., Holloman W. Homologous pairing promoted by Ustilago RecI protein. Cold Spring Harb Symp Quant Biol. 1984;49:675–682. doi: 10.1101/sqb.1984.049.01.076. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Evans D. H., Morrison P. T. Purification and characterization of an activity from Saccharomyces cerevisiae that catalyzes homologous pairing and strand exchange. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5560–5564. doi: 10.1073/pnas.84.16.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Parsons C. A., West S. C. Resolution of model Holliday junctions by yeast endonuclease is dependent upon homologous DNA sequences. Cell. 1988 Feb 26;52(4):621–629. doi: 10.1016/0092-8674(88)90474-6. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Goodman S. D., Scocca J. J. Nucleotide sequences and properties of the sites involved in lysogenic insertion of the bacteriophage HP1c1 genome into the Haemophilus influenzae chromosome. J Bacteriol. 1987 Jan;169(1):238–246. doi: 10.1128/jb.169.1.238-246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]