Abstract
Metnase (SETMAR) is a SET-transposase fusion protein that promotes non-homologous end joining (NHEJ) repair in humans. Although both SET and the transposase domains were necessary for its function in DSB repair, it is not clear what specific role Metnase plays in the NHEJ. In this study, we show that Metnase possesses a unique endonuclease activity that preferentially acts on ssDNA and ssDNA-overhang of a partial duplex DNA. Cell extracts lacking Metnase poorly supported DNA end joining, and addition of wt-Metnase to cell extracts lacking Metnase markedly stimulated DNA end joining, while a mutant (D483A) lacking endonuclease activity did not. Given that Metnase overexpression enhanced DNA end processing in vitro, our finding suggests a role for Metnase's endonuclease activity in promoting the joining of non-compatible ends.
Keywords: Metnase, SETMAR, DSB damage, Non-homologous end joining repair, DNA cleavage, Transposase, DNA endonuclease
DNA double-strand breaks (DSB) are the primary cytotoxic lesions repaired by non-homologous end joining (NHEJ) in mammals (1, 2). NHEJ repair involves processing and ligation of two free DNA ends, requiring Ku70/80 heterodimer, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Artemis, and XRCC4/DNA Ligase IV (3-13). Upon DSB damage, the Ku70/80 complex first binds to the DNA ends, and recruits DNA-PKcs (3, 4, 6, 7, 11, 12, 14, 15). Autophosphorylation of DNA-PKcs is required for NHEJ, and Artemis, WRN, and XRCC4 are also phosphorylated in a DNA-PKcs-dependent manner (16). Artemis possesses nuclease activity and interacts with DNA-PKcs, and may be required for DNA end processing prior to DNA end joining (16). The recruitment of the XRCC4-DNA Ligase IV (Lig4) complex is essential for the final ligation step (17, 18). XLF, also known as Cernunnos, is a newly identified NHEJ factor known to stimulate Lig4 through its interaction with XRCC4 (12, 19, 20).
In vitro NHEJ repair analysis (21-25) has been a useful tool for identification of additional repair factors, but was limited to joining of linearized plasmid DNA with compatible ends (21). A PCR-based intermolecular end joining assay (22, 26, 27), on the other hand, allows to measure joining of both compatible and non-compatible ends (27). Recent studies with in vitro end joining approach suggested that, although NHEJ repair is dependent on the Ku complex, DNA-PKcs, and XRCC4/Ligase4, it also requires additional factors for end joining (22, 28, 29). Metnase, also known as SETMAR, is a novel SET [Su(var)3-9, Enhancer-of-zeste, Trithorax] and transposase fusion protein (30, 31). The chimeric fusion of the Hsmar1 transposase with a SET domain is a unique event that occurred in the evolution of anthropoid primates approximately 50 million years ago and is not found in prosimian monkeys or other mammals (30). Metnase possesses many but not all of the activities of a transposase, including sequence-specific DNA binding and DNA looping, the assembly of a paired end complex (PEC), the cleavage of a 5′-end of the TIR element, and the promotion of integration at a TA dinucleotide target site (30, 32-36). A transposase domain containing the DDE acidic motif conserved among retroviral integrase and transposase families (37, 38). It also possesses histone lysine methyltransferase (HLMT) activity at histone 3 lysine 4 and lysine 36 (39, 40) associated with chromatin opening (41-43) at DNA damage sites (40). Metnase's involvement in NHEJ repair came from an in vivo study showing that overexpression of Metnase increased NHEJ repair, while it did not produce any significant changes in homologous recombination repair (HRR) (39). Similarly, cells treated with Metnase-siRNA showed a significant reduction for in vivo NHEJ repair activity (39). Metnase over-expression resulted in a 3-fold survival advantage after ionizing radiation compared to vector controls (39), further evidence of a linkage between Metnase and NHEJ. Metnase is also involved in genomic integration of foreign DNA (39, 44) that depends on some of the NHEJ factors (45, 46). A deletion of either SET or the transposase domain abrogated Metnase's function in DNA repair, indicating that both domains are required for this function (39). However, whether Metnase plays a direct role in NHEJ, or enhanced the activity of other NHEJ components, is not well defined.
In this study, we show that Metnase possesses a unique endonuclease activity that preferentially acts on ssDNA and ssDNA overhang of a partial duplex DNA. Cell extracts lacking Metnase exhibited significantly lowered end joining activity, which was comparable to those seen in extracts lacking DNA-PKcs or Ku80. Addition of wt-Metnase but not the mutant (D483A) restored DNA end joining activity with cell extracts lacking Metnase. These data imply that Metnase plays a direct role in the joining of both compatible and non-compatible ends.
Materials and Methods
Cells, enzymes, oligonucleotides, and antibodies
HEK-293 cells, mouse Ku80-/-, DNA-PK-/-and ATM-/- cells were previously described (39, 47). Restriction enzymes (BamH I, Hind III, Kpn I, EcoR V, and Pst I) were obtained from Promega (Madison, WI). The oligonucleotides were obtained from the Integrated DNA Technologies (Coralville, IA). An anti-Metnase antiserum (polyclonal) was generated from rabbits using two peptides representing amino acids 483-495 and 659-671 (39). An anti-FLAG antibody was obtained from Sigma (St. Louis, MO). The oligonucleotides and the 5′-fluoresent labeled DNA were obtained from the Integrated DNA Technologies (Coralville, IA).
Chemicals and DNA substrates
The following suppliers provided the listed items: [γ-32P]-ATP (3000 Ci/mmol) from Perkin-Elmer and Analytical Science (Boston, MA), DE81 filters from Whatman Bio System (Maidstone, England), heparin-Sepharose from Amersham Biosciences (Piscataway, NJ), and Bradford reagents and protein molecular weight markers were purchased from Bio-Rad (Hercules, CA). Closed-circular pBluescript (pBS) II SK+ duplex phagemid DNA (3.0 kbps) was linearized with indicated restriction enzyme(s), and purified on agarose gel twice using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). Concentrations of recovered DNA were determined by spectrophotometer.
Generation of Metnase over- and under-expressing cell lines
Human embryonic kidney (HEK-293) cells expressing either wt-Metnase or the mutant (D483A) were generated by a stable transfection of HEK-293 cells with either pcDNA3.1-V5/His (Invitrogen) for the control or pcDNA3.1-Metnase-V5/His for the over-expression of Metnase (39). Transfectants were selected in 800ug/ml G418 for 14-21 days, and individual colonies were expanded and analyzed for Metnase expression by Western blot using a polyclonal antibody specific for Metnase (39). Metnase underexpressors were generated by stable transfection of HEK-293 cells with either pRNA-U6.1/Hyg (Genescript) for the control or pRNA-U6.1/Hyg-siRNA-Metnase to reduce the expression of endogenous Metnase. Transfectants were selected in 150 μg/ml hygromycin for 10-14 days. Reverse transcription-PCR was used as the initial screen for clones that had a reduced expression of Metnase compared to the control U6 clone. The primers for Metnase and 18S and the sequence for the Metnase siRNA were described previously (39).
