Abstract
The development of responsive nanomaterials—nanoscale systems that actively respond to stimuli—is one general goal of nanotechnology. Here we develop nanoparticles that can be controllably triggered to synthesize proteins. The nanoparticles consist of lipid vesicles filled with the cellular machinery responsible for transcription and translation, including amino acids, ribosomes, and DNA caged with a photo-labile protecting group. These particles served as nanofactories capable of producing proteins including green fluorescent protein (GFP) and enzymatically-active luciferase. In vitro and in vivo, protein synthesis was spatially and temporally controllable, and could be initiated by irradiating micron-scale regions on the timescale of milliseconds. The ability to control protein synthesis inside nanomaterials may enable new strategies to facilitate the study of orthogonal proteins in a confined environment and for remotely-activated drug delivery.
Keywords: protein, nanoparticle, molecular nanotechnology, drug delivery
Reconstituting protein synthesis at the nanoscale requires the integration of a large number of cellular components.1–3 Though DNA encodes the complete information required to specify the amino acid sequence of a protein, several cellular machines are required to decode this information in order to generate functional proteins.4, 5 Here, we encapsulated the machinery required to achieve transcription and translation inside of artificial membranes, producing micro- and nano-particles capable of synthesizing functional proteins. These systems were subsequently engineered to induce expression in response to an external stimulus, allowing temporal and spatial control of protein production in vitro and in vivo.
To create particles capable of synthesizing proteins, phospholipid vesicles were formed around a minimal E. coli S30 extract and plasmid DNA template encoding a reporter protein (Fig.1a). The cell extract was used as the source of biochemical machinery, energy, ions, and T7 RNA polymerase.6 cDNA was used to eliminate complexities associated with mRNA splicing.7 To form vesicles, a phospholipid that self-assembles into soft lipid bilayers at physiological temperature was used (1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC); a phosphatidylcholine with two 14-carbon tails). Micro-scale vesicles were formed spontaneously after mixing the extract and DNA with DMPC. The particles were collected by centrifugation, and non-entrapped extract, DNA, and protein were removed by repeated washing.
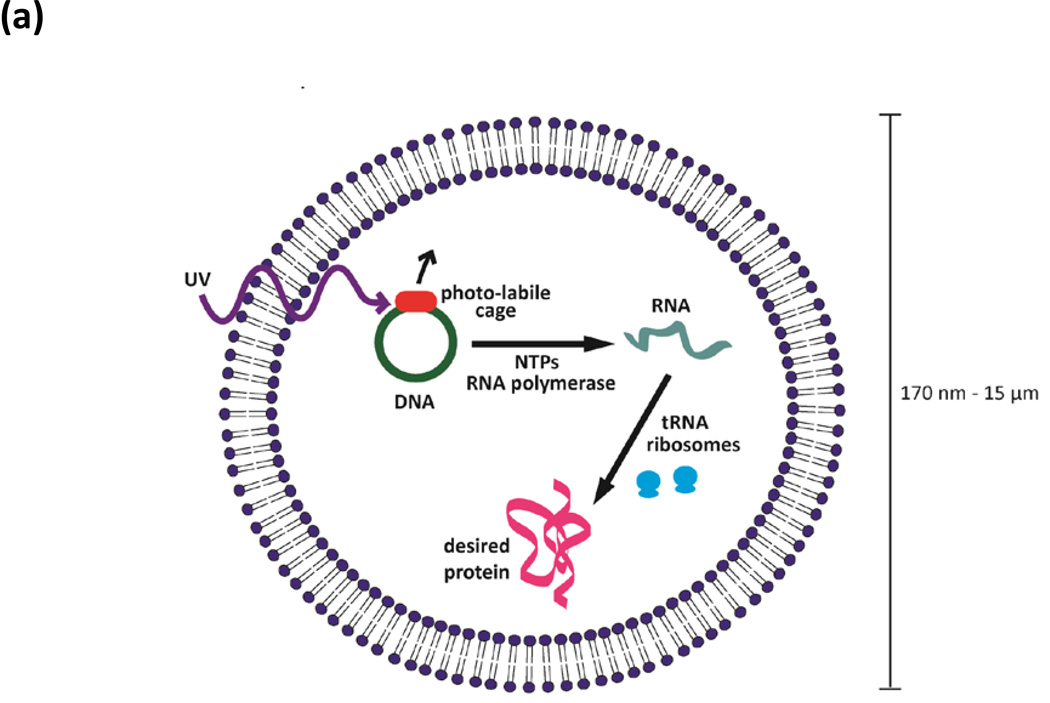
FIGURE 1. Protein producing particles.
