Abstract
In the peripheral nervous system, Schwann cells (SCs) surrounding damaged axons undergo an injury response that is driven by an intricate transcriptional program and is critical for nerve regeneration. To examine whether these injury-induced changes in SCs are also regulated posttranscriptionally by miRNAs, we performed miRNA expression profiling of mouse sciatic nerve distal segment after crush injury. We also characterized the SC injury response in mice containing SCs with disrupted miRNA processing due to loss of Dicer. We identified 87 miRNAs that were expressed in mouse adult peripheral nerve, 48 of which were dynamically regulated after nerve injury. Most of these injury-regulated SC miRNAs were computationally predicted to inhibit drivers of SC dedifferentiation/proliferation and thereby re-enforce the transcriptional program driving SC remyelination. SCs deficient in miRNAs manifested a delay in the transition between the distinct differentiation states required to support peripheral nerve regeneration. Among the miRNAs expressed in adult mouse SCs, miR-34a and miR-140 were identified as functional regulators of SC dedifferentiation/proliferation and remyelination, respectively. We found that miR-34a interacted with positive regulators of dedifferentiation and proliferation such as Notch1 and Ccnd1 to control cell cycle dynamics in SCs. miR-140 targeted the transcription factor Egr2, a master regulator of myelination, and modulated myelination in DRG/SC cocultures. Together, these results demonstrate that SC miRNAs are important modulators of the SC regenerative response after nerve damage.
Introduction
Neuronal regeneration in adult vertebrates is largely limited to the peripheral nervous system (PNS). Unlike neurons in the CNS, PNS neurons are capable of long-distance axonal regeneration. Central to the differential plasticity of peripheral neurons are Schwann cells (SCs), the glial cells of the PNS. SCs have the striking ability to dedifferentiate and revert back to an immature-like state following axonal loss (Jessen and Mirsky, 2008). After a transient phase of proliferation, dedifferentiated SCs help support axonal regeneration by clearing myelin debris, forming tubular structures known as bands of Bungner, and upregulating the synthesis of a number of neurotrophic factors. As peripheral axons regenerate, SCs redifferentiate to form mature Remak bundles or new myelin sheaths, helping to restore, in this way, peripheral nerve function (Geuna et al., 2009). Greater insight into this highly orchestrated process of SC-mediated support of axonal regeneration could help improve the treatment of peripheral and central neurodegenerative diseases characterized by axonopathy, including diabetic neuropathy and amyotrophic lateral sclerosis (Coleman, 2005).
MicroRNAs (miRNAs) are small (∼22 nt) noncoding RNAs capable of posttranscriptionally regulating protein expression. miRNAs have emerged as ubiquitous regulators of developmental timing and cellular differentiation (Flynt and Lai, 2008). The central role of miRNAs in cell fate determination suggests that they could modulate the transcriptional program responsible for the dedifferentiation and redifferentiation of SCs following nerve injury (Jessen and Mirsky, 2008). This idea is supported by zebrafish studies implicating miRNAs in the regulation of tissue regeneration (Thatcher et al., 2008; Yin et al., 2008; Ramachandran et al., 2010). Moreover, miRNAs are essential for the normal development of SCs, as miRNA-deficient SCs are unable to differentiate past an immature state (Bremer et al., 2010; Pereira et al., 2010; Yun et al., 2010). The behavior, identity, and potential role of miRNAs in the regenerative response of adult SCs after nerve injury, however, remain unexplored.
In this study, we examined the dynamics of miRNA expression in mouse sciatic nerve following crush-induced axonal loss. We identified 87 miRNAs consistently expressed in adult peripheral nerve. Most of these SC miRNAs were computationally predicted to inhibit drivers of SC dedifferentiation/proliferation and their expression was dynamically regulated after injury to potentially re-enforce the transcriptional program driving the SC regenerative response. Accordingly, miRNA-deficient SCs had delayed transitions between the distinct differentiation states involved in the SC injury response. Among the miRNAs consistently expressed in SCs, miR-34a and miR-140 were characterized as functional regulators of two distinct processes performed by SCs following peripheral nerve damage: proliferation and myelination. Together, our results demonstrate that miRNAs are important modulators of the SC regenerative response after nerve damage.
Materials and Methods
Animals and matings.
All animal experiments were performed in compliance with institutional animal protocols. For all nerve-lesion studies, except those involving mice lacking Dicer specifically in SCs (Dicer-SCKOs), C57BL6 mice of either gender were used. Dicer-SCKO mice were generated by crossing DicerloxP/loxP mice (Cobb et al., 2005) to PLP-CreERT+/− mice (Doerflinger et al., 2003). Compound heterozygotes (PLP-CreERT+/−, Dicer+/loxP) were then backcrossed to homozygous DicerloxP/loxP to generate PLP-CreERT+/−, DicerloxP/loxP mice. One-month-old PLP-CreERT+/−, DicerloxP/loxP mice of either gender were injected daily with 100 μg/g tamoxifen (10 mg/ml stock dissolved in a 10:1 peanut oil/ethanol mixture) for a total of 10 d (with a 2 d break after day 5) to generate Dicer-SCKOs. Control (Ctrl) mice were injected in the same way with vehicle alone (10:1 peanut oil/ethanol mixture). PLP-CreERT+/− were also crossed to Rosa26-YFP+/− mice to generate PLP-CreERT+/−, Rosa26-YFP+/− mice, which were treated with tamoxifen in an analogous manner.
Surgical procedures.
For lesion studies, adult mice (2–3 months of age) were anesthetized by intraperitoneal injection of Avertin. The sciatic nerve was exposed at the hip and either crushed using No. 5 jeweler's forceps for 30 s or transected using small scissors. In the case of transection, the distal stump was deflected onto an adjacent muscle to prevent regeneration. When necessary, the site of injury was marked with a single 10-0 nylon epineural suture. Nerve lesions were produced on the right side and the intact contralateral nerve served as the uninjured control. At the appropriate time points, nerves were harvested and processed for histology or immediately frozen in liquid nitrogen for subsequent extraction of RNA.
RNA preparation and quantitative real-time PCR.
Total RNA was isolated after homogenization (for sciatic nerves) or lysis (for cultured SCs) in Qiazol lysis reagent (Qiagen) using a miRNeasy Minikit (Qiagen) according to the manufacturer's protocol. RNA concentration was quantified using an ND-1000 spectrophotometer (Nanodrop Technologies). mRNA was reverse transcribed from 100 ng of total RNA using qScript cDNA SuperMix (Quanta Biosciences) while miRNA was reverse transcribed from 100 ng of total RNA using NCode Vilo miRNA cDNA synthesis kit (Invitrogen). mRNA qRT-PCR was performed using a SYBR green-based detection system on a 7900 HT Sequence Detector instrument (Applied Biosystems), as described previously (Nagarajan et al., 2001). GAPDH expression was used to normalize samples and obtain relative expression values that were used to calculate percentage changes. miRNA qRT-PCR was performed in an analogous manner, but cycling conditions were adapted as per NCode Vilo miRNA qRT-PCR kit (Invitrogen) recommendations. U6 snRNA was used for normalization. The sequences of the qRT-PCR primers used are as follows (5′-3′): Dicer: forward, AATTGGCTTCCTCCTGGTTAT; reverse, GTCAGGTCCTCCTCCTCCTC; mir-34a: forward, GGCAGTGTCTTAGCTGG TTGT; mir-26a: forward, GATTTCAAGTAATCCAGGATAGGCT; mir-140: CAGTGGTTTTACCC TATGGTAG; all miRs reverse, NCode Vilo Universal Primer; Egr-2: forward, CATGGGCAAATTCTCCATTGA; reverse, TTGCAAGATGCCCGCAC; MPZ: forward, CCCTGGCCATTGTGGTTTAC; reverse, CCATTCACTGGACCAGAAGGAG. MBP: forward, CCAGTAGTCCATTTCTTCAAGAACATT; reverse, AGCTAAATCTGCTGAGGGACAGG. Ccnd1: forward, GCGTACCCTGACACCAATCT; reverse, GAACCGGTCCAGGTAGTT CA; Notch1: forward, TGACCTGCTCACTCTCACAGA GTAC; reverse, CGACAGATGTATGAAGACT CAAAGG.
Microarray and computational analysis.