SDS-PAGE and Western blot analysis
Protein fractions were analyzed by 10 % SDS-polyacrylamide gel electrophoresis (SDS-PAGE). For western blot, proteins were transferred to polyvinylidene difluoride (PVDF) membrane, probed with an anti-FLAG (monoclonal mouse IgG, Sigma) or an anti-Metnase antibody (polyclonal rabbit IgG) followed by horseradish peroxidase-conjugated secondary antibody. Protein was visualized by using the ECL system (Amersham Biosciences).
Preparation of cell extracts
Whole cell extracts were prepared from HEK-293 cells as described previously (21). Briefly, HEK-293 cells expressing different levels of Metnase were harvested at 80-90% confluency, and washed three times in ice-cold PBS and once in hypotonic lysis buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 5 mM DTT). Cells were resuspended in 500 μl of hypotonic buffer, incubated on ice for 20 min and homogenized after addition of protease inhibitors (0.17 μg/ml phenylmethyl-sulfonyl fluoride, 1 μg/ml aprotinin, 0.01 units/ml trypsin inhibitor, 1 μg/ml pepstatin, 1 μg/ml chymostatin, 1 μg/ml leupeptin). Following 20 min incubation on ice, 0.5 volume of high-salt buffer (50 mM Tris-HCl, pH 7.5, 1 M KCl, 2 mM EDTA, 2 mM DTT) was added to the cell lysates prior to centrifugation for 3 hr at 42,000 rpm in a Beckman SW50.1 rotor. The supernatant was dialyzed against E buffer (20 mM Tris-HCl pH 8.0, 0.1 M KOAc, 20% glycerol (v/v), 0.5 mM EDTA, 1 mM DTT) for 3 hours and was fast-frozen and stored at −80°C.
DNA cleavage assay
DNA cleavage assay was carried out using the previously described procedure with modification (35). Briefly, reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5% glycerol, BSA (10 μg), and 2 mM MgCl2 were incubated with indicated amounts of wt-Metnase or the mutant in the presence of 200 fmol of 5′-32P-DNA. After incubation at 37°C for indicated amount of time, reaction mixtures were analyzed by 12 % polyacrylamide gel electrophoresis containing 8M urea for DNA cleavage.
Glycerol gradient centrifugation
Immunopurified wt-Metnase was sedimented through a linear 10-35 % (vol/vol) glycerol gradient at 45,500 rpm for 26 hrs at 4°C. Fractions (175 ul/each) were collected from the bottom, and the aliquots were examined for DNA cleavage activity and also run on 10% SDS-PAGE for western-blot analysis.
DNA end joining assay coupled to E. coli colony formation
Reactions mixtures (100 μl) contained 50 mM Tris-HCl (pH 7.5), 0.5 mM Mg-acetate, 60 mM potassium acetate, 2 mM ATP, 1 mM DTT, and 100 μg/ml BSA. Where indicated, 2.5 mM dNTPs were included in the reaction mixtures. Cell-free extracts were pre-incubated for 5 min at 37°C before addition of 1.0 μg of DNA substrate. Following incubation for 1 hr at 37°C, DNA products were deproteinized, purified by QIAprep Kit (Qiagen, Valencia, CA), and transformed into E. coli for colony formation. For the transformation, we used the high efficiency DH5α competent cells (> 1.0 × 108 cfu/μg). Where indicated, PCR amplification of end joining products was performed using Taq DNA polymerase (Promega, Madison, WI) and the two primers (M13 Reverse and T7 primers).
Results
Metnase prefers ssDNA to dsDNA for its DNA cleavage activity
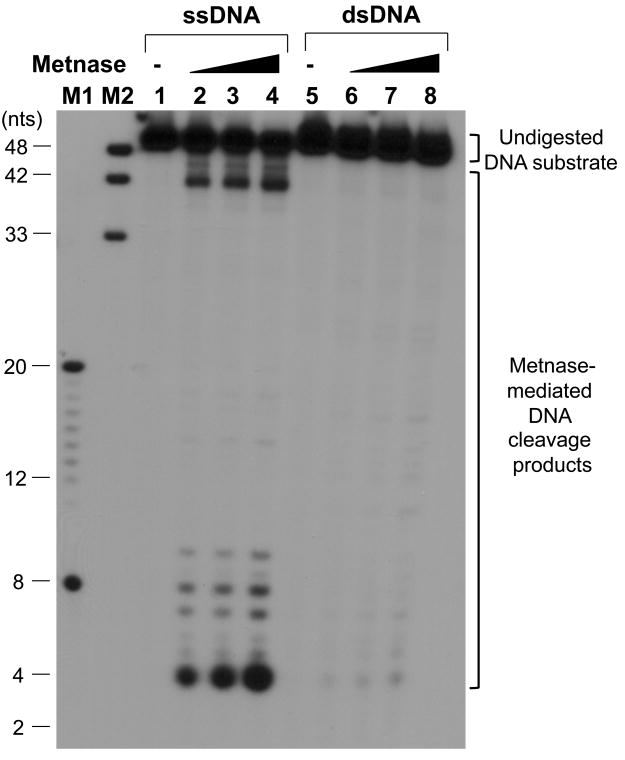
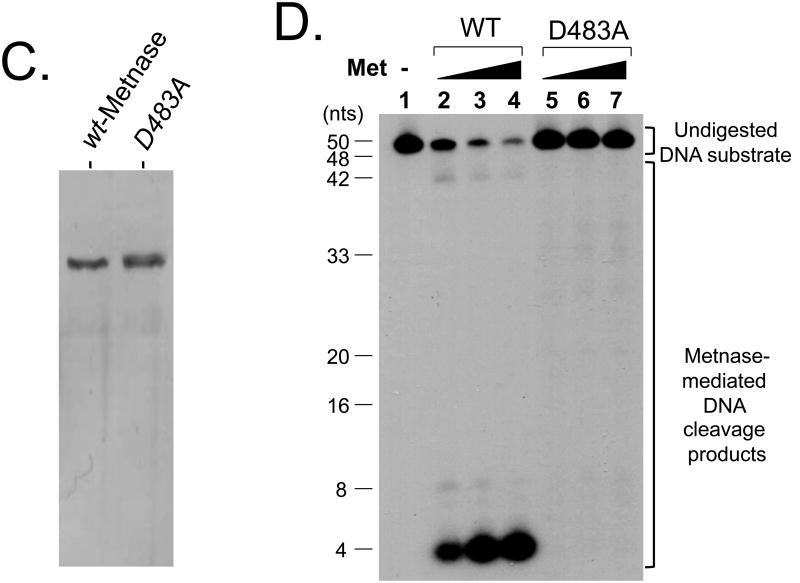
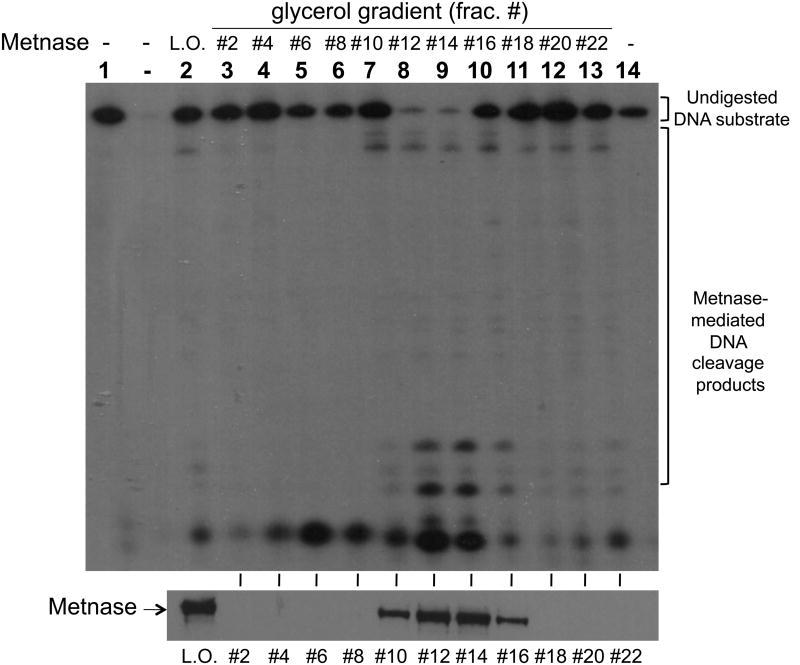