(a) A schematic of an encapsulated in vitro transcription/translation nano- and micro-scale particulate system. DNA, tRNA, ribosomes, amino acids, ribonucleotide tri-phosphates (rNTPs) and ions were loaded into lipid vesicles. Protein production can be triggered by irradiating caged DNA with light. (b) Overlaid transmitted light and fluorescence images of a GFP-producing particle. (c) Confocal images of four sections from a single particle. GFP is seen throughout the inside of the vesicle. Bar 5 µm.
The lipid vesicles were capable of producing GFP, imaged using fluorescence microscopy (Fig. 1b). GFP only fluoresces if folded properly,8 confirming that the fluorescent particles contained all components necessary for transcription, translation, and folding. The particles assumed a vesicular structure, with GFP encapsulated in the aqueous core. Confocal microscopy showed that the protein was dispersed throughout the inner core of the vesicles and not bound to the outside of the membrane (Fig. 1c).
To test if the particles were capable of producing proteins that are enzymatically active, a template encoding Renilla luciferase was incorporated into the particles. After the particles were collected and thoroughly washed, they were lysed, and the internal contents were examined by the complementary assays of immunoblotting (Supplementary Fig. S1) and bioluminescence (Fig. 2a). When luciferin was added to the lysate, the solution emitted robust luminescence (1.01 × 107 AU), indicating that the luciferase produced in the particles was enzymatically active.9
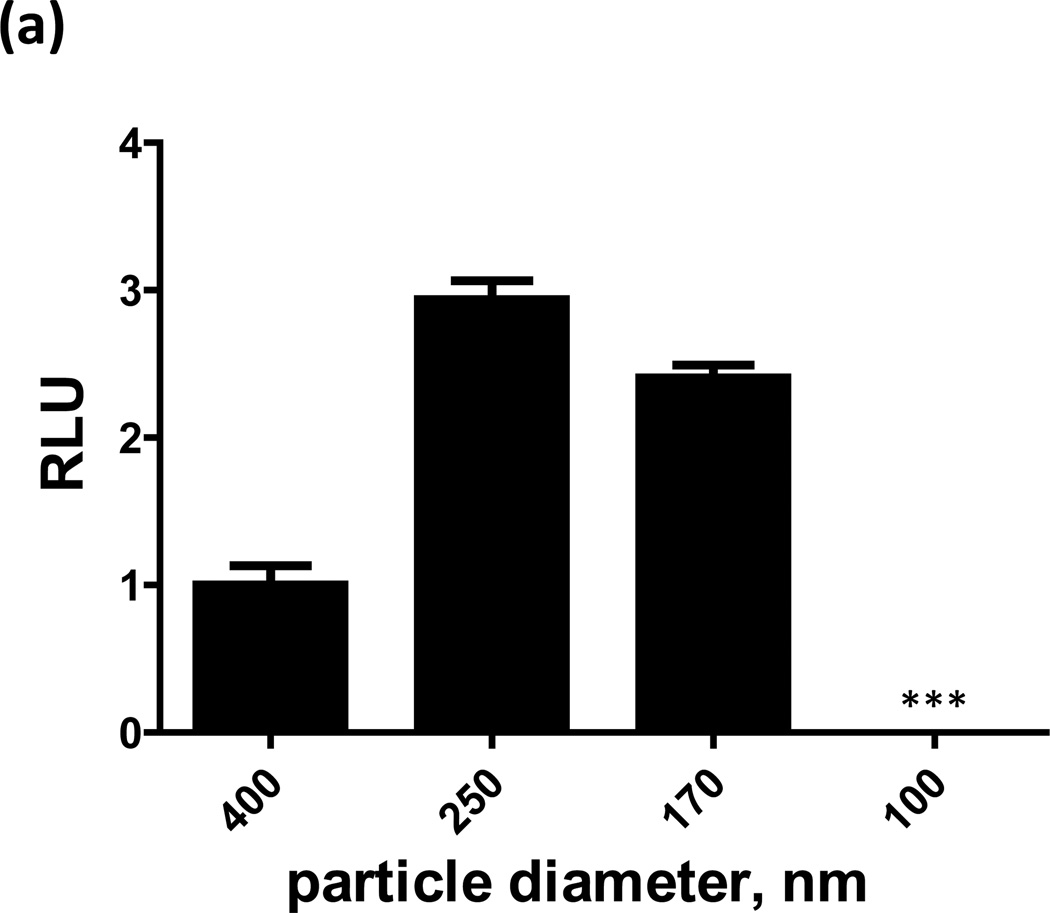
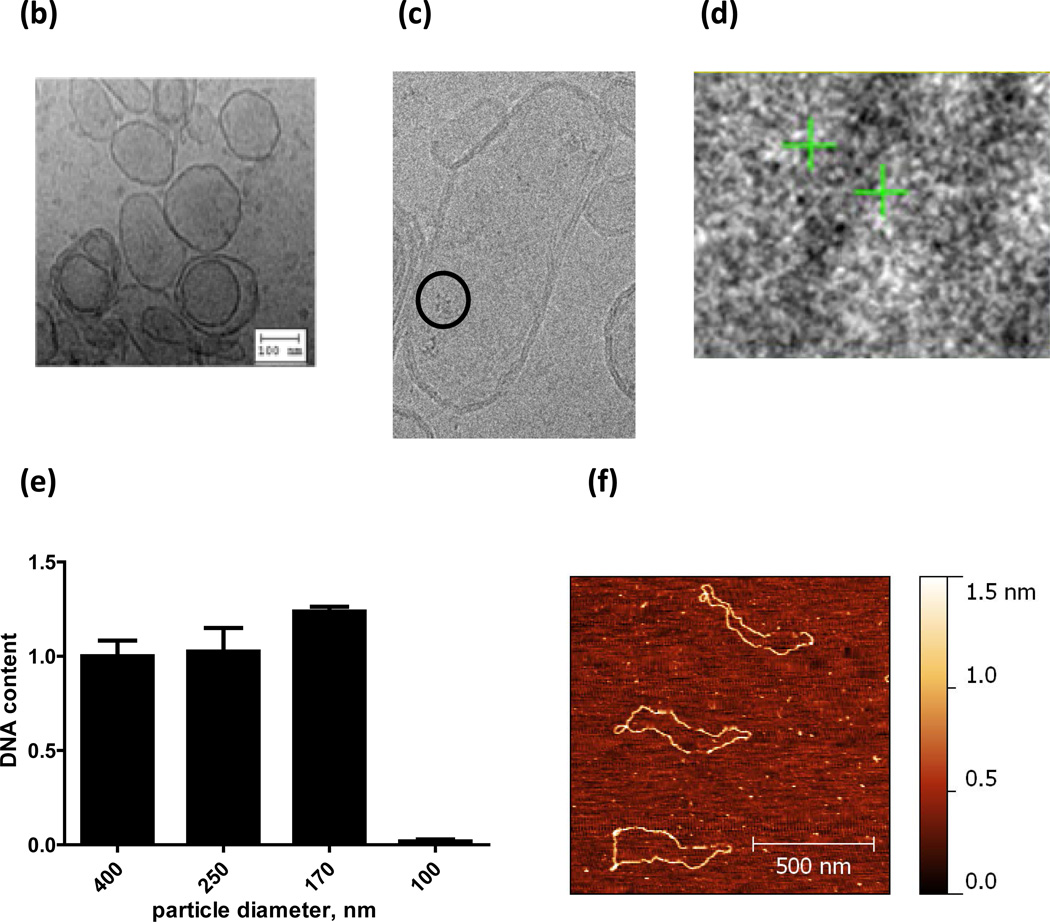
FIGURE 2. Particles are functional at the nano-scale but have a minimum size constraint.