SC microarray gene expression profiling experiments were performed in previous studies using Affymetrix MU74v2 chips (Nagarajan et al., 2002). Expression data were processed and normalized using Affymetrix MAS5 algorithm. An SC-expressed gene was defined as one that was called present in at least one time point during SC regeneration. k-Means clustering was used to cluster genes based on their expression profiles; gene clusters that had an expression pattern similar to the previously identified myelination or proliferation profile (Nagarajan et al., 2002) based on the Pearson correlation coefficient were identified and merged. The average expression profile, i.e., the centroid, was calculated as the final myelination or proliferation profile.
For miRNA expression profiling, total RNA samples were prepared by isolating and pooling RNA from at least three different mice at the appropriate times after nerve crush. Replicates were prepared entirely independently from two separate pools of at least three animals each. Two replicates were used for hybridization onto qNPA miRNA microarrays (HTG Molecular) according to the manufacturer's protocol, and microarray images were obtained and background subtracted as per the manufacturer's recommendations. Scanned miRNA microarray expression data were first processed using the software provided by the manufacturer. This microarray platform includes duplicate probe sets for 1046 miRNAs. miRNA expression data were further filtered using the following criteria: (1) the average of the expression of control miRNA probe sets on the array was used as a cutoff to remove miRNAs that have low or no expression, and (2) miRNAs for which the expression of one duplicate probe set at all time points were significantly higher than that of the other duplicate probe set were also removed for further analysis, based on a Mann–Whitney U test. Expression data were then z-score normalized for each miRNA and the average expression of the two duplicate probe sets was calculated at each time point. These miRNA expression profiles were compared with the previously identified myelination or proliferation profiles in the following target bias analysis.
TargetScanS, PicTar, and miRanda algorithms were used to predict potential miRNA regulatory targets. miRNA targets predicted by these programs were downloaded from the program websites. miRNA target bias analysis was performed as follows: for each miRNA, SC-expressed genes that had an miRNA seed sequence match in the 3′UTR were identified using TargetScanS. The Pearson's correlation coefficient of gene and miRNA expression profiles was calculated for each target and nontarget. The Mann–Whitney test p value was calculated to test whether miRNA target genes tend to have higher expression correlations with miRNA than nontarget genes, i.e., a positive target bias.
Immunohistochemistry.
The following primary and secondary antibodies were used: chicken anti-GFP (1:1000; Aves), rabbit anti-βIII tubulin (1:1000; Covance), rabbit anti-phosphohistone 3 (1:200; Millipore), rat anti-MBP (1:100; Millipore), anti-rabbit Cy3 (1:500; Jackson ImmunoResearch Laboratories), anti-rat Cy3 (1:500; Jackson ImmunoResearch Laboratories), and anti-chicken Alexa 488 (1:500; Jackson ImmunoResearch Laboratories).
For immunohistochemical analysis of sciatic nerves, nerves were dissected, immersion fixed in 4% paraformaldehyde for 2 h, rinsed with PBS, and cryoprotected in 30% sucrose. Sciatic nerves were then embedded in Tissue-Tek OCT compound (Sakura Finetek) and sectioned at 6 μm. All frozen sections were immunostained by postfixing in ice-cold acetone at −20°C for 10 min and blocking in 5% fish skin gelatin in PBS 0.2% Triton for 1 h at room temperature. Sections were then incubated with primary antibody diluted in blocking buffer overnight at 4°C. Secondary antibody incubation was performed at room temperature for 1 h also in blocking buffer. After all stainings, sections were mounted with Vectashield mounting medium with DAPI (Vector Laboratories) for microscopic visualization. Images were captured using an upright microscope equipped for epifluorescence microscopy (Nikon 80i, and CoolSnapES camera; Photometrics) and were processed using MetaMorph, ImageJ, and Gimp software using global adjustments for brightness and contrast. All cell number quantifications were performed by counting the cells of interest in four randomly selected regions in longitudinal nerve sections from each animal assessed (at least three mice per genotype at each time point) at a 20× magnification. The area covered by MBP immunostaining was determined using ImageJ after automated background adjustment and binarization (Sasaki et al., 2009) from four randomly selected fields at a 20× magnification in longitudinal nerve sections. MBP area was normalized to total nerve area; results represent the average of at least three mice per genotype.
Nerve light and electron microscopy and morphometry.
Sciatic nerve segments from Dicer-SCKO or Ctrl mice were dissected at the appropriate times after injury and placed in 3% glutaraldehyde overnight. The isolated nerve segments were ∼15 mm in length and, in the case of crush-injured nerves, included the distal regenerated stump. After washing with phosphate buffer, nerves were postfixed in 1% osmium tetraoxide in phosphate buffer overnight at 4°C. Specimens were then dehydrated in graded alcohols and embedded in 100% epoxy (Araldite 502). One-micrometer-thick plastic embedded sections were prepared and stained with toluidene blue for light microscopy. For electron microscopy, thin sections were prepared, stained with uranyl acetate and lead citrate, and photographed with a JEOL 1200 electron microscope. All nerves underwent qualitative assessment of neural architecture followed by detailed histomorphometric analysis performed in a blinded fashion, as previously described (Hunter et al., 2007), using at least three mice per genotype at each time point. For injured nerves, histomorphometric analysis was performed 5 mm distal to the crush site. Total regenerated axon counts were determined by counting both remyelinated and non-remyelinated axons in Ctrl and Dicer-SCKO nerves 14 d after injury from electron micrographs taken at 5000× magnification.
Plasmids.
miRNAs were obtained by PCR amplifying individual miRNA precursors from genomic DNA. The resulting fragments were cloned between the BamHI and NheI sites in the miRNASelect pEP-MIR Cloning and Expression Vector (Cell Biolabs) using the InFusion HD cloning system (Clonetech) according to the manufacturer's recommendations. Pre-miRNAs included the miRNA stem loop and ∼100 nt of flanking sequence on either side and were amplified using the following primers: pre-mir-34a: forward, TAGTTCTCGAGGATCCCCAGCTGAATCCCGACTA GT; reverse, AAAAGCTCGAGCTAGCTCTCTTAGCCAGAAGTGCTC; pre-mir-140: forward, TAGTTC TCGAGGATCCGCCTCTGACCCTGCTTGCTGGTGGTGTA; reverse, AAAAGCTCGAGCTAGC CCAACACCCACCCAATAGACGCCT. pEP-mir Null Vector (Cell Biolabs) was used as Ctrl.
A lentiviral miRNA expression vector was made by PCR amplifying the pEP-MIR expression cassette (promoter, miR processing portion, and mCherry reporter: forward, AGATCCAGTTTGGTTAATTAAGTGAGG CTCCGGTGCCCGTCA; reverse, TAGAG TCGCGGCCGCTTTACTTGTACAGCTCGTCCATGCCGCCGGTGGA) and subcloning it between the PacI and BsrgI sites of an FCIV lentiviral vector (Araki et al., 2004) using the InFusion HD cloning system (Clonetech) according to the manufacturer's recommendations. For luciferase assays, the 3′UTR regions of Egr2, Notch1, and Ccnd1 were PCR amplified from genomic DNA using the following primers: Ccnd1 3′UTR: forward, AAAGCTGCGCACTAGTGGG CCACCGGGCAGGCGGGAGCCA; reverse, ATCCTTTATTAAGCTTTGAGATTTTACCAATTT TATTT; Notch1 3′UTR: forward, AAAGCTGCGCACTAGTACAGAGATGTGGGATGCAGGACC; reverse, ATCCTTTATTAAGCTTGTTTAGCAAAATGTGGACAACCA; Egr2 3′UTR: forward, AAAGCT GCGCACTAGTGATGAAGCTCTGGCTGACACACCA; reverse, ATCCTTTATTAAGCTTACCA TAGTCAATAAGCCATCCAT. DNA fragments were cloned downstream of the luciferase gene between the HindIII and SpeI sites in the pMIR-REPORT miRNA Expression Reporter Vector (Ambion). The 3′UTR of Egr2 lacking the miR-140 landing pad was cloned in an analogous manner using the following sets of overlapping primers: F1, AAAGCTGCGCACTAGTGATGAAGCTCTGGCTGACACACCA R1, ACAGCCATACTTA AACCCAAGTCTCGAGCTCGCGGCGCCTCTAGAAAGGCATCCTGTACACATGCA; F2, TGCATGTGTACAGGATGCCTTTCTAGAGGCGCCGCGAGCTCGAGACTTGGGTTTAAGTATGGCTGT; R2, AAAGATCCTTTATTAAGCTTACCATAGTCAATAAGCCATCCATTAT CTGAACTCCAGTTTCAAAGCTTGGCGCGCCAGAATAGATTGTTTCTCTCT. pRL-CMV Renilla Luciferase Reporter Vector (Promega) was used as a transfection control.