Metnase, a SET-transposase fusion protein, is a DSB repair factor that possesses most of the transposase's activities, including 5′-terminal inverted repeat (TIR)-specific DNA binding, DNA looping via the assembly of a paired end complex (PEC), the cleavage of a 5′-end of the TIR element, and the promotion of integration at a TA dinucleotide target site (30, 32-36). We previously showed that Metnase, unlike mariner transposase (48), possesses TIR-independent endonuclease activity that converts circular duplex DNA to nicked circular and/or linearized duplex DNA and also cleaves duplex DNA (35). To understand the role of Metnase's TIR-independent endonuclease, we examined its non-TIR DNA cleavage with ss-and dsDNA. When Metnase was incubated with 50mer of either ssDNA or dsDNA (see Fig. 1A for details), Metnase exhibited its endonucleolytic activity that targets several key cleavage sites as shown previously [35]. Metnase showed a significantly higher DNA cleavage activity with ssDNA, as compared to dsDNA (Fig. 1B). This was a surprising result because Metnase has a conserved DDE motif in the transposase domain that targets TIR-specific dsDNA (33, 49). We then compared wt-Metnase with a mutant (D483A) defective in DNA cleavage (35) for ssDNA cleavage to see whether ssDNA cleavage activity observed in Fig. 1B belongs to Metnase. Although we saw no difference in protein purity between flag-wtMetnase and the mutant (flag-D483A) on SDS-PAGE (Fig. 1C), the mutant showed little or no cleavage activity under the conditions where wt-Metnase exhibited >90% cleavage of ssDNA substrate (Fig. 1D). To further confirm Metnase-mediated ssDNA cleavage activity, immunopurified wt-Metnase was subjected to a glycerol gradient centrifugation (see Materials & Methods for the details), and the fractions were examined for wt-Metnase and ssDNA cleavage activity. The peak fractions (#12-14) of wt-Metnase (Fig. 1E, bottom panel) correlated well with ssDNA cleavage activity (Fig. 1E, top panel), suggesting that ssDNA cleavage activity observed in Fig. 1B was mediated by Metnase.
Figure 1. Metnase prefers ssDNA over dsDNA for its DNA cleavage activity.
(A) DNA substrates (50-mer of ss- and dsDNA) used in this study. (B) Reaction mixtures (20 μl) containing 50 fmol of 5′-32P-labeled ssDNA (lanes 1-4) or dsDNA (lanes 5-8) were incubated with 0 ng (lanes 1 & 5), 25 ng (lanes 2 & 6), 50 ng (lanes 3 & 7), and 100 ng (lanes 4 & 8) of wt-Metnase in the presence of 2 mM MgCl2. After incubation for 60 min at 37°C, cleavage products were analyzed by 12% PAGE containing 8M urea. DNA size makers were indicated on the left (lanes M1 & M2). (C) 10% SDS-PAGE (silver staining) of of purified wt-Metnase and D483A (50 ng each) used in this study. (D) The mutant (D483A) lacks ssDNA cleavage activity. Reaction mixtures (20 μl) containing 50 fmol of 5′-32P-labeled 50-mer ssDNA were incubated with 0 ng (lanes 1), 50 ng (lanes 2 & 5), 100 ng (lanes 3 & 6), and 200 ng (lanes 4 & 7) of either wt-Metnase (lanes 2-4) or D483A (lanes 5-7). After incubation at 37°C for 60 min, reaction mixtures were analyzed by 12% PAGE containing 8M urea for ssDNA cleavage. DNA size markers were indicated on the left. (E) Biochemical analysis of glycerol gradient fractions of wt-Metnase for ssDNA cleavage activity. Fractions collected from a glycerol gradient centrifugation (see Materials & Methods' for details) were analyzed by western blot of wt-Metnase (bottom panel) and ssDNA cleavage activity (top panel). Lane 1 contains only DNA substrate. L.O. represents load-on of glycerol gradient centrifugation (immunoaffinity purified wt-Metnase)
Metnase preferentially cleaves ssDNA overhang of a partial duplex DNA
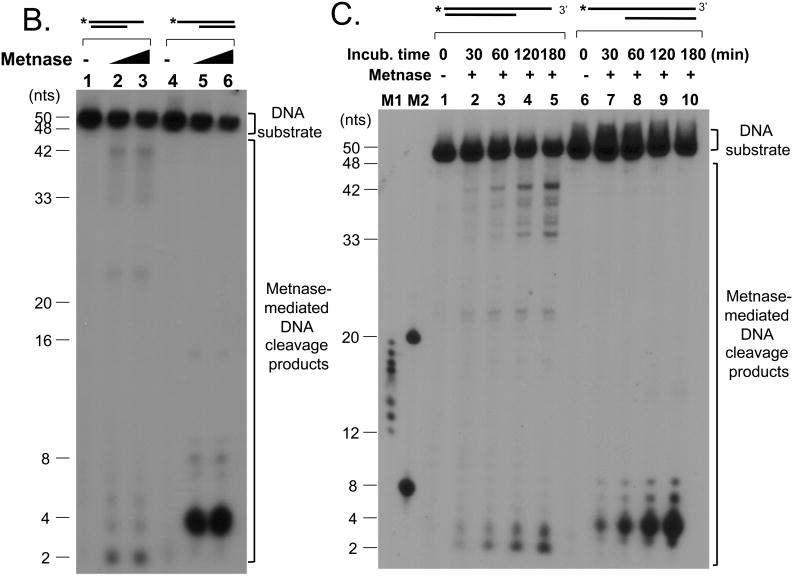
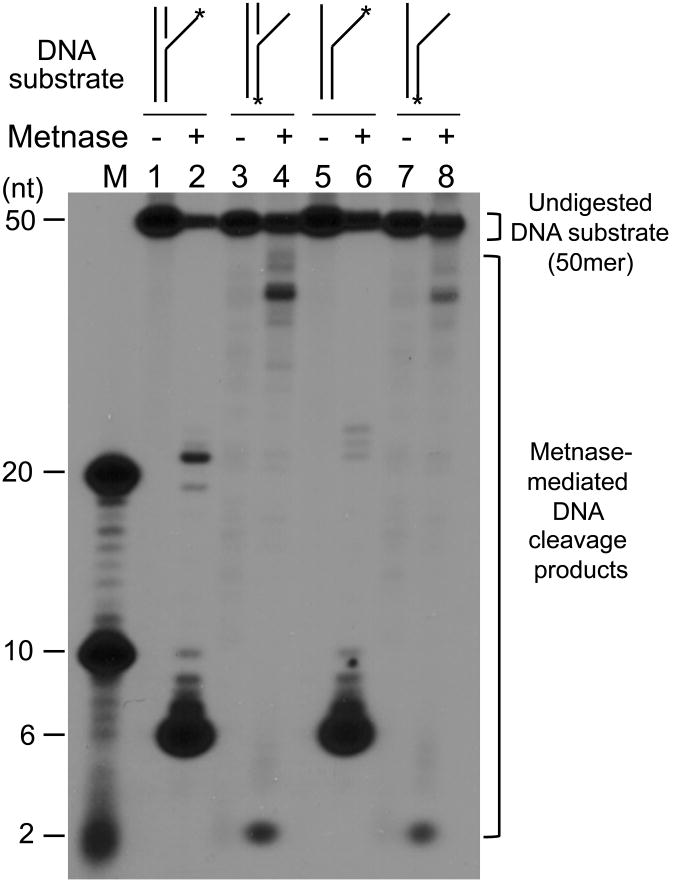
To find out physiologic relevance of Metnase's ssDNA cleavage activity, we next examined Metnase for cleavage of partial duplex DNA (30mer of oligonucleotides annealed to either 3′- or 5′-end of 5′-32P-labeled 50mer DNA) that mimics DSB damage (Fig. 2A). If Metnase possesses a preferential ssDNA overhang cleavage activity, we would expect to see >30 nts cleavage products with a partial duplex DNA with 3′-overhang (Fig. 2B, lanes 1-3), while <20 nts cleavage products would be generated in the presence of a partial duplex DNA with 5′-overhang (Fig. 2B, lanes 4-6). Indeed, Metnase showed a