(a) Total luciferase activity is shown for particles extruded to various sizes. No signal is observed for particles with a diameter of 100 nm. (b) A cryoTEM micrograph of 170 nm particles, with structures resembling ribosomes (c), and DNA (d) inside them. (d) The cross-section between the two green arrows is ~2.3 nm, that of DNA. While DNA is detected in particles 170 nm or larger (e) it remains at baseline levels in the 100 nm particles; explaining why the smaller particles are dysfunctional. AFM images (f) of the 4kb, luciferase-encoding, plasmid in solution reveal its unfolded dimensions (~1360 nm long) and that the plasmid must supercoil (g) in order to fit into the nanoparticles. To fit into a 100-nm particle the supercoiled plasmid would need at least 4 writhes in its configuration, while only 2 writhes are needed to fit into a 170-nm particle.
Graph bars are the mean±S.D. of 3 separate experiments with 4 experimental points each.
To test if protein-producing particles have a minimum size constraint, nanoparticles of various sizes were formed by extrusion. Particles with average diameters of ~400, 250, and 170 nm were all capable of producing functional luciferase; however, 100 nm particles were not (Fig. 2a). CryoTEM images (Fig. 2b) suggest that the 170 nm extruded nanoparticles display an elliptical morphology with the ribosomes and code inside. Testing the internal content of the 100 nm particles showed they lack DNA (Fig. 2e), thus explaining why they are dysfunctional. In these studies we used a luciferase-encoding 4 kb (~1360 nm long) plasmid, Fig. 2f. For such a plasmid to fit into a nanoparticle it must supercoil (Fig. 2g). One possibility is that the spontaneous supercoiling to fit the dimensions of the smallest particle (<100 nm) is inefficient10, 11 and may require additional condensing agents, such as cationic lipids.
Interestingly, when comparing the total amount of protein produced, dispersions with smaller particles (170 or 250 nm) produced more protein than dispersions with larger (400 nm) particles, Fig. 2. Comparing the internal content of the different particles showed that while 170 nm particles produced 81±9 proteins/particle, the 400 nm particles, which have a 13-times greater volume, produced only 190±15 proteins/particle. This observation that smaller particles are more efficient in producing proteins has also been suggested in other studies that tested protein production at the nanoscale.12–15 While the exact reason for this phenomenon is unclear, it seems possible that the close proximity of the ‘reacting’ components (e.g., nucleoside 5´-triphosphates (NTPs) and amino acids) to the machinery (e.g., RNA polymerase and ribosomes) plays a role in more efficient use of resources during these processes.
We next sought to develop nanoparticles wherein protein production can be remotely triggered in vitro and in vivo. To control the production of proteins temporally, a photo-labile protecting group was conjugated to the DNA.16 Luciferase-encoding DNA was caged with 1-(4,5-dimethoxy-2-nitrophenyl) diazoethane (DMNPE) to block transcription. UV irradiation at 365 nm uncaged the DNA, and luciferase was produced.