Lentivirus production.
Lentiviruses expressing miR-34a, miR-140, or a Ctrl miRNA were produced in HEK293T cells as previously described (Sasaki et al., 2006).
Luciferase assays.
HEK293T cells were seeded at a density of 50,000 cells/well in 24-well plates in DMEM media (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 2 mm l-glutamine. Cell were transfected 24 h later, with either a pEP-MIR vector expressing a pre-miRNA or with the pEP-mir null control and pMIR-REPORT luciferase reporter vector containing the appropriate 3′UTR linked to luciferase. pRL-CMV renilla luciferase reporter vector (Promega) was used as a transfection control. A total of 200 ng of plasmid DNA per well were transfected at a ratio of 50:1:0.5 (miRNA:luciferase reporter:transfection Ctrl). Cells were harvested 48 h posttransfection and assayed using a dual-luciferase reporter assay system (Promega) according to the manufacturer's protocol.
Primary SC cultures and lentiviral infection.
Primary rat SCs were cultured from postnatal days 1–3 rat pups sciatic nerves as previously described (Nagarajan et al., 2001). Established SC cultures were then transduced at an early passage (<passage 3) with a lentiviral vector expressing miR-34a, miR-140, or a Ctrl miRNA. Efficiency of infection was confirmed by visualization of expression of mCherry fluorescent reporter in the pEP-MIR cassette in almost 100% cells.
SC in vitro proliferation, differentiation, or myelination assays.
For SC in vitro differentiation assays, Ctrl or miR-140 overexpressing SCs were initially seeded onto collagen-coated 24-well plates (∼75,000 cells/well) in 10% FBS-DMEM media; 24 h later, cells were switched to 1% FBS-DMEM media for 2 d. SCs were then induced to differentiate by adding 1% FBS-DMEM media containing 250 μm 8-(4-chlorophenylthio)adenosine-3′,5′ cyclic monophosphate (cpt-cAMP; Biolog). RNA was isolated 72 h later. Reported results are from duplicate wells from two independent assays. For SC proliferation and in vitro myelination assays, dorsal root ganglion neurons were dissected from E14 rat embryos, dissociated, and seeded onto Matrigel-coated (BD Biosciences) four-well plates in Neurobasal media (Invitrogen) containing 2% B27 (Invitrogen) and 50 ng/ml NGF. Glia were eliminated by addition of 20 μm 5-fluoro-2-deoxyuridine (Sigma) and 20 μm uridine (Sigma) to the media for 4 d. Seven days after seeding, DRGs were switched to F-12/DMEM media supplemented with 15% FBS, 2 mm l-glutamine, and 50 ng/ml NGF, and Ctrl or miR-overexpressing SCs were added to each well. To examine SC proliferation, 25,000 Ctrl or miR-34a-overexpressing SCs per well were plated onto DRG neurons and growth curves were generated by monitoring changes in cell number over 5 d. mCherry-positive cells expressing miR-34a were visualized at 20× magnification and quantified from five randomly selected fields. Results are averages from triplicate wells from two independent assays. For in vitro myelination assays, 50,000 Ctrl of miR-140-overexpressing SCs per well were plated onto DRG neurons and allowed to proliferate and ensheath axons for 7 d. At this point, myelination was induced by adding 50 μg/ml ascorbic acid. Myelination was allowed to proceed for 14 d and myelinated segments were visualized through MBP immunostaining at 20× magnification as previously described (Ryu et al., 2008). The number of myelinated segments was quantified by averaging five different fields from triplicate wells in three independent assays.
Western blotting.
Ctrl and miRNA-overexpressing SCs were lysed in 200 μl of buffer containing 150 mm sodium chloride, 50 mm HEPES, 1% NP-40, 1 mm EDTA, 1 mm sodium fluoride, 1 mm sodium orthovanodate, and complete protease inhibitor mixture (Roche Applied Science). Lysates were clarified by centrifugation at 14,000 rpm for 10 min and quantified using the MicroBCAProtein assay kit (Pierce). For Western blotting, the proteins were separated by SDS-PAGE and transferred to a PVDF membrane (Millipore). Membranes were blocked in 5% milk in 0.1% TBS-Tween and incubated overnight with the appropriate primary antibody. Following incubation with secondary antibodies conjugated to HRP (GE Healthcare), membranes were developed with SuperSignal West Dura substrate (Pierce). The primary antibodies used were mouse anti-Ccnd1 (1:1000; Cell Signaling Technology), anti-Notch1 (1:1000; Cell Signaling Technology), and rabbit anti-Egr2 (1:1000; Covance). The secondary antibodies used were anti-mouse and anti-rabbit HRP (1:5000; Jackson ImmunoResearch Laboratories).
Statistical analysis.
All values are expressed as mean ± SEM. If not stated otherwise, p values were determined by unpaired, two-tailed Student's t test. All statistical analyses were performed using Microsoft Excel 2007.
Results
Dynamics of SC miRNA expression after injury are consistent with a modulatory role in the SC regenerative response
To study the potential regulatory role of miRNAs in the SC regenerative response after peripheral nerve injury, we performed miRNA microarray expression profiling of mouse sciatic nerves after crush-induced axonal degeneration. In previous work examining mRNA expression profiles during the SC injury response (Araki et al., 2001; Nagarajan et al., 2002; Bosse et al., 2006), we and others found that expression of regulators of SC dedifferentiation/proliferation and myelination peaks 4 (C4) and 14 (C14) d after nerve crush, respectively. In the present study, we therefore profiled miRNA expression in the distal stump of injured nerves at C4 and C14, as miRNAs that are differentially expressed at these two time points are likely to be involved in the regulation of the SC regenerative response.
We identified 87 miRNAs that were reproducibly detected in the adult peripheral nerve (Fig. 1). Given that SCs constitute >80% of the cells in mouse sciatic nerves (King, 2005), the detected miRNAs primarily represent the microRNAome of these glia. Indeed, over one-third of the miRNAs detected by our microarray analysis were previously found to be expressed in cultured isolated SCs (Verrier et al., 2009). Moreover, several of the most highly expressed miRNAs identified, including let-7f, miR-138, miR-145, and miR-16, have been previously related to the postnatal development of SCs (Bremer et al., 2010; Verrier et al., 2010; Yun et al., 2010). Remarkably, the expression of over half of the detected miRNAs was upregulated or downregulated in the distal nerve segment following crush injury (48 of 87; C4 vs Ctrl, p < 0.05), suggesting that miRNAs may indeed play a role in modulating the processes of SC dedifferentiation and remyelination after injury.
Figure 1.
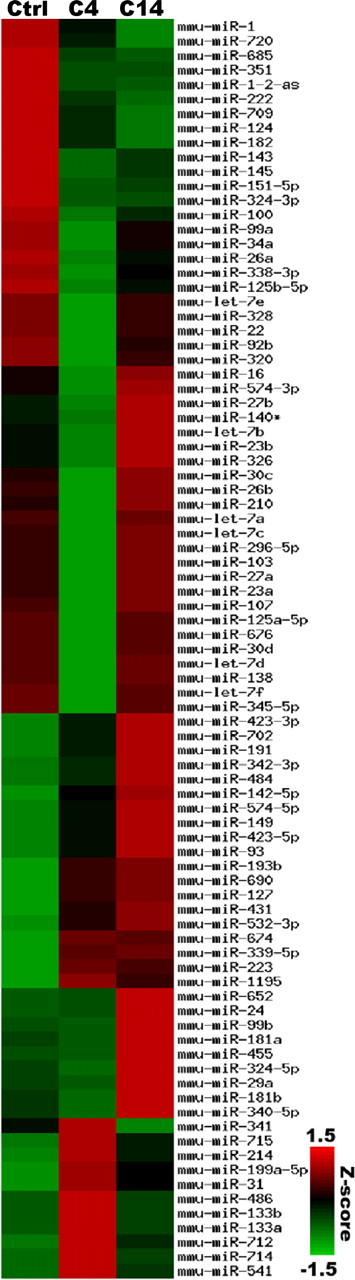
Expression profiles of 87 miRNAs expressed in SC. z-Score-normalized miRNA expression levels 4 d (C4) and 14 d (C14) after nerve crush injury as well as in uninjured nerves (Ctrl) are shown in a heat map. Red indicates higher expression than the average across all samples; green indicates lower expression. mmu refers to the standard abbreviation used to designate miRNAs of mouse origin.