preferential cleavage of ssDNA region of a partial duplex DNA with 5′-overhang, while it acted on both ss- and dsDNA regions of a 3′-overhang DNA (Fig. 2B). A kinetic analysis also showed that Metnase preferentially cleaves on ssDNA overhang of a partial duplex DNA (Fig. 2C). To see functional implication of Metnase's DNA cleavage activity in DSB repair, we also examined Metnase for cleavage of various DNA substrates (listed in Table 1) that can be generated after DSB damage and/or during DNA repair (7, 11, 13, 14, 16, 28). Although all DNA substrates used here contain ssDNA region in their structures, Metnase showed a preferred ssDNA overhang cleavage activity with flap and pseudo Y DNA substrates (Fig. 3A, lanes 2 & 8). When Metnase was incubated with hairpin DNA, loop DNA, and duplex DNA with ssDNA gap (see Table 1 for DNA structures in details), it did not show any DNA cleavage at the ssDNA region (Fig. 3A, lanes 6, 12, & 14). This result, along with the observation shown in Fig. 2, suggests that Metnase possesses a unique endonuclease activity that preferentially acts on ssDNA overhang of a partial duplex DNA. The finding that Metnase has no hairpin or DNA loop opening activity is an important functional difference between Metnase and Artemis since the latter has a preferred hairpin opening activity (50, 51), and suggests that Metnase does not have a role in opening hairpin intermediates in V(D)J recombination. We also examined whether Metnase has a directional preference (5′-vs. 3′-) in its ssDNA overhang cleavage activity with flap and/or pseudo Y DNA. Metnase showed ssDNA overhang cleavage with both 5′- and 3′-flap/pseudo Y DNA (Fig. 3B, lanes 4 & 8), although intensity of 3′-overhang cleavage products (>30 nts) was somewhat diminished probably due to an additional DNA cleavage towards 5′-end, producing 2 nts cleavage product. Metnase-mediated DNA cleavage with various DNA substrates summarized in Fig. 3C suggests that Metnase has a unique endonuclease activity that preferentially acts on ssDNA overhang of a partial duplex DNA.
Figure 2. Metnase has a preferred DNA cleavage activity at single-strand region of a partial duplex DNA.
(A) Partial duplex DNA with either 5′- or 3′-ssDNA overhang used in this study. (B) Reaction mixtures (20 μl) containing either 50 fmol of 5′-32P-labeled 3′-overhang DNA (lanes 1-3) or 5′-overhang DNA (lanes 4-6) were incubated with 0 ng (lanes 1, 4), 50 ng (lanes 2, 5), and 100 ng (lanes 3, 6) of wt-Metnase. After incubation for 90 min at 37°C, reaction mixtures were analyzed by 12% PAGE containing 8M urea. (C) Kinetic analysis of Metnase's DNA cleavage activity with 3′- (lanes 1-5) or 5′-overhang (lanes 6-10) partial duplex DNA in the presence of wt-Metnase (100 ng). M1 & M2 represent two DNA markers used in this study.
Table 1.
DNA substrates used in this study

|
represents the position of 5′-32 labeling.
Figure 3. Characterization of Metnase's DNA cleavage activity with different DNA substrates.
(A) Metnase preferentially cleaves ssDNA overhang of flap and pseudo Y DNA. Wt-Metnase (0 or 50 ng) was incubated with 100 fmol of 5′-P32-labeled DNA substrate indicated on the top (see Table 1 for the details of individual substrates) for 60 min prior to 12% denatured PAGE (+ 8M urea) analysis. Lanes “M” represent DNA markers. (B) Metnase exhibits both 3′- and 5′-ssDNA overhang cleavage activity. Metnase (0 or 50 ng) was incubated with 5′-flap DNA (lanes 1-2), 3′-flap DNA (lanes 3-4), 5′-pseudo Y DNA (lanes 5-6), and 3′-pseudo Y DNA (lanes 7-8). M represents DNA markers. (C) Diagrams of the major DNA cleavage sites catalyzed by Metnase. Arrows mark the major cleavage sites from the 32P-labeled (*) 5′-end (calculated from the cleavage products shown in Fig. 3). The length of the arrows approximately was proportional to the intensity of resulting cleavage products.
