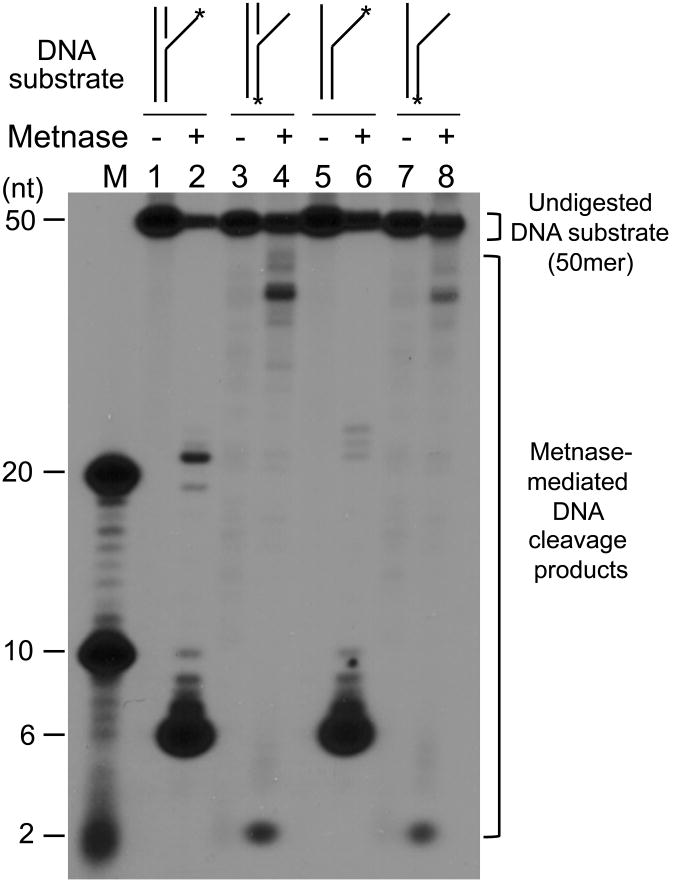
Figure 3. Characterization of Metnase's DNA cleavage activity with different DNA substrates.
(A) Metnase preferentially cleaves ssDNA overhang of flap and pseudo Y DNA. Wt-Metnase (0 or 50 ng) was incubated with 100 fmol of 5′-P32-labeled DNA substrate indicated on the top (see Table 1 for the details of individual substrates) for 60 min prior to 12% denatured PAGE (+ 8M urea) analysis. Lanes “M” represent DNA markers. (B) Metnase exhibits both 3′- and 5′-ssDNA overhang cleavage activity. Metnase (0 or 50 ng) was incubated with 5′-flap DNA (lanes 1-2), 3′-flap DNA (lanes 3-4), 5′-pseudo Y DNA (lanes 5-6), and 3′-pseudo Y DNA (lanes 7-8). M represents DNA markers. (C) Diagrams of the major DNA cleavage sites catalyzed by Metnase. Arrows mark the major cleavage sites from the 32P-labeled (*) 5′-end (calculated from the cleavage products shown in Fig. 3). The length of the arrows approximately was proportional to the intensity of resulting cleavage products.