Genes involved in the dedifferentiation/proliferation and remyelination of SCs following crush-induced axonal degeneration are dynamically regulated and have reciprocal patterns of expression (Nagarajan et al., 2002). We reasoned that miRNAs involved in the SC injury response would be dynamically regulated in a manner similar to genes known to be important for the regulation of this process. We applied k-means clustering to our previous SC mRNA expression profiling datasets (see Materials and Methods, above) and identified gene clusters containing known regulators of SC dedifferentiation/proliferation (proliferation profile) or differentiation/myelination (myelination profile) after injury (Fig. 2a) (Nagarajan et al., 2002). Of the 48 miRNAs differentially regulated following nerve crush injury, 31 were expressed in a pattern that was highly correlated with either the myelination or proliferation profile (with a Pearson's correlation coefficient >0.8). Interestingly, 21 of these miRNAs were regulated similarly to genes in the myelination cluster: their expression was inhibited immediately after injury and increased as remyelination ensued (Fig. 2a). The expression of a significant portion of SC miRNAs, therefore, is dynamically regulated after crush injury in a manner consistent with them playing a critical role in modulating the SC regenerative response.
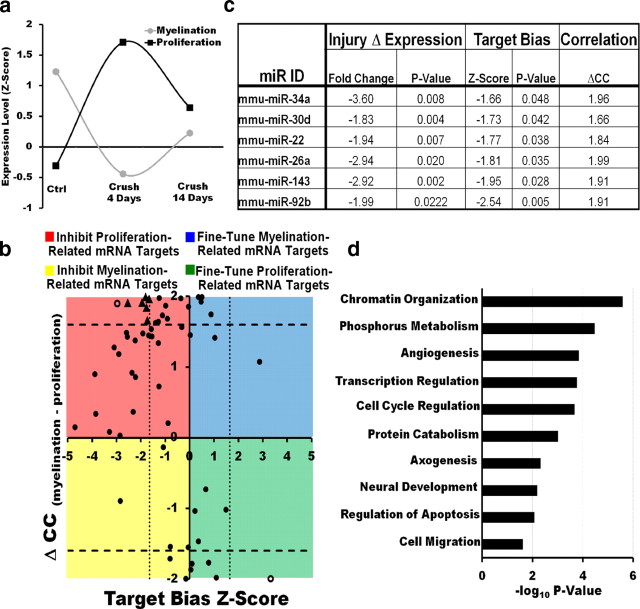
Figure 2.
Target bias analysis suggests that SC miRNAs modulate the injury response by repressing positive regulators of SC dedifferentiation/proliferation. a, mRNA expression profiles of gene clusters containing known regulators of SC dedifferentiation/proliferation (the proliferation profile) or differentiation/myelination (the myelination profile) after injury. b, Graph plotting the target bias for each miRNA in subset 1 (measured by a Mann–Whitney z-score) against the correlation of its expression with the myelination or proliferation profile (measured by ΔCC, the difference in correlation coefficients with the proliferation profile and the myelination profile). Biological functions of miRNAs in regulating the SC injury response may be inferred from their correlation with the proliferation or myelination gene clusters and by their positive or negative target bias. The majority of SC miRNAs cluster in the top left quadrant of the graph (negative target bias, correlation with the myelination profile), suggesting that SC miRNAs are likely to modulate the injury response primarily through their inhibition of positive regulators of SC dedifferentiation/proliferation. Dotted and dashed lines represent statistical significance cutoffs. miRNAs differentially regulated after injury (C4 vs Ctrl p < 0.05), with a statistically significant target bias and a high correlation with the myelination or proliferation clusters (ΔCC > 1.6), are represented by triangles. Empty circles represent miRNAs that met statistical significance cutoffs for target bias and high correlation with the myelination or proliferation clusters, but were not differentially regulated after injury. c, miRNAs likely to be direct regulators of the SC injury response (triangles in b) based on their differential regulation after injury, significant target bias, and a high correlation with the myelination or proliferation clusters. d, Predicted targets for miRNAs in c are enriched for genes involved in cellular processes that are normally inhibited in adult or differentiating SCs but are active in immature or proliferating SCs.
Target bias analysis suggests that SC miRNAs modulate the injury response by repressing positive regulators of SC dedifferentiation/proliferation
miRNAs often act in concert with transcriptional control to regulate target gene expression more efficiently (Hornstein and Shomron, 2006). For this reason, biologically relevant miRNAs tend to be either coexpressed (positive target bias) or negatively correlated (negative target bias) with their predicted targets to either fine-tune or inhibit their activity, respectively (Tsang et al., 2007). We hypothesized that miRNAs could modulate the SC injury response in a similar manner by cooperating with the transcriptional regulatory program that controls this process. An miRNA whose expression profile is correlated with that of myelination-associated genes (Fig. 2a) could thus reinforce the transcriptional program driving remyelination by inhibiting positive regulators of SC dedifferentiation/proliferation (demonstrating a negative target bias) or by modulating myelination-associated genes (demonstrating a positive target bias). Conversely, an miRNA whose expression is correlated with that of proliferation/dedifferentiation-associated genes could coordinate with the transcriptional regulatory program by inhibiting drivers of myelination (negative target bias) or by fine-tuning proliferation-related genes (positive target bias). To begin to address the biological function of SC miRNAs in modulating the injury response, we therefore studied their target bias as well as examined their correlation with the proliferation and myelination expression profiles.
We first used the TargetScanS algorithm (Grimson et al., 2007) to identify SC-expressed genes that were predicted targets of the 87 miRNAs we detected in SCs. We found that 59 of the 87 miRNAs detected by our microarray analysis had predicted targets among the ∼8000 transcripts detected in our previous SC mRNA expression profiling datasets (Nagarajan et al., 2002; our unpublished observations). TargetScanS is a relatively stringent miRNA target prediction algorithm, which requires an exact match between the miRNA seed sequence and any predicted sites. The 59 miRNAs we identified (subset 1) therefore represent a high-confidence set of modulators of the SC injury response and were further examined in our functional analysis.
We then used a Mann–Whitney U test to determine whether the expression of each miRNA in subset 1 had a higher positive or negative correlation with the expression of its predicted targets than with that of predicted nontarget genes (indicative of a target bias; see Materials and Methods, above). We found that 25 miRNAs in subset 1 had a significant target bias. Remarkably, 23 of these 25 miRNAs had a negative target bias, meaning that when the expression of a given miRNA was high, the mRNA level of its putative targets tended to be low and vice versa. This prevalence of a negative target bias is in agreement with previous studies (Farh et al., 2005; Lau et al., 2008) and suggests that the majority of SC miRNAs reinforce rather than fine-tune the concurrent transcriptional regulatory processes (Tsang et al., 2007).
Finally, the extent of target bias for each miRNA in subset 1 (measured by a Mann–Whitney z-score; see Materials and Methods, above) was plotted against the correlation of its expression with the myelination or proliferation profile (measured by ΔCC, the difference in correlation coefficients with the proliferation profile and the myelination profile). Interestingly, when plotted in this manner, the majority of SC miRNAs clustered in the top left quadrant of the graph, indicative of a negative target bias and an expression pattern that correlated with that of genes in the myelination cluster (Fig. 2b). Only six miRNAs had a negative target bias and a high correlation with the expression pattern of genes in the proliferation cluster. Similarly, very few miRNAs showed a positive target bias (Fig. 2b). Importantly, these results suggest that SC miRNAs are likely to modulate the injury response primarily through their inhibition of positive regulators of SC dedifferentiation/proliferation.