A Metnase mutant defective in DNA cleavage failed to support DNA end joining in vitro
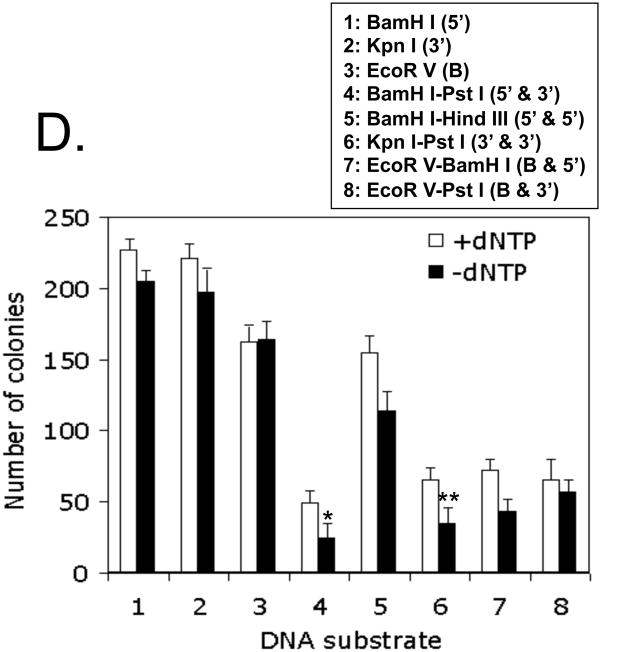
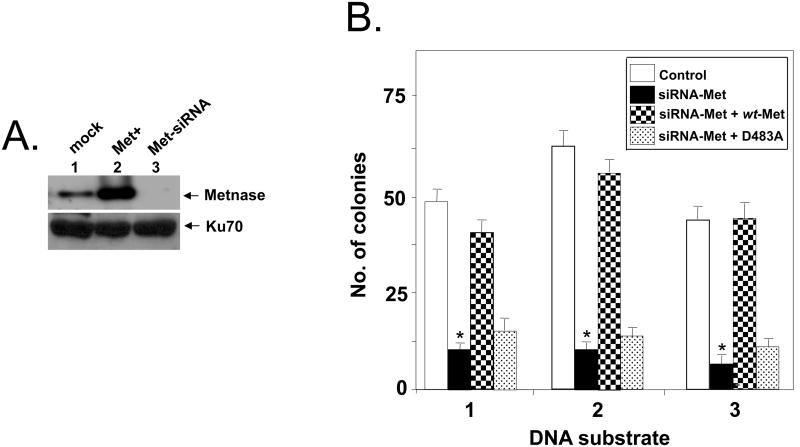
Since Metnase stimulates NHEJ repair (32, 39, 44), we next examined whether Metnase's DNA cleavage activity is involved in DNA end joining. For this, cell extracts containing different level of Metnase were examined for end joining activity using an intramolecular end joining coupled to E. coli colony formation, where circular duplex DNA generated from end joining of linear plasmid DNA in the presence of cell extracts was quantitatively measured by ampicillin-resistant E. coli colonies following DNA isolation and transformation (Fig. 4A) (52). As shown previously (21, 22), colony formation requires the presence of ATP and Mg+2, but was completely inhibited in the presence of wortmannin, a specific inhibitor of PI-3 kinase family (Fig. 4B). DNA end joining activity was supported by cell-free extracts prepared from wild-type mouse fibroblast and ATM-/- but not those from DNA-PKcs-/- and Ku80-/- cells (Fig. 4C), suggesting that the intramolecular end joining measured by a coupled to E.coli colony formation occurs via NHEJ repair pathway. In keeping with the previous observation (22), joining of non-complementary ends was less effective than the compatible end joining but was stimulated in the presence of dNTPs (Fig. 4D), suggesting that DNA polymerase action facilitates intramolecular joining of non-compatible ends in vitro (22, 53, 54). We then examined whether Metnase expression level influences joining of non-compatible ends in a cell-free system. Cell-free extracts prepared from HEK-293 cells overexpressing wt-Metnase stimulated joining of non-compatible ends by 25-50%, while extracts prepared from cells transfected with Metnase-specific siRNA showed 8-20 fold lower DNA end joining activity than the control extracts (Fig. 5B). More importantly, addition of purified wt-Metnase to cell extracts restored DNA end joining activity, while addition of the mutant protein (D483A) lacking DNA cleavage activity (Fig. 1D) (35) poorly restored end joining activity (Fig. 5B), suggesting that Metnase's DNA cleavage activity is involved in DNA end joining in vitro.
Figure 4. Intramolecular joining of linearized DNA in a cell-free system.
(A) Intramolecular end joining assay that measures joining of linearized plasmid DNA in a cell-free system. Following incubation of linearized pBS with cell extracts at 37°C, DNA is isolated and transformed into E. coli for colony counts. (B) The assay was carried out in the presence or absence of 1 mM ATP, 1 mM MgCl2, or 10 uM wortmannin for 60 min prior to DNA isolation and E. coli transformation for colony counts. Where indicated, cell extracts were pre-incubated for 30 min on ice with wortmannin. Wortmannin was dissolved in DMSO at a high concentration (10 mM) to limit the final DMSO concentration in the reaction mixtures to less than 1%. Values are average (±SEM) of 6 independent assays. *P < 0.005; **P = 0.01. (C) Intramolecular end joining is dependent on Ku80 and DNA-PKcs. Normal mouse fibroblast (black bar) or cells lacking Ku80 (checker bar), DNA-PKcs (white bar), or ATM (grey bar) (47) were used to prepare cell extracts. DNA end joining assay coupled to E. coli colony formation was carried out with linearized pBS with containing 5′-overhangs, 3′-overhangs, or blunt ends. Values are averages (±SEM) of 3 distinct determinations. P < 0.01; *P = 0.05. (D) Intramolecular end joining of linearized DNA in the presence and absence of dNTPs. Linearized pBS DNA (1.0 μg) with different compatible or non-compatible ends was incubated with 30 μg of HEK-293 cell extracts, 1 mM MgCl2, and 1 mM ATP for 60 min. Where indicated, 25 μM dNTPs were included. Following incubation, DNA was isolated and transformed into E. coli for colony counts. Numbers 1-8 represent DNA substrates used for DNA end joining assay. B, 5′, and 3′ represent DNA ends with blunt ends, 5′-overhang, and 3′-overhangs, respectively. Values are the average (±SEM) of 3 separate experiments. P < 0.01; *P = 0.05; **P = 0.07.
Figure 5. Wt-Metnase stimulated intramolecular joining of linearized DNA in a cell-free system, while a mutant (D483A) lacking DNA cleavage activity did not.