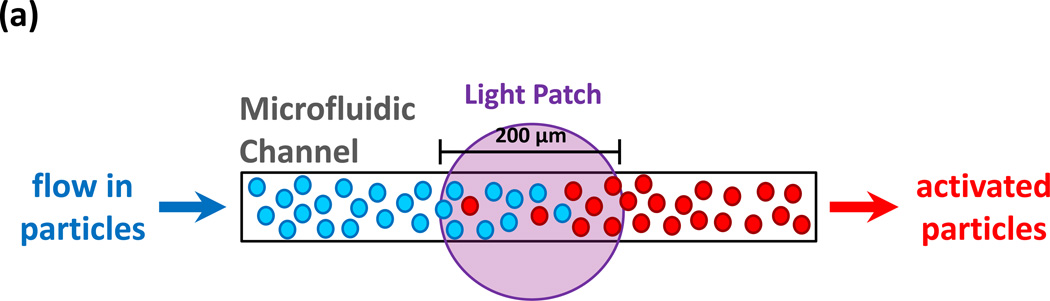
To better understand the timescale of irradiation necessary to activate the particles and to test if they could be activated when flowing at the speed of physiological blood flow,17 particles were irradiated while flowing through a 100 µm-wide microfluidic channel (Fig. 3a) using 300–400 nm light at 120 mW/cm2. Although these wavelengths have tissue penetration of ~10 mm,18 two photon microscopy could be used to deepen penetration significantly in the future.19, 20 Flow velocity was varied, and the rate of luciferase production decreased as velocity increased (Fig. 3b). This was expected, as the amount of time that the particles were in the patch of light decreased as flow velocity increased. When light was irradiated through a microscope objective, the particles were activated in as short as 10 ms at physiological flow velocity (2 cm/s). Notably, luciferase production in the flowing channel could be activated in a pulsatile manner by modulating the light source (Fig. 3c).
FIGURE 3. Protein production can be remotely triggered.
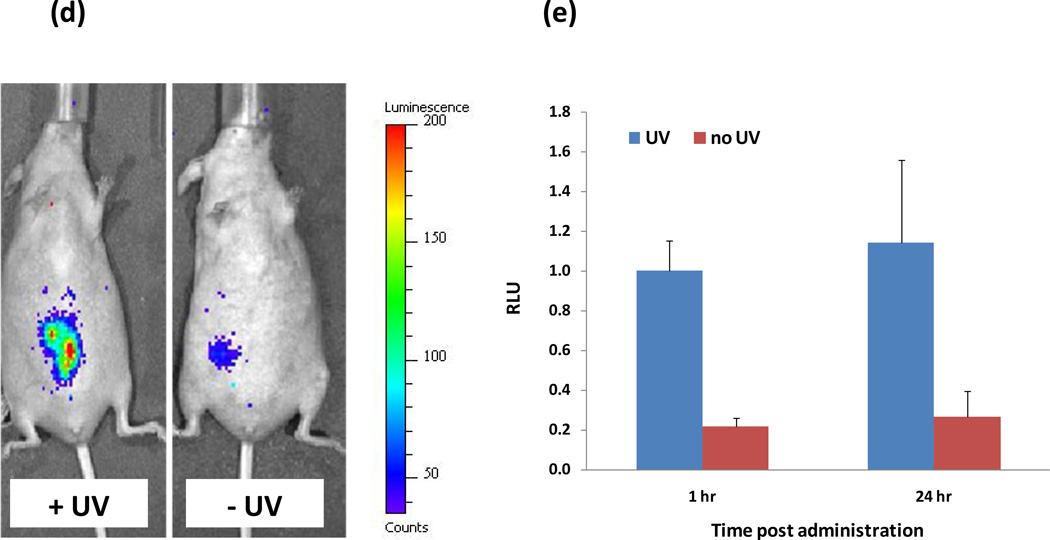
(a) A schematic of the inducible system in microfluidic channels. The formulated DNA is caged by DMNPE, which is released by irradiation with UV light, permitting transcription to occur. Following UV exposure, the particles were incubated for 1 hr at 37 °C to ensure completion of the protein production process. (b) Protein production in 200 nm nanoparticles is inversely correlated with flow velocity in a microfluidic channel (black bars). The quantum efficiency, i.e., the number proteins produced per absorbed photons, is proportional to flow velocity (b, grey bars) (c) The rate of luciferase production in a solution of 200 nm nanoparticles flowing through a microfluidic channel at a flow velocity of 2 cm/s, with and without irradiation. (d–e) Uncaging of DNA by local UV irradiation induces the production of luciferase in vivo. (d) Representative whole body bioluminescence imaging of mice injected locally with particles containing caged luciferase-encoding DNA. After injection the mice were either administered UV (400 mW/cm2) at the site of injection or left untreated. (e), Quantitation of the whole body bioluminescence imaging shown in (d).