To further validate this conclusion, we performed gene ontology enrichment analysis for the putative targets of miRNAs that were as follows: (1) differentially regulated after injury (C4 vs Ctrl, p < 0.05), (2) demonstrated a statistically significant target bias, and (3) had a high correlation with the myelination or proliferation clusters (ΔCC > 1.6). We reasoned that miRNAs that met these three criteria were more likely to be direct regulators of the SC injury response. Using these rigorous criteria, six miRNAs were identified. Interestingly, these miRNAs all had a negative target bias and an expression pattern that correlated with the myelination profile (Fig. 2c). Gene ontology enrichment analysis showed that the predicted targets of these miRNAs were enriched for genes involved in cellular processes that are normally more active in immature or proliferating SCs. These processes included cell cycle regulation, cell migration, and regulation of transcription or chromatin organization (Fig. 2d), and genes such as SC mitogens (e.g., fibroblast growth factor), oncogenes (e.g., Kras), or cyclins (e.g., cyclin E2). These results indicate that SC miRNAs regulate the injury response primarily by inhibiting positive regulators of SC dedifferentiation/proliferation, and reinforce the transcriptional program that keeps these dedifferentiation/proliferation genes quiescent in mature or differentiating SCs. This reinforcement likely facilitates rapid and robust transitions between the distinct differentiation states involved in the SC injury response (Farh et al., 2005; Stark et al., 2005).
SCs with disrupted miRNA processing show delays in cycling between differentiation states after peripheral nerve injury
The results described above suggest that miRNAs work in concert with transcriptional controls to ensure the correct extent and timing of the SC dedifferentiation and subsequent remyelination following nerve injury. To more directly examine this idea, we performed nerve lesion studies on mice in which mature miRNA synthesis was disrupted in SCs through the targeted deletion of Dicer1, a gene encoding an essential enzyme in miRNA biosynthesis (Bartel and Chen, 2004). Mice harboring loxP-flanked Dicer1 alleles (Cobb et al., 2005) were crossed to mice expressing a tamoxifen-inducible form of Cre-recombinase in both myelinating and nonmyelinating SCs under the control of the proteolipid protein promoter (PLP-CreERT) (Doerflinger et al., 2003). The resulting PLP-CreERT:Dicer fl/fl mice were aged to 1 month and then injected daily with either 100 μg/g tamoxifen (Dicer-SCKOs) or vehicle (Ctrl) for 10 consecutive days (see Materials and Methods, above). Eight weeks after exposure to tamoxifen, a majority of SC nuclei (82% ± 5.1%, identified morphologically) in PLP-CreERT mice carrying a copy of a Cre-inducible Rosa26-YFP reporter exhibited YFP fluorescence (Fig. 3a). Accordingly, Dicer mRNA levels as well as the basal levels of a number of highly expressed miRNAs were significantly reduced in the sciatic nerves of Dicer-SCKO versus Ctrl mice (Fig. 3b and data not shown). However, at 8 weeks and for as long as 14 weeks after tamoxifen treatment, nerves from Dicer-SCKO mice showed no apparent pathology and were indistinguishable from Ctrl nerves, indicating that disruption of miRNA processing in SCs after peripheral nerve myelination was fully completed was well tolerated (Fig. 3g,h and data not shown).
Figure 3.
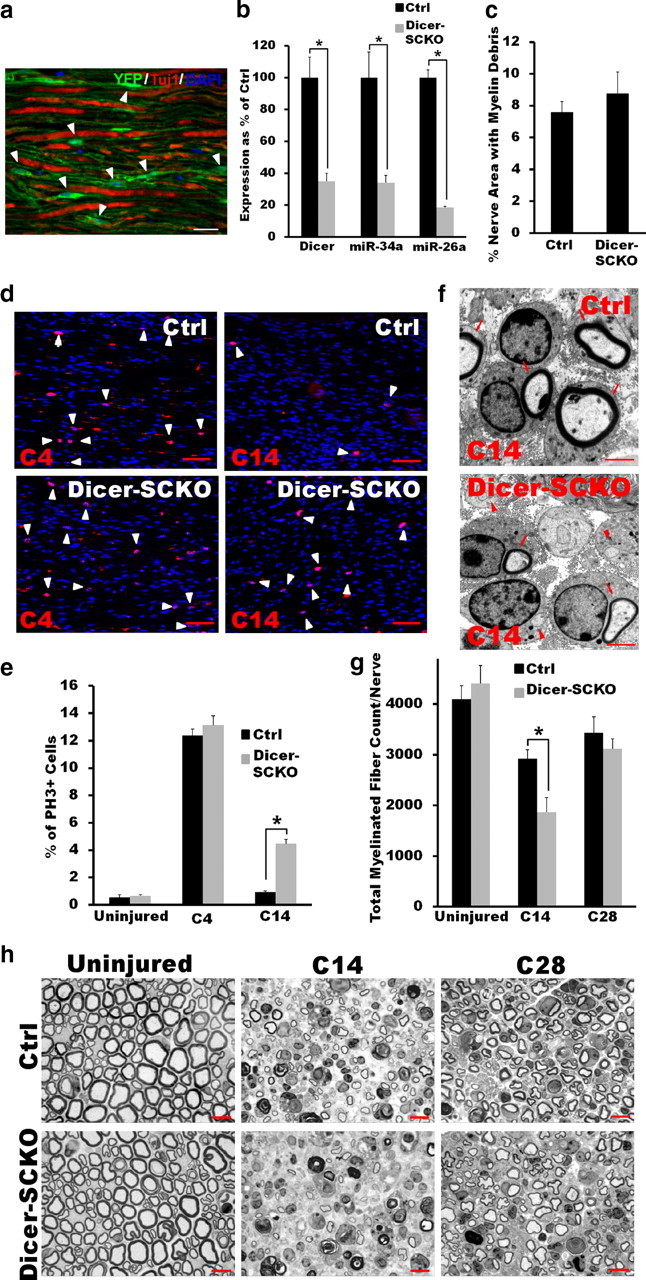
SCs with disrupted miRNA processing have delayed cycling between differentiation states after peripheral nerve injury. a, YFP fluorescence in SCs (arrowheads) of longitudinal sciatic nerve sections of Rosa26-YFP/PLP-CreERT mice 8 weeks after injection with tamoxifen. PLP-CreERT induces specific and efficient recombination in most SCs and excision-dependent YFP fluorescence is localized to regions of SC cytoplasm, but not to axons (Tuj1; note non-overlapping Tuj1 and YFP staining). Scale bar, 50 μm. b, qRT-PCR results show efficient depletion of Dicer1 mRNA and of mature miRNAs in Dicer-SCKO sciatic nerves 8 weeks after tamoxifen-induced recombination. Dicer mRNA levels and the levels of miR-34a and miR26a in the sciatic nerve of Dicer-SCKO mice at this time were decreased by >60% (p < 0.001 in all three cases, two-tailed Student's t test) compared with Ctrl littermates. Reported values are normalized to GAPDH for mRNA and U6 for miRNAs; error bars represent SEM; n = 5 mice per genotype. c, Quantification of the amount of myelin debris present in Ctrl and Dicer-SCKO nerves 14 d after crush injury shows no impairment in the ability of Tfam-deficient SCs to break down existing myelin. Error bars represent SEM; n = 3 mice per genotype. d, e, Pictures (d) and quantification (e) of PH3 immunostaining between sciatic nerves of Ctrl and Dicer-SCKO mice collected 4 (C4) or 14 (C14) days after crush injury. No difference in the number of proliferating SCs between Ctrl and Dicer-SCKO sciatic nerves was observed 4 d after crush injury. At C14, however, there were significantly more PH3-positive SCs in the sciatic nerves of Dicer-SCKO versus Ctrl mice (p < 0.05, two-tailed Student's t test), indicating exit from cell cycle was delayed in SCs with disrupted miRNA processing. Scale bar, 25 μm. Error bars represent SEM; n = 3 mice per genotype. f, At C14, electron micrographs of sciatic nerves from Ctrl and Dicer-SCKO mice show equivalent numbers of regenerated axons (arrows, remyelinated; arrowheads, non-remyelinated). Note the increased number of non-remyelinated axons in Dicer-SCKO nerves. Scale bars, 2 μm. g, h, Pictures (h) and quantification (g) of myelinated profiles in nerves from Ctrl and Dicer-SCKO mice before crush injury, or at C14 or C28. There is no difference in total number of myelinated axons in nerves from Dicer-SCKO versus Ctrl mice before crush injury. However, examination of toluidene-blue-stained plastic sections revealed a significant decrease (p < 0.05, two-tailed Student's t test) in the number of myelinated profiles at C14 in nerves from Dicer-SCKO mice. By C28, there was no longer a difference in total number of myelinated profiles between Dicer-SCKO and Ctrl nerves; however, nerves in Dicer-SCKO mice are more thinly myelinated. Scale bar, 25 μm. Error bars represent SEM; n = 3 or 4 mice per genotype at each time point. *p < 0.05 statistical significance.