(A) A targeted inhibition of Metnase expression by a Metnase-specific siRNA (Met-siRNA). HEK-293 cells harboring Flag vector (control; lane 1), HEK293 cells stably expressing Flag-Metnase (Met+; lane 2), or HEK293 cells treated with Metnase-siRNA (Met-siRNA; lane 3) were incubated for 48 hrs prior to western blot analysis using an anti-Metnase polyclonal antibody. Expression of Ku70 was included as a loading control. (B) Addition of purified wt-Metnase but not the mutant (D483A) to cell extracts lacking Metnase restores DNA end joining activity. Reaction mixtures (100 μl) containing 30 μg of cell extracts, linearized pBS DNA (1.0 ug), 1 mM MgCl2 and 1 mM ATP were incubated for 60 min. Where indicated, 0.4 ug of purified wt-Metnase or the mutant protein (D483A) was added to the extracts lacking Metnase (siRNA-Met) prior to DNA end joining reactions. Following incubation, DNA was isolated and transformed into E. coli for colony counts. Numbers 1, 2, & 3 at the bottom of the figure represent pBS DNA digested with Bam HI-Hind III (5′-5′), Kpn I-Pst I (3′-3′), and Bam HI-Pst I (5′-3′), respectively, which produced linearized DNA with different non-compatible ends. The figure is representative of three DNA end joining assay. Values are the average (±SEM) of 3 separate experiments (P < 0.01, *P = 0.05).
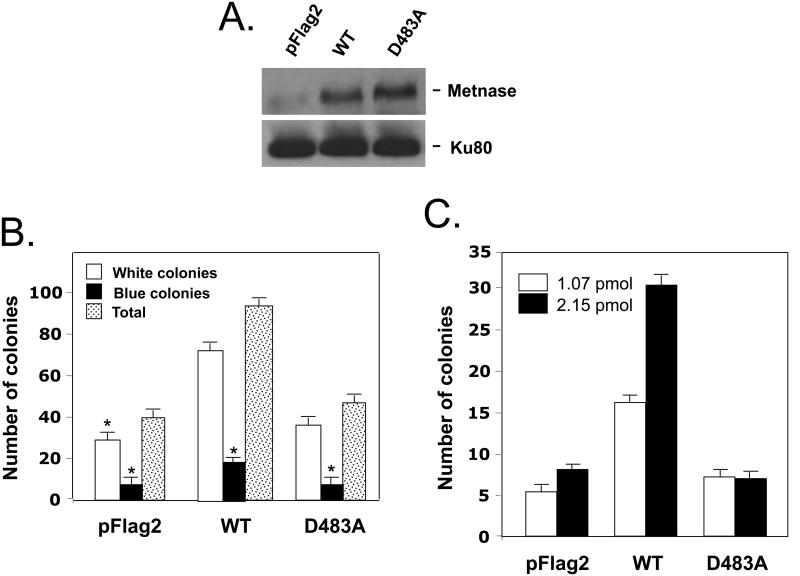
To see in vivo relevance of Metnase's DNA cleavage activity, we next examined cells stably expressing the mutant (D483A) for the joining of linearized plasmid DNA in vivo. DNA end joining activity was measured by rejoining of a plasmid DNA that was linearized within the β-galactosidase gene (39). Overexpression of wt-Metnase increased precise and total end joining by 2.3 and 2.6 fold, respectively, whereas cells expressing the mutant (D483A) showed a very little influence on DNA end joining (Fig. 6B). Cells expressing wt-Metnase or the mutant (D483A) were also examined for genomic integration of exogenous DNA by measuring the assimilation and passage to progeny of a selective marker. A stable expression of wt-Metnase in HEK-293 cells did not affect plating efficiency (data not shown), however, it increased genomic integration of a plasmid DNA by 4-5 fold, while over-expression of the mutant (D483A) had little or no effect on genomic integration (Fig. 6C). Together, our results suggest that DNA cleavage activity of Metnase has a positive role in DNA end joining in vivo.
Figure 6. A Metnase mutant (D483A) defective in DNA cleavage activity fails to stimulate DNA end joining in vivo.
(A) Western blot analysis of HEK-293 cells stably transfected with pFlag vector (pFlag2), over-expressing wt-Metnase (WT), or over-expressing the mutant (D483A) using an anti-Metnase polyclonal antibody (top panel). Expression of Ku80 was included as a loading control (bottom panel). (B) Effect of wt-Metnase and D483A on intramolecular end joining of linearized (Eco RI-digested) pBS. Stable expression of either wt-Metnase or D483A in HEK-293 cells was examined for their imprecise repair (white colonies), precise repair (blue colonies), and total NHEJ repair (blue + white colonies) as described previously (39). Values are averages (±SEM) of 3 distinct determinations. P < 0.01; *P = 0.05. (C) The mutant (D483A) lacking DNA cleavage activity failed to promote integration of foreign DNA into chromosomes. HEK-293 cells stably transfected with pFlag2, pFLAG2-wt-Metnase (WT), or pFLAG2-D483A (D483A) were transfected with 2 ug (1.07 pmol) & 4 ug (2.15 pmol) of pRNA/U6-Hygro, and the number of hygromycin-resistant colonies was a measure of genomic integration as described previously (32). Values are the average (±SEM) of 5 separate experiments (P < 0.01).
Metnase enhances processing of non-compatible ends in NHEJ repair in vitro
To further understand the role for Metnase's DNA cleavage activity in NHEJ, we examined whether Metnase directly influences DNA end processing during NHEJ. For this, linearized plasmid DNA containing different non-compatible ends [Kpn I-Pst I (3′ & 3′), Bam HI-Hind III (5′ & 5′), and Bam HI-Pst I (3′ & 5′)] was incubated with cell extracts containing different levels of Metnase. Following in vitro end joining reactions coupled to E. coli colony formation, 18 colonies were randomly selected and analyzed for DNA end processing by PCR amplification and DNA sequencing analysis. The PCR product of a control plasmid DNA in the absence of cell extracts was 210 nts in size, whereas most, if not all, of the end joining products showed smaller than 210 nts of PCR products (Supplemental data to Table 2). Minimal base-pair loss (0-15 nts) was observed in end joining products with cell extracts underexpressing Metnase (siRNA-Met), whereas end joining products with cell extracts overexpressing Metnase showed base-pair loss of up to 85 nts (Table 2). This result suggests a role for Metnase in DNA end processing, which likely promotes joining of non-compatible ends in a cell-free system.
Table 2.
Effect or Metnase expression on DNA end processing and Joining of linearized plasmid DNA in a cell-free system*
| Metnase expressiona | Type of non-compatible endsb | No. of coloniesc | Deleted bpsd (average ±SD) | 5′ Deletion (average ±SD) | 3′ Deletion (average ±SD) |
|---|---|---|---|---|---|
| Normal | 5′&5′ | 32 ± 5 | 40 ± 18 | 27 ± 12 | 13 ± 9 |
| Low | 5′&5′ | 7 ± 2 | 10 ± 5 | 5 ± 5 | 10 ± 5 |
| High | 5′&5′ | 68 ± 7 | 74 ± 38 | 49 ± 12 | 25 ± 11 |
| Normal | 3′&3′ | 86 ± 6 | 57 ± 18 | 22 ± 7 | 35 ± 15 |
| Low | 3′&3′ | 6 ± 3 | 15 ± 6 | 10 ± 8 | 5 ± 5 |
| High | 3′&3′ | 114 ± 8 | 85 ± 28 | 32 ± 21 | 53 ± 28 |
| Normal | 5′&3′ | 39 ± 4 | 32 ± 11 | 27 ± 7 | 20 ± 10 |
| Low | 5′&3′ | 3 ± 1 | 14 ± 4 | 10 ± 4 | 4 ± 2 |
| High | 5′&3′ | 72 ± 5 | 84 ± 27 | 52 ± 17 | 32 ± 10 |
Experimental details were described in the Materials & Methods section.
