The aminoacylation of RNA structures generally is considered the starting point for the emergence of the theater of proteins from the RNA world. Once an aminoacyl ester linkage is established with an RNA acceptor, peptide bond formation is thermodynamically favored. In contemporary protein biosynthesis, aminoacylation is directed to the 3′-end of the tRNA structure, which, in turn, brings the activated amino acid to the ribosomes for addition to the growing polypeptide chain. But during the last 9 years a variety of artificial RNA substrates for aminoacylation has been reported (1–8). And in this issue of the Proceedings, the list of examples is expanded in a significant way. Felden and Giegé (9) describe for the first time an aminoacylation system based on a circular RNA substrate. The results obtained with this novel substrate add further support to the idea that the contemporary system of tRNA aminoacylation could have grown out of early aminoacylation systems that used diverse RNA oligonucleotide substrates. Interactions between some of these aminoacylated substrates could have led to a primitive system of peptide synthesis.
All of these substrates for aminoacylation are based, in one way or another, on one of the two domains within the tRNA structure. The amino acid attachment site is at the end of a long helix (the acceptor-TψC minihelix) that terminates at the 3′-end in the universal sequence NCCAOH (Fig. 1, Top). The second domain of the tRNA structure is at right angles (in three dimensions) and consists of a 10-bp helix joined to a hairpin loop that harbors the anticodon triplet of the genetic code. The resulting two-domain L-shaped structure places the amino acid attachment site of the minihelix about 75 Å from the anticodon triplet.
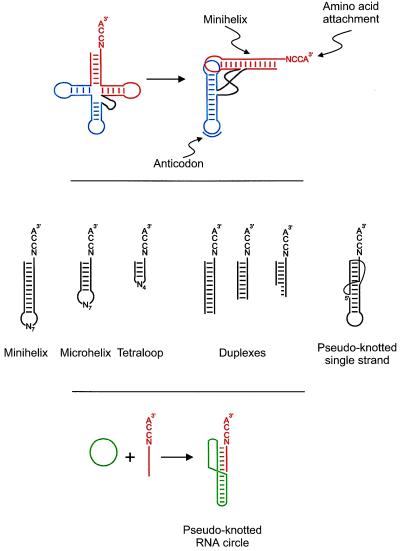
Figure 1.
The diversity of RNA oligonucleotide substrates for aminoacylation. (Top) The tRNA cloverleaf secondary structure (Left) that folds into an L-shape (Right). The minihelix domain is shaded in red, and the second domain containing the template reading head is shown in blue. (Middle) The variety of oligonucleotide substrates for aminoacylation. All substrates terminate at the 3′-end in the single-stranded NCCA tetra nucleotide, where the identity of N depends on the specific aminoacylation system. The four nucleotides (N4) of the loop of the tetraloop structure are specifically chosen to impart stability to the short (4 bp) helix joined to that loop (37). (Bottom) The pseudo-knotted RNA circle described by Felden and Giegé (9).
Significantly, the anticodon itself is not used in every case by the cognate tRNA synthetase to identify the correct tRNA for its amino acid. Two prominent examples are alanyl- and seryl-tRNA synthetases, where the respective enzyme makes no contact whatsoever with the anticodon triplet of the cognate tRNA (10–12). Instead, the minihelix domain contains critical determinants for aminoacylation. These minihelix determinants exist even in those tRNAs where the cognate synthetase makes an important contact with the anticodon triplet (2, 4–6, 13–15). The most rigorous proof of the “information content” in the minihelix domain was the direct demonstration that, for alanyl-tRNA synthetase, the minihelix in isolation was a robust substrate. The critical determinant for aminoacylation of tRNAAla is a G3:U70 base pair in the acceptor helix (16, 17). Transfer of this base pair into other tRNAs conferred alanine acceptance on them. The same properties can be demonstrated for the minihelix—that is, charging depends on the G3:U70 pair and transfer of the pair into other minihelices converted them to alanine acceptors (1). This result showed clearly that the relationship between alanine and the triplets of the genetic code was indirect.
Since this demonstration of minihelix aminoacylation with alanine, a total of 10 different amino acids have been enzymatically charged to minihelix-like substrates. Some of these systems are more robust than others, but in all cases charging is sequence specific. Moreover, nucleotide substitutions in an isolated minihelix that affect charging efficiency also affect charging when placed in the minihelix domain of the corresponding full tRNA. Thus, nucleotide determinants for specific aminoacylation of minihelices have been retained in the contemporary tRNA. Because the minihelix domain is thought to be the more ancient part of the tRNA structure (18–20), aminoacylation of minihelices is imagined to reflect an earlier system of charging. Eventually, the minihelix domain was combined with a second, newer domain that brought in a template reading head containing the anticodon triplets of the genetic code (21, 22).