To examine whether the particles could be remotely activated to produce luciferase in vivo, particles were injected locally into mice. The animals were either irradiated at the site of the injection or left untreated. Whole body bioluminescence imaging confirmed robust production of luciferase expression, which persisted for at least 24 hours (Fig. 3d,e). While the quantum efficiency in vitro is in the range of 10–40 photons per protein produced (Fig. 3b), in vivo this ratio grows to approx. 6500 photons per protein. This is explained by the attenuation of UV rays in tissue.21, 22
Such an approach, in which autonomous nanoscale production units are located in the body and can be remotely activated to synthesize a potent compound from inert precursors, may find utility in the localized delivery of therapeutics.23–27 To date, this objective has been met for therapeutic applications only with live bacteria that were predesigned to produce proteins in disease sites.28–30 Unlike bacterial systems, artificial systems are modular, and their physical/chemical properties can be modified. Incorporating mechano-synthesis functions in nanomaterials may have applications in both basic biophysics and in medicine.
Supplementary Material
ACKNOWLEDGMENT
We thank Phillip Sharp for his support. AS was supported by the Misrock Foundation. CK was a Johnson & Johnson fellow of the Life Sciences Research Foundation. This work was supported by MIT-Harvard Center for Cancer Nanotechnology Excellence Grant U54 CA151884 from the National Cancer Institute, by a generous gift from the Marie D. & Pierre Casimir-Lambert Fund to Phillip Sharp and partially by the Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. This work was supported by NIH grant # EB000244 to RL and DGA. The cryoTEM imaging was performed by NanoImaging Services, Inc. (La Jolla, CA). Donald Gantz, Boston University, also helped with cryoTEM imaging (Fig. 2b).
Footnotes
The authors declare no competing financial interests.
Contributions
A.S. and M.S.G. and C.K performed experiments. A.S., M.S.G., C.K. D.G.A, and R.L. wrote and revised the manuscript.
REFERENCES
- 1.Jain RK, Stylianopoulos T. Nat Rev Clin Oncol. 2010;7(11):653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szostak JW, Bartel DP, Luisi PL. Nature. 2001;409(6818):387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 3.Hulme SE, Whitesides GM. Angew Chem Int Ed Engl. 2011;50(21):4774–4807. doi: 10.1002/anie.201005461. [DOI] [PubMed] [Google Scholar]
- 4.Bustamante C, Cheng W, Mejia YX. Cell. 2011;144(4):480–497. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Nat Biotechnol. 2001;19(8):751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 6.Nevin DE, Pratt JM. FEBS Lett. 1991;291(2):259–263. doi: 10.1016/0014-5793(91)81297-l. [DOI] [PubMed] [Google Scholar]