Normal, Metnase expression in HEK-293 cells transfected with empty vector (U6); Low. HEK-293 cells stably transfected with U6-siRNA-Metnase; High, HEK-293 cells stably transfected with pFlag-wt-Metnase.
Three linearized DNA with non-compatible ends [3′ & 3′ (Kpn I & Pst I), 5′ & 5′ (Bam HI & Hind III), and 5′ & 3′ (Bam HI & Pst 1)] were used for the experiment.
End joining products were isolated and transformed into E. coli for colony counts.
Eighteen colonies were picked from each group and analyzed for base-pair loss by PCR amplification and DNA sequencing.
Discussion
NHEJ repair involves a direct rejoining of two separated DNA ends and comprises the major DSB repair pathway in mammals. Earlier studies identified Ku70/80 heterodimer, DNA-PKcs, and XRCC4/Ligase4 are required for NHEJ repair in vitro (16, 22, 27, 28, 54-58). However, given that DNA end joining often requires processing of non-compatible ends additional factors are necessary for completion of NHEJ in a cell-free system (22, 28, 29). We previously reported that Metnase forms DSB foci with other repair factors and stimulates DSB repair and genomic integration of foreign DNA in vivo (32, 39, 44, 59). In this study, we showed that Metnase possesses a unique endonuclease activity that plays a positive role in the joining of non-compatible ends in cell-free system.
Upon DNA damage, Metnase is introduced to the DSB damage sites via physical interaction with a binding partner, human Pso4, and colocalizes with other DSB repair factors (32). Although it is not clear what role Metnase plays in DNA end joining, Metnase-mediated stimulation of NHEJ repair in vivo requires both the SET and the transposase domains (39). Metnase directly mediates dimethylation of H3K36 at DSB sites, and enhanced the association of early DNA repair factors such as NBS1 and Ku70 (40), suggesting that Metnase's HLMT activity is involved in NHEJ. Given that thousands of potential Metnase binding (TIR and TIR-like) sites present in human chromosomes (30, 33, 60), a possibility exists that over- or underexpression of Metnase affects DNA end joining simply by influencing the expression of other repair-related genes in vivo. The in vitro end joining experiments presented in this study, however, showed extracts prepared from cells treated with Metnase-siRNA failed to support DNA end joining in vitro, while addition of wt-Metnase fully restored DNA end joining activity (Fig. 5), suggesting that Metnase has a critical role in NHEJ repair.
Cell extracts overexpressing wt-Metnase not only stimulated DNA end joining (Fig. 6) (32, 39, 61, 62), but also enhanced DNA end processing based on DNA sequencing analysis of end joining products (Table 2). In contrast, cell extracts lacking Metnase showed opposite results, indicating that Metnase stimulates DNA end joining via processing of non-compatible ends. DNA end processing facilitates end joining by increasing the chance for partial annealing between two non-compatible ends. Several DSB repair factors such as the Mre11/Rad50/Nbs1 (MRN) complex, Artemis, and the Werner syndrome protein (WRN) have been suggested to be involved in the processing of non-compatible ends (50, 51, 63, 64). The Mre11/Rad50/Nbs1 (MRN) complex and Artemis possess a 3′-5′ exonuclease (and endonuclease) activity and ssDNA-specific 5′-3′ exonuclease, respectively. The Werner syndrome protein (WRN) is a RecQ-like DNA helicase that also possesses 3′-5′ exonuclease activity (65, 66). Furthermore, DNA-PKcs phosphorylates WRN (67), and the Ku complex stimulates WRN's exonuclease activity (68), suggesting that WRN may also participate in DNA end processing. MRN's exonuclease activity is for mismatched DNA ends and pauses at sites of microhomology (69), while its endonuclease is to open fully paired hairpin DNA (64). Artemis possesses an endonuclease activity specific for hairpins and 5′ or 3′ overhangs following phosphorylation by DNA-PKcs (50, 51), suggesting that it plays a role in V(D)J recombination repair and perhaps in removing the 5′ and 3′ overhangs of non-compatible ends during NHEJ repair. Given that Metnase possesses no hairpin or loop opening activity (Fig. 3), it does not play a role in V(D)J recombination.
While Metnase contributes to DNA end joining through an enhanced processing of non-compatible ends, its DNA cleavage activity cannot explain Metnase's stimulatory role in the joining of compatible ends. Similar to DNA-PK- and Ku80-defective cells (Fig. 4C), cell extracts lacking Metnase failed to support joining of compatible ends (data not shown) (39), suggesting that Metnase also has a role in the joining of compatible ends, perhaps by promoting recruitment of the XRCC4-Lig4 complex (59), an essential player in the ligation step through a physical interaction upon DNA damage. The DNA binding property of Metnase may assist in the localization of DNA Ligase IV at the free DNA ends. In this case, Metnase is epistatically above end-processing and subsequent joining, but perhaps below free end recognition and protection, in the NHEJ cascade.
Supplementary Material
Acknowledgments
We would like to thank Dr. M. Oshige for his help in earlier studies.
This work was supported by grants from NIH (CA92111 & CA140422), NIH predoctoral training grants (T32 DK0075-20), the Walther Cancer Institute, and the IU Simon Cancer Center.