The contemporary minihelices are not likely identical to the earliest substrates. Rather, other structures—precursors to the minihelix—were probably prominent. The list of laboratory examples of minihelix-like structures and pieces of minihelices has grown steadily, and these examples illustrate the diverse possibilities for early aminoacylation systems that could have developed into the modern minihelix motif. Demonstrated substrates include: RNA duplexes of various lengths; short hairpin oligonucleotides made up of the first 7 bp of the minihelices (designated as microhelices); hairpin duplexes with only 4 bp that are stabilized by a specific tetraloop motif; and a pseudoknot that is created by the hydrogen bonding of nucleotides in a hairpin loop to sequences that are distal to the stem of the hairpin (Fig. 1, Middle). In the latest example, Felden and Giegé (9) design and synthesize a knotted RNA circle. The design is a clever modification of a pseudoknot motif found near the 3′-end of brome mosaic virus RNA (23, 24). The heart of the modification includes the introduction of a single-stranded circle that rearranges into a minihelix-like pseudoknot upon hybridization with a short linear fragment. The hybridized linear fragment is charged specifically with histidine (Fig. 1, Bottom).
The key to success in all of these examples lies in the localization of nucleotide determinants for aminoacylation to the region near the amino acid attachment site. Typically, one or more of the first 4 bp of the helix and specific 2′-hydroxyl groups play an important role in determining charging efficiency and specificity, as does G3:U70 for tRNAAla (7). In addition, all substrates end in the universal NCCAOH, where “N” is the so-called discriminator base. [This nucleotide is usually an important determinant of aminoacylation efficiency. Its specific identity originally was proposed to correlate with the type of amino acid (aliphatic, hydrophilic, aromatic, etc.) that was attached to the associated tRNA (25).] These specific minihelix sequences/structures constitute an operational RNA code for amino acids that is distinct from the genetic code. The idea, therefore, is to present each of these determinants in such a way that the synthetase has unobstructed access to them. In the latest study, the knotted circle was designed so that nucleotides near the amino acid attachment site were directly accessible to histidyl-tRNA synthetase. Specific nucleotide substitutions in the knotted circle were made at locations known to be important for charging with histidyl-tRNA synthetase. These mutations affected the efficiency of aminoacylation in the ways expected, based on results with the full tRNA and with a conventional minihelix structure. So, the lesson in general seems to be that the RNA structural format in which the determinants are presented does not make much difference, as long as those determinants are accessible.
Aminoacylation systems are imagined to have started with ribozymes (26, 27) that gradually gave birth to ribonucleoproteins (like RNase P; refs. 28–30) and then to the contemporary synthetases (31). These early catalysts may have interacted with just the nucleotides at the end of the RNA substrate, as evidenced by the location of contemporary determinants near the amino acid attachment site. Indeed, the primordial, historical tRNA synthetase is thought to be represented by the domain of the contemporary enzyme that contains the catalytic site. This domain has insertions that enable docking of the 3′-end of the tRNA substrate at the active site. A second domain of synthetases, which is believed to be a later addition, interacts with more distal sites in the tRNA structure, including (in many instances) the anticodon.
The diverse artificial substrates studied so far raise the possibility that aminoacylation of RNAs other than tRNAs may be found in contemporary cellular systems. The large plant viral RNAs that are aminoacylated with specific amino acids are a prominent class of novel substrates (3, 23, 32). Here the 3′-end of the viral RNA recreates a tRNA-like structure through the formation of a pseudoknot that recapitulates the minihelix domain. Another novel structure is 10Sa RNA (33, 34), which has a complex secondary structure (35) that is charged with alanine (the 10Sa RNA contains the G3:U70 base pair that is essential for aminoacylation with alanine). This RNA is both an amino acid acceptor and an mRNA (36). We suspect that many more natural examples of novel RNAs that can be aminoacylated will be discovered. These may give the best insights yet into the development of early systems of aminoacylation, especially if they can be related to the tRNA structure in some way. The challenge is to find them.
Footnotes
A commentary on this article begins on page 10431.