To determine whether miRNA deficiency led to deficits in the SC injury response and remyelination, sciatic nerves of Dicer-SCKO and Ctrl littermates were crushed 8 weeks after tamoxifen administration. We first examined the response of SCs to axonal degeneration by assessing changes in SC proliferation and in their ability to break down existing myelin (Geuna et al., 2009). The rate of myelin loss, a measure of SC dedifferentiation, was similar between Dicer-SCKO and Ctrl mice, with no differences observed in the amount of myelin debris (Fig. 3c) or of MBP immunofluorescence in the nerves after injury (data not shown). Similarly, immunostaining of the nerve distal stump 4 d after injury using an antibody against the proliferation marker phosphohistone 3 (PH3) showed that, following axonal loss, miRNA-deficient SCs were able to reenter the cell cycle to the same extent as Ctrl SCs (percentage of PH3-positive cells: Ctrl = 12.37 ± 0.47; Dicer-SCKO = 13.12 ± 0.69; Fig. 3d,e). Depletion of miRNAs, therefore, did not interfere with the ability of SCs to dedifferentiate and proliferate after injury.
We next studied the late phase of the SC-injury response, during which these glia exit the cell cycle and redifferentiate to restore peripheral nerve function. PH3 immunostaining was initially used to assess the proportion of proliferating SCs 14 d after crush, a time when SCs have normally exited cell cycle and are actively differentiating. Interestingly, PH3 immunoreactivity showed that while the number of proliferating SCs in Ctrl nerves had returned to preinjury levels, significantly more SCs in Dicer-SCKO nerves were still actively proliferating at this time (percentage of PH3-positive cells: Ctrl = 0.93 ± 0.08; Dicer-SCKO = 4.47 ± 0.32; p < 0.001; Fig. 3d,e). Consistent with the reduced ability of miRNA-deficient SCs to exit cell cycle in a timely manner, nerves from Dicer-SCKO mice showed reduced remyelination compared with nerves from Ctrl mice, as indicated by a decrease in the total number of myelinated axons 14 d after injury (Fig. 3g,h). Note that this difference in remyelination after injury in Dicer-SCKO mice was not due to changes in the overall number of SCs (SC number/50K μm2: Ctrl = 183.4 ± 33.7; Dicer-SCKO = 203.5 ± 18.9; p = 0.6). An examination of electron micrographs of injured sciatic nerves from Ctrl and Dicer-SCKO mice revealed equivalent total numbers of regenerated axons (remyelinated and non-remyelinated axons/10K μm2: Ctrl = 525 ± 46.2; Dicer-SCKO = 576.9 ± 2.5; p = 0.3; Fig. 3f). Interestingly, 28 d after nerve injury, the total number of myelinated axons in nerves from Dicer-SCKO and Ctrl mice were similar (Fig. 3g,h), but the axons in Dicer-SCKO nerves remained more thinly myelinated [G-ratio (axon area/fiber area): Ctrl = 0.41 ± 0.016; Dicer-SCKO = 0.46 ± 0.012; p = 0.05; Fig. 3h]. Together, these results support the theory that SC miRNAs modulate the injury response by cooperating with transcriptional controls to ensure the correct extent and timing of SC dedifferentiation, cell cycle reentry/exit, and subsequent differentiation and remyelination after nerve injury.
miR-34a is a potential regulator of SC dedifferentiation and proliferation after peripheral nerve injury
Cellular differentiation is tightly linked to exit from the cell cycle. Therefore, the influence of miRNAs on cell fate decisions is often linked to their ability to modulate cell cycle regulators (Ivey and Srivastava, 2010). Given that SCs need to dedifferentiate and temporarily reenter the cell cycle to support axonal regrowth after injury (Geuna et al., 2009), we hypothesized that the regulation of SC cell cycle entry/exit by miRNAs could underlie their influence on the injury response. Among the miRNAs identified as most likely play a role in the SC injury response (Fig. 2c), we identified miR-34a as a particularly good candidate to regulate the competency of SCs to proliferate or differentiate. miR-34a is a tumor suppressor gene whose ectopic expression can induce cell cycle arrest (Hermeking, 2010). Moreover, Notch1 and cyclin D1 (Ccnd1), two important mediators of SC dedifferentiation, proliferation, and cell cycle entry/exit after peripheral nerve injury (Kim et al., 2001; Woodhoo et al., 2009), are targets of miR-34a in cancer cells (Sun et al., 2008; Pang et al., 2010). In addition, we identified a negative target bias for miR-34a in SCs for genes in the proliferation cluster (Fig. 2c), suggesting that miR-34a is a modulator of cell cycle regulators in SCs.
To verify a potential role of miR-34a in the regulation of cell cycle entry in SCs, we first examined the expression of this miRNA in more detail. Quantitative RT-PCR confirmed that miR-34a is highly expressed in adult peripheral nerves (Ctrl), but that its expression level drops sharply after nerve crush (C4) or transection (Trx14) when SCs dedifferentiate and reenter the cell cycle. miR-34a expression, however, is restored to preinjury levels as SCs exit the cell cycle and redifferentiation ensues (C7, C14; Fig. 4a). The expression pattern of miR-34a before and after injury is consistent with a role in the modulation of cell cycle regulators in SCs.
Figure 4.