- 7.Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, Hood L. Cell. 1980;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- 8.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Biochemistry. 1993;32(5):1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 9.Paulmurugan R, Gambhir SS. Anal Chem. 2003;75(7):1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vologodskii A, Cozzarelli NR. Biophys J. 1996;70(6):2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnier Y, Dorier J, Stasiak A. Nucleic Acids Res. 2008;36(15):4956–4963. doi: 10.1093/nar/gkn467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siuti P, Retterer ST, Choi CK, Fowlkes JD, Doktycz MJ. Annual ORNL Biomedical Science and Engineering Center Conference ORNL Biomedical Science and Engineering Center Conference. 2009;2009:1–4. doi: 10.1109/BSEC.2009.5090477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siuti P, Retterer ST, Doktycz MJ. Lab Chip. 2011;11(20):3523–3529. doi: 10.1039/c1lc20462a. [DOI] [PubMed] [Google Scholar]
- 14.Angenendt P, Nyarsik L, Szaflarski W, Glokler J, Nierhaus KH, Lehrach H, Cahill DJ, Lueking A. Anal Chem. 2004;76(7):1844–1849. doi: 10.1021/ac035114i. [DOI] [PubMed] [Google Scholar]
- 15.Mei Q, Fredrickson CK, Simon A, Khnouf R, Fan ZH. Biotechnol Prog. 2007;23(6):1305–1311. doi: 10.1021/bp070133p. [DOI] [PubMed] [Google Scholar]
- 16.Monroe WT, McQuain MM, Chang MS, Alexander JS, Haselton FR. J Biol Chem. 1999;274(30):20895–20900. doi: 10.1074/jbc.274.30.20895. [DOI] [PubMed] [Google Scholar]
- 17.Drew PJ, Shih AY, Driscoll JD, Knutsen PM, Blinder P, Davalos D, Akassoglou K, Tsai PS, Kleinfeld D. Nat Methods. 2010;7(12):981–984. doi: 10.1038/nmeth.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ran C, Zhang Z, Hooker J, Moore A. Mol Imaging Biol. 2011 doi: 10.1007/s11307-011-0489-z. [DOI] [PubMed] [Google Scholar]
- 19.Collins HA, Khurana M, Moriyama EH, Mariampillai A, Dahlstedt E, Balaz M, Kuimova MK, Drobizhev M, Yang V, Phillips D, Rebane A, Wilson BC, Anderson HL. Nature Photonics. 2008;2:420–424. [Google Scholar]
- 20.Gagey N, Neveu P, Jullien L. Angew Chem Int Ed Engl. 2007;46(14):2467–2469. doi: 10.1002/anie.200604598. [DOI] [PubMed] [Google Scholar]
- 21.Meinhardt M, Krebs R, Anders A, Heinrich U, Tronnier H. J Biomed Opt. 2008;13(4) doi: 10.1117/1.2957970. 044030. [DOI] [PubMed] [Google Scholar]
- 22.Crippa R, Cristofoletti V, Romeo N. Biochim. Biophys. Acta. 1978;538:164–170. doi: 10.1016/0304-4165(78)90260-x. [DOI] [PubMed] [Google Scholar]
- 23.Schrum JP, Zhu TF, Szostak JW. Cold Spring Harb Perspect Biol. 2010;2(9) doi: 10.1101/cshperspect.a002212. a002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A, Heath JR. Nat Med. 2011;17(6):738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang G, Ren H, Rao J. Nat Chem. 2010;2(1):54–60. doi: 10.1038/nchem.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB, Carroll NJ, Jiang X, Dunphy DR, Willman CL, Petsev DN, Evans DG, Parikh AN, Chackerian B, Wharton W, Peabody DS, Brinker CJ. Nat Mater. 2011;10(5):389–397. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noireaux V, Libchaber A. Proc Natl Acad Sci U S A. 2004;101(51):17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes NS. Nat Rev Cancer. 2010;10(11):785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Min JJ, Nguyen VH, Kim HJ, Hong Y, Choy HE. Nat Protoc. 2008;3(4):629–636. doi: 10.1038/nprot.2008.32. [DOI] [PubMed] [Google Scholar]
- 30.Loeffler M, Le'Negrate G, Krajewska M, Reed JC. Proc Natl Acad Sci U S A. 2007;104(31):12879–12883. doi: 10.1073/pnas.0701959104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.