References
- 1.Francklyn C, Schimmel P. Nature (London) 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 2.Frugier M, Florentz C, Giegé R. Proc Natl Acad Sci USA. 1992;89:3990–3994. doi: 10.1073/pnas.89.9.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudinger J, Florentz C, Dreher T, Giegé R. Nucleic Acid Res. 1992;20:1865–1870. doi: 10.1093/nar/20.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nureki O, Niimi T, Muto Y, Kanno H, Kohno T, Muramatsu T, Kawai G, Miyazawa T, Giege R, Florentz C, Yokoyama S. In: The Translation Apparatus. Nierhaus K H, Franceschi F, Subramanian A R, Erdmann V A, Wittmann-Liebold B, editors. New York: Plenum; 1993. pp. 59–66. [Google Scholar]
- 5.Frugier M, Florentz C, Giegé R. EMBO J. 1994;13:2218–2226. doi: 10.1002/j.1460-2075.1994.tb06499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamann C S, Hou Y-M. Biochemistry. 1995;34:6527–6532. doi: 10.1021/bi00019a034. [DOI] [PubMed] [Google Scholar]
- 7.Martinis S A, Schimmel P. In: tRNA: Structure, Biosynthesis and Function. Soll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 349–370. [Google Scholar]
- 8.Saks M E, Sampson J R. EMBO J. 1996;15:2843–2849. [PMC free article] [PubMed] [Google Scholar]
- 9.Felden B, Giegé R. Proc Natl Acad Sci USA. 1998;95:10431–10436. doi: 10.1073/pnas.95.18.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park S J, Schimmel P. J Biol Chem. 1988;263:16527–16530. [PubMed] [Google Scholar]
- 11.Cusack S, Berthet-Colominas C, Biou V, Borel F, Fujinaga M, Hartlein M, Krikliviy I, Nassar N, Price S, Tukalo M A, et al. In: The Translation Apparatus. Nierhaus K H, Franceschi F, Subramanian A R, Erdmann V A, Wittmann-Liebold B, editors. New York: Plenum; 1993. pp. 1–12. [Google Scholar]
- 12.Cusack S, Yaremchuk A, Tukalo M. EMBO J. 1996;15:2834–2842. [PMC free article] [PubMed] [Google Scholar]
- 13.Francklyn C, Shi J-P, Schimmel P. Science. 1992;255:1121–1125. doi: 10.1126/science.1546312. [DOI] [PubMed] [Google Scholar]
- 14.Martinis S A, Schimmel P. Proc Natl Acad Sci USA. 1992;89:65–69. doi: 10.1073/pnas.89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felden B, Florentz C, Westhof E, Giegé R. Biochimie (Paris) 1993;75:1143–1157. doi: 10.1016/0300-9084(93)90014-j. [DOI] [PubMed] [Google Scholar]
- 16.Hou Y-M, Schimmel P. Nature (London) 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 17.McClain W H, Foss K. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 18.Weiner A M, Maizels N. Proc Natl Acad Sci USA. 1987;84:7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maizels N, Weiner A M. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 577–602. [Google Scholar]
- 20.Weiner A M, Maizels N. Curr Biol. 1994;4:560–563. doi: 10.1016/s0960-9822(00)00126-3. [DOI] [PubMed] [Google Scholar]
- 21.Schimmel P, Giegé R, Moras D, Yokoyama S. Proc Natl Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noller H F. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, New York: Cold Spring Harbor Lab. Press; 1993. pp. 137–156. [Google Scholar]
- 23.Giegé R, Puglisi J D, Florentz C. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- 24.Felden B, Florentz C, Giegé R, Westhof E. J Mol Biol. 1994;235:508–531. doi: 10.1006/jmbi.1994.1010. [DOI] [PubMed] [Google Scholar]
- 25.Crothers D M, Seno T, Söll D G. Proc Natl Acad Sci USA. 1972;69:3063–3067. doi: 10.1073/pnas.69.10.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piccirilli J A, McConnell T S, Zaug A J, Noller H F, Cech T R. Science. 1992;256:1420–1424. doi: 10.1126/science.1604316. [DOI] [PubMed] [Google Scholar]
- 27.Illangasekare M, Sanchez G, Nickles T, Yarus M. Science. 1995;267:643–647. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]
- 28.Altman S, Baer M F, Bartkiewicz M, Gold H, Guerrier-Takada C, Kirsebom L A, Lumelsky N, Peck K. Gene. 1989;82:63–64. doi: 10.1016/0378-1119(89)90030-9. [DOI] [PubMed] [Google Scholar]
- 29.Brown J W, Pace N R. Biochimie (Paris) 1991;73:689–697. doi: 10.1016/0300-9084(91)90049-7. [DOI] [PubMed] [Google Scholar]
- 30.Altman S, Kirsebom L, Talbot S. FASEB J. 1993;7:7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- 31.Henderson B S, Schimmel P. Bioorg Med Chem. 1997;5:1071–1079. doi: 10.1016/s0968-0896(97)00043-6. [DOI] [PubMed] [Google Scholar]
- 32.Dreher T W, Tsai C-H, Florentz C, Giegé R. Biochemistry. 1992;31:9183–9189. doi: 10.1021/bi00153a010. [DOI] [PubMed] [Google Scholar]
- 33.Oh B K, Apirion D. Mol Gen Genet. 1991;229:52–56. doi: 10.1007/BF00264212. [DOI] [PubMed] [Google Scholar]
- 34.Ushida C, Himeno H, Watanabe T, Muto A. Nucleic Acid Res. 1994;22:3392–3396. doi: 10.1093/nar/22.16.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felden B, Himeno H, Muto A, McCutcheon J P, Atkins J F, Gesteland R F. RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- 36.Keiler K C, Waller P R H, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 37.Shi J-P, Martinis S A, Schimmel P. Biochemistry. 1992;31:4931–4936. doi: 10.1021/bi00136a002. [DOI] [PubMed] [Google Scholar]