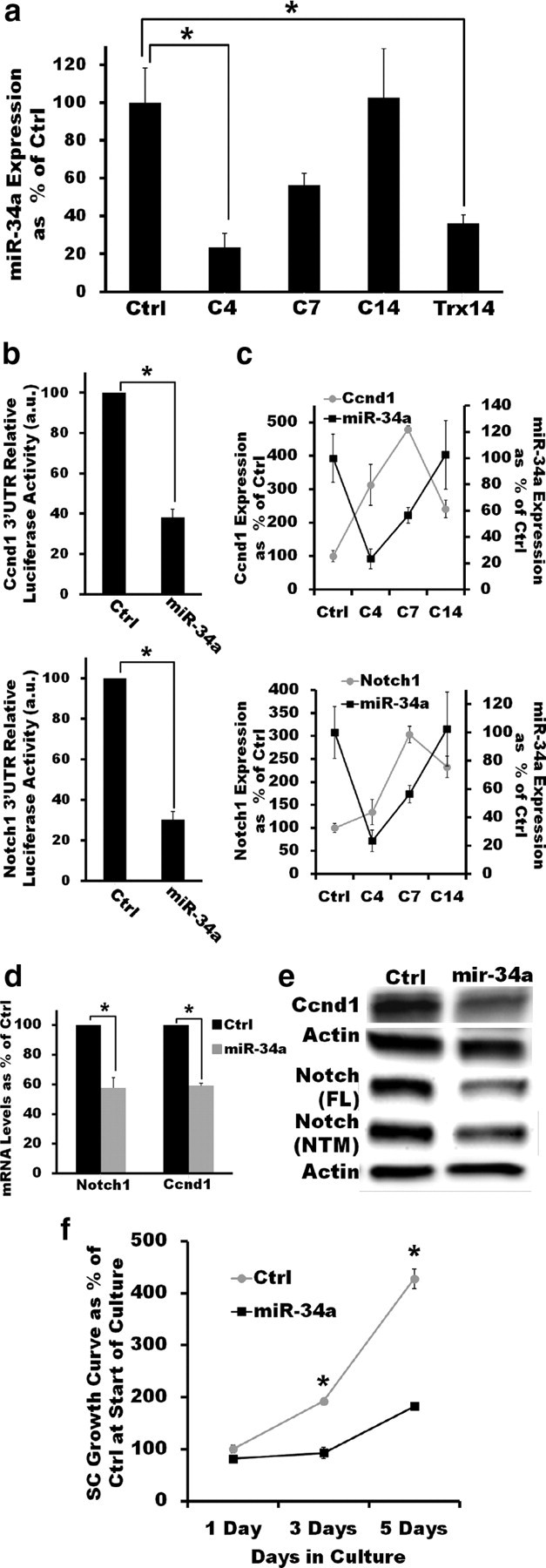
miR-34a targets Notch-1 and Ccnd1 to regulate SC proliferation in vitro. a, The dynamic regulation of miR-34a expression in sciatic nerves after crush injury suggests a potential role in the modulation of cell cycle regulators. miR-34a levels are significantly decreased 4 d after crush injury (p < 0.01, two-tailed Student's t test) but recover as redifferentiation ensues (C14). After nerve transection (Trx), an injury paradigm in which remyelination does not occur, mir-34a levels remain low compared with uninjured nerves even 14 d after injury (p < 0.05, two-tailed Student's t test). Reported values are normalized to U6; error bars represent SEM; n = 3–6 mice at each time point. b, miR-34a directly interacts with the 3′UTRs of Ccnd1 (top) and Notch1 (bottom). Overexpression of miR-34a but not of Ctrl miRNA in HEK293T cells expressing a luciferase reporter construct carrying the 3′UTR of either Notch1 or Ccnd1 results in significantly decreased luciferase activity (p < 0.01 in both cases, two-tailed Student's t test). Error bars represent SEM; n = 3 independent assays c, qRT-PCR measurements show that the expression pattern of Ccnd1 (top) and Notch1 (bottom) after injury is inversely correlated with that of miR-34a, suggesting a direct targeting of these transcripts by miR-34a. Reported values are normalized to GAPDH for mRNA and U6 for miRNAs; error bars represent SEM; n = 3–6 mice at each time point. d, e, Lentivirus-mediated mir-34a overexpression (≈3-fold) in cultured SCs significantly reduces Notch1 and Ccnd1 mRNA (d; p < 0.05, in both cases, two-tailed Student's t test) and protein levels (e, ≈2.5-fold decreas). FL, Full-length; NTM, cleaved transmembrane/intracellular region. qRT-PCR reported values are normalized to GAPDH; error bars represent SEM; n = duplicate wells from three independent assays. f, Lentivirus-mediated mir-34a overexpression (≈3-fold) impairs the ability of SCs to proliferate in the presence of DRG axons. miR-34a-expressing SCs have a significantly decreased axon-induced proliferation in vitro compared with control SCs (no significant difference at day 1; p < 0.01 at days 3 and 5; two-tailed Student's t test). Error bars represent SEM; n = triplicate wells from two independent assays. *p < 0.05 statistical significance.
We next assessed the effect of the differential expression of miR-34a after nerve injury on Notch1 and Ccnd1, two putative targets relevant to SC dedifferentiation/proliferation and the injury response (Atanasoski et al., 2001; Kim et al., 2001; Woodhoo et al., 2009). We overexpressed miR-34a in HEK293T cells together with a luciferase construct containing the 3′-UTR of either Notch1 or Ccnd1 and confirmed that, as previously reported (Sun et al., 2008; Pang et al., 2010), miR-34a directly recognized and interacted with the transcripts of these two genes (Fig. 4b). Consistent with direct targeting of Notch1 and Ccnd1 by miR-34a, the expression pattern of these two genes was reciprocally correlated with that of miR-34a following axonal degeneration (i.e., miR-34a expression decreased after injury as Notch1 and Ccnd1 expression increased). The converse expression pattern was observed as SC redifferentiation took place (Fig. 4c). Furthermore, when miR-34a was overexpressed in isolated SCs, Ccnd1 and Notch1 were downregulated at both the transcript and protein levels (Fig. 4d,e). In SCs, therefore, miR-34a directly targets at least two positive regulators of SC dedifferentiation and proliferation (Notch1 and Ccnd1). In addition to Notch1 and Cnd1, other potentially relevant targets to SC dedifferentiation and remyelination (e.g., YY1) could also mediate some of the effects of miR-34a on the injury response (He et al., 2010; Chen et al., 2011).
Finally, to directly determine whether miR-34a regulates cell cycle entry and exit in SCs, we assessed the effect of overexpressing this miRNA on the axon-induced proliferation of SCs in vitro. When isolated SCs are cultured in the presence of DRG neurons, contact with axons initially induces SCs to enter the cell cycle and proliferate. We found that ectopic expression of miR-34a did not affect SC health and viability, but severely inhibited DRG axon-induced proliferation of SCs in vitro (Fig. 4f). Together, our results indicate that mir-34 can directly interact with positive regulators of dedifferentiation and proliferation in SCs to control cell cycle dynamics in these peripheral glial cells, thus providing a mechanism by which miRNAs could modulate proper cycling of SCs between the distinct differentiation states that support nerve regeneration after injury.
miR-140 targets Egr2, a master regulator of myelination, to help modulate SC redifferentiation after nerve injury
The differentiation status of SCs after injury is determined by the balance between signaling systems that drive SC maturation/myelination versus those that induce SC dedifferentiation/proliferation (Jessen and Mirsky, 2008). While we initially found that most SC miRNAs interact with targets involved in SC dedifferentiation/proliferation (see above), we also searched for miRNAs that targeted positive regulators of myelination. To this end, we focused on identifying miRNAs that interact with Egr2, a transcription factor that is required for the formation and maintenance of myelin and is often mutated in patients with peripheral myelinopathies (Svaren and Meijer, 2008).
In silico analysis using PicTar (Krek et al., 2005), TargetScanS (Grimson et al., 2007), and miRanda (John et al., 2004) algorithms identified a putative binding site for miR-140 in the 3′-UTR of Egr2 (Fig. 5a). This binding site has an exact match to the 2–8 bp seed sequence of mouse miR-140 (AGUGGUU), which is highly conserved in human, rat, dog, and horse; thus, this miRNA was selected for further testing. Although our microarray analysis detected expression of miR-140* (which originates from the opposite arm of the pre-miRNA for mir-140) rather than miR-140 in nerve samples (Fig. 1), qRT-PCR confirmed that miR-140 is also expressed in adult peripheral nerve. Moreover, we found that miR-140 expression was dynamically regulated after peripheral nerve injury, i.e., its expression level was significantly decreased immediately after nerve crush (C4) and remained low until at least C7 (Fig. 5b). miR-140 expression was restored to preinjury levels once SC redifferentiation ensued (C14; Fig. 5b). Interestingly, the identified expression pattern of miR-140 after peripheral nerve injury is positively correlated with that of its putative target Egr2 (Fig. 5c), suggesting that miR-140 influences the injury response by fine-tuning the steady-state level of Egr2 in SCs that are actively forming or maintaining myelin (Tsang et al., 2007).
Figure 5.