Abbreviations used
- ATM
Ataxia Telangiectasia Mutated
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- DSB
DNA double-strand break
- DTT
dithiothreitol
- HLMT
histone lysine methyltransferase
- HRR
homologous recombination repair, HEK293, Human embryonic kidney cells
- NHEJ
non-homologous end joining
- pBS
pBluescript
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SET
Su(var)3-9, Enhancer-of-zeste, Trithorax
- TIR
5′-terminal inverted repeat
Footnotes
Supporting Information Available: Supplemental data to Table 2 is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bennett CB, Lewis AL, Baldwin KK, Resnick MA. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc Natl Acad Sci U S A. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett CB, Westmoreland TJ, Snipe JR, Resnick MA. A double-strand break within a yeast artificial chromosome (YAC) containing human DNA can result in YAC loss, deletion or cell lethality. Mol Cell Biol. 1996;16:4414–4425. doi: 10.1128/mcb.16.8.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 4.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 5.Dudasova Z, Dudas A, Chovanec M. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol Rev. 2004;28:581–601. doi: 10.1016/j.femsre.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 10.Poplawski T, Blasiak J. [Non-homologous DNA end joining] Postepy Biochem. 2005;51:328–338. [PubMed] [Google Scholar]
- 11.Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- 12.Sekiguchi JM, Ferguson DO. DNA double-strand break repair: a relentless hunt uncovers new prey. Cell. 2006;124:260–262. doi: 10.1016/j.cell.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Weterings E, van Gent DC. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair (Amst) 2004;3:1425–1435. doi: 10.1016/j.dnarep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 15.Kuhne C, Tjornhammar ML, Pongor S, Banks L, Simoncsits A. Repair of a minimal DNA double-strand break by NHEJ requires DNA-PKcs and is controlled by the ATM/ATR checkpoint. Nucleic Acids Res. 2003;31:7227–7237. doi: 10.1093/nar/gkg937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 17.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 18.Grawunder U, Zimmer D, Kulesza P, Lieber MR. Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J Biol Chem. 1998;273:24708–24714. doi: 10.1074/jbc.273.38.24708. [DOI] [PubMed] [Google Scholar]
- 19.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 20.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 21.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc Natl Acad Sci U S A. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budman J, Chu G. Processing of DNA for nonhomologous end-joining by cell-free extract. Embo J. 2005:849–860. doi: 10.1038/sj.emboj.7600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic acids research. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pospiech H, Rytkonen AK, Syvaoja JE. The role of DNA polymerase activity in human non-homologous end joining. Nucleic acids research. 2001;29:3277–3288. doi: 10.1093/nar/29.15.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason RM, Thacker J, Fairman MP. The joining of non-complementary DNA double-strand breaks by mammalian extracts. Nucleic acids research. 1996;24:4946–4953. doi: 10.1093/nar/24.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Budman J, Chu G. Assays for nonhomologous end joining in extracts. Methods Enzymol. 2006;408:430–444. doi: 10.1016/S0076-6879(06)08027-X. [DOI] [PubMed] [Google Scholar]
- 27.Budman J, Kim SA, Chu G. Processing of DNA for nonhomologous end-joining is controlled by kinase activity and XRCC4/ligase IV. J Biol Chem. 2007;282:11950–11959. doi: 10.1074/jbc.M610058200. [DOI] [PubMed] [Google Scholar]
- 28.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 29.Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA. Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci U S A. 2003;100:2462–2467. doi: 10.1073/pnas.0437964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson HM, Z KL. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205:203–217. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
- 32.Beck BD, Park SJ, Lee YJ, Roman Y, Hromas RA, Lee SH. Human Pso4 is a metnase (SETMAR)-binding partner that regulates metnase function in DNA repair. J Biol Chem. 2008;283:9023–9030. doi: 10.1074/jbc.M800150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol. 2007;27:1125–1132. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miskey C, Papp B, Mates L, Sinzelle L, Keller H, Izsvak Z, Ivics Z. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol Cell Biol. 2007;27:4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roman Y, Oshige M, Lee YJ, Goodwin K, Georgiadis MM, Hromas RA, Lee SH. Biochemical characterization of a SET and transposase fusion protein, Metnase: its DNA binding and DNA cleavage activity. Biochemistry. 2007;46:11369–11376. doi: 10.1021/bi7005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beck BD, L SS, Hromas RA, Lee SH. Metnase Binding Partner hPso4 negatively Regulates the Metnase’ TIR-Specific DNA Binding Activity. Archive Biophys Biochem. 2010;498:89–94. doi: 10.1016/j.abb.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu TK, Davies DR. Structure and function of HIV-1 integrase. Curr Top Med Chem. 2004;4:965–977. doi: 10.2174/1568026043388547. [DOI] [PubMed] [Google Scholar]
- 38.Polard P, Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 39.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci U S A. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fnu S, Williamson EA, De Haro L, Wray J, Brenneman M, Lee S-H, Nickoloff J, Hromas R. A histone code for non-homologous end joining DNA repair. Proc Natl Acad Sci U S A. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 42.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 44.Williamson EA, Farrington J, Martinez L, Ness S, O'Rourke J, Lee SH, Nickoloff J, Hromas R. Expression levels of the human DNA repair protein metnase influence lentiviral genomic integration. Biochimie. 2008 doi: 10.1016/j.biochi.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daniel R, Greger JG, Katz RA, Taganov KD, Wu X, Kappes JC, Skalka AM. Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. J Virol. 2004;78:8573–8581. doi: 10.1128/JVI.78.16.8573-8581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daniel R, Katz RA, Skalka AM. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 47.Park SJ, Ciccone SL, Freie B, Kurimasa A, Chen DJ, Li GC, Clapp DW, Lee SH. A positive role for the Ku complex in DNA replication following strand break damage in mammals. The Journal of biological chemistry. 2004;279:6046–6055. doi: 10.1074/jbc.M311054200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Dawson A, Finnegan DJ. DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res. 2001;29:3566–3575. doi: 10.1093/nar/29.17.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodwin KD, He H, Imasaki T, Lee SH, Georgiadis MM. Crystal structure of the human Hsmar1-derived transposase domain in the DNA repair enzyme Metnase. Biochemistry. 2010;49:5705–5713. doi: 10.1021/bi100171x. [DOI] [PubMed] [Google Scholar]
- 50.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 51.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 52.Pfeiffer P, Vielmetter W. Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res. 1988;16:907–924. doi: 10.1093/nar/16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SH, Kim CH. DNA-dependent protein kinase complex: a multifunctional protein in DNA repair and damage checkpoint. Mol Cells. 2002;13:159–166. [PubMed] [Google Scholar]
- 56.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 57.Lieber MR. NHEJ and its backup pathways in chromosomal translocations. Nat Struct Mol Biol. 2010;17:393–395. doi: 10.1038/nsmb0410-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lieber MR, Wilson TE. SnapShot: Nonhomologous DNA end joining (NHEJ) Cell. 2010;142:496–496. e491. doi: 10.1016/j.cell.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 59.Hromas R, Wray J, Lee SH, Martinez L, Farrington J, Corwin LK, Ramsey H, Nickoloff JA, Williamson EA. The human set and transposase domain protein Metnase interacts with DNA Ligase IV and enhances the efficiency and accuracy of non-homologous end-joining. DNA repair. 2008 doi: 10.1016/j.dnarep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park JS, Wang M, Park SJ, Lee SH. Zinc finger of replication protein A, a non-DNA binding element, regulates its DNA binding activity through redox. J Biol Chem. 1999;274:29075–29080. doi: 10.1074/jbc.274.41.29075. [DOI] [PubMed] [Google Scholar]
- 61.Hromas R, Wray J, Lee SH, Martinez L, Farrington J, Corwin LK, Ramsey H, Nickoloff JA, Williamson EA. The human set and transposase domain protein Metnase interacts with DNA Ligase IV and enhances the efficiency and accuracy of non-homologous end-joining. DNA Repair (Amst) 2008;7:1927–1937. doi: 10.1016/j.dnarep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williamson EA, Rasila KK, Corwin LK, Wray J, Beck BD, Severns V, Mobarak C, Lee SH, Nickoloff JA, Hromas R. The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation. Nucleic Acids Res. 2008;36:5822–5831. doi: 10.1093/nar/gkn560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 64.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner syndrome protein. II. Characterization of the integral 3′ --> 5′ DNA exonuclease. J Biol Chem. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- 66.Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 67.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 68.Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 69.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci U S A. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.