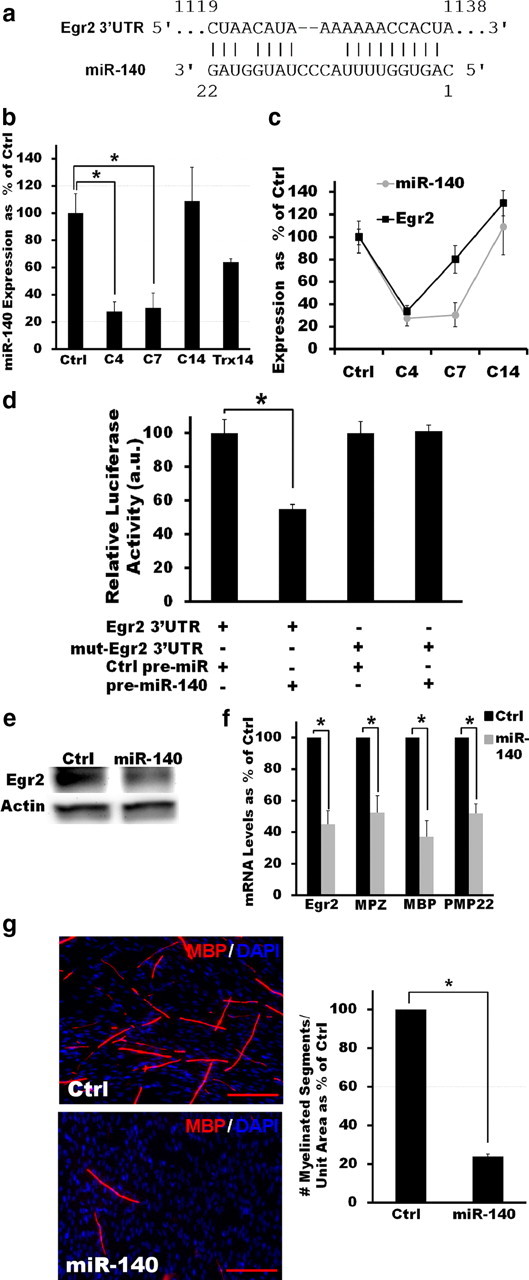
miR-140 directly interacts with Egr2 to regulate SC myelination in vitro. a, PicTar, TargetScanS, and miRanda algorithms all identify a putative binding site for miR-140 in the 3′-UTR of Egr2. b, qRT-PCR results show that miR-140 is expressed in mouse sciatic nerves and that its expression is dynamically regulated after injury. miR-140 expression is significantly decreased after injury (C4 and C7, p < 0.05, two-tailed Student's t test) but recovers as redifferentiation ensues (C14, no significant difference). Reported values are normalized to U6; error bars represent SEM; n = 3–6 mice at each time point. c, qRT-PCR measurements show that the expression pattern of Egr2 is correlated with that of miR-140. Reported values are normalized to GAPDH for mRNA and U6 for miRNAs; error bars represent SEM; n = 3–6 mice at each time point. d, Luciferase assays confirm a direct interaction between miR-140 and the 3′-UTR of Egr2. Overexpression of miR-140, but not of Ctrl miRNA, in HEK293T cells expressing a luciferase reporter construct carrying the 3′-UTR of Egr2 results in significantly decreased luciferase activity (p < 0.05, two-tailed Student's t test). Mutating the predicted landing pad for miR-140 in the 3′-UTR of Egr2 disrupts the interaction between miR-140 and the Egr2 3′-UTR luciferase construct and restores luciferase activity. e, Western blot analysis shows that overexpression of miR-140 in cultured SCs reduces the levels of Egr2 protein ≈2.8-fold. f, Overexpression of miR-140 in SCs significantly decreases the levels of Egr2 as well as of a number of myelin markers when these cells are cocultured with DRGs and induced to myelinate (in all cases p < 0.05, two-tailed Student's t test). Reported values are normalized to GAPDH; error bars represent SEM; n = duplicate wells from two independent assays. g, Similarly, miR-140 overexpression significantly inhibits the ability of SCs to form myelin in vitro in cocultures with DRG neurons as visualized by MBP immunostaining (p < 0.05, two-tailed Student's t test). Error bars represent SEM; n = triplicate wells from three independent assays. *p < 0.05 statistical significance.
The dynamic regulation of miR-140 during active myelin remodeling prompted us to further examine this miRNA in SCs. To confirm that Egr2 is a target of miR-140, we first cotransfected HEK293T cells with plasmids encoding either pre-mir-140 or a control pre-miRNA, together with a luciferase reporter construct carrying the 3′UTR of Egr2. Coexpression of this reporter plasmid with miR-140 decreased luciferase activity in these cells, indicating that miR-140 can directly recognize and interact with the Egr2 3′UTR (Fig. 5d). This interaction was dependent on the computationally predicted miR-140 binding site in this region, as a 3′UTR fragment containing a mutant site was no longer responsive to coexpression of pre-mir-140 (Fig. 5d). Consistent with results from these luciferase reporter assays, Egr2 protein levels were reduced 2.8-fold in cultured SCs overexpressing miR-140 (Fig. 5e). These results identify Egr2 as a new target of miR-140 and indicate that this miRNA directly interacts with Egr2 via a highly conserved site in its 3′UTR to regulate the expression of this transcription factor in SCs.
Finally, we examined whether miR-140 affected the ability of SCs to differentiate and myelinate in vitro. In the absence of neurons, prolonged exposure to cAMP-stimulating agents can promote SC growth arrest and differentiation and induce expression of genes typically associated with myelinating SCs (e.g., Mpz and Pmp22) (Jessen et al., 1991; Monje et al., 2009). Using this system, we evaluated the effect of miR-140 overexpression on SC differentiation. Consistent with the downregulation of Egr2 protein by mir-140 in SCs (Fig. 5e), cAMP-induced, Egr2-mediated upregulation of myelin markers in cultured SCs was inhibited in the presence of this miRNA (cAMP-induced gene expression fold induction, MPZ: Ctrl miR = 4.7 ± 0.51; miR-140 = 2.09 ± 0.35; p < 0.05; PMP22: Ctrl miR = 1.5 ± 0.06; miR-140 = 0.9 ± 0.06; p < 0.05).
When SCs are cultured together with DRGs in the presence of ascorbic acid, DRG axons will support the differentiation of SCs and the formation of myelin (Wood et al., 1990). Myelination can be monitored in this system by using MBP immunostaining as a marker. We found that overexpression of miR-140 in SCs cocultured with DRGs inhibited the expression of Egr2 as well as a number of myelination markers (Fig. 5f). Most importantly, miR-140 overexpression inhibited myelination in this in vitro assay, as evidenced by the decreased number of myelin segments detected by MBP immunostaining (Fig. 5g). Together, our results indicate that miR-140 can directly interact with Egr2 and modulate the level of this transcription factor to regulate myelination. This suggests another mechanism by which miRNAs may modulate the injury-induced regenerative process of SCs in peripheral nerves.
Discussion
Following axonal loss, SCs mount a regenerative response involving coordinated dedifferentiation, proliferation, and redifferentiation that helps restore peripheral nerve function. Extensive work has established that this SC injury response is driven by an intricate transcriptional program controlling the balance between positive and negative regulators of SC differentiation (Jessen and Mirsky, 2008). The present study describes a new regulatory layer of the SC injury response involving posttranscriptional regulation by miRNAs. We show that SC miRNAs modulate the injury response by targeting primarily positive regulators of SC dedifferentiation/proliferation, cooperating with transcriptional controls to ensure rapid and robust transitions between the distinct differentiation states that support nerve regeneration. Moreover, we also identify miR-34a and miR-140 as important functional regulators of SC proliferation and myelination, two central processes of the SC regenerative response after injury. miRNAs therefore appear to be important modulators of the SC regenerative response after peripheral nerve injury.
miRNAs are thought to have evolved to canalize the genetic network and confer robustness to gene expression programs (Hornstein and Shomron, 2006). miRNAs achieve this by integrating into the preexisting genetic network in two ways. On the one hand, miRNAs can wire into coherent feedforward loops (FFLs) such that high expression of a given miRNA is induced at a time or location when the transcription of its predicted targets is repressed (negative target bias) (Hornstein and Shomron, 2006; Tsang et al., 2007). This type of network motif functions as a posttranscriptional failsafe and results in mutually exclusive expression domains for the miRNA and its putative targets (Mansfield et al., 2004; Stark et al., 2005). On the other hand, miRNAs can wire into incoherent FFLs, which would promote high expression of a given miRNA at a time or location where its predicted targets are also abundant (positive target bias) (Hornstein and Shomron, 2006; Tsang et al., 2007). Biological examples of incoherent FFLs (Karres et al., 2007) have shown that in this type of network motif miRNAs have a fine-tuning action and keep protein levels in the correct functional range. Our work demonstrates that SC miRNAs show a prevalence of negative target biases during the SC injury response. This indicates that SC miRNAs primarily participate in coherent FFLs. Moreover, the expression of most SC miRNAs after injury was negatively correlated with that of positive regulator of SC dedifferentiation/proliferation. Interestingly, we also found that predicted targets of SC miRNAs are enriched for genes active in immature or proliferating SCs that are quiescent in adult, differentiated SCs. Together, these observations indicate that SC miRNAs regulate the injury response primarily by inhibiting positive regulators of SC dedifferentiation/proliferation and thereby reinforcing a preexisting transcriptional silencing mechanism that keeps dedifferentiation/proliferation-related genes quiescent in differentiated SCs yet allows their induction following axonal degeneration.
The above results imply that the majority of miRNAs expressed in mature SCs are involved in maintaining the differentiated state. In accordance with this, we found that immediately after injury, most SC miRNAs are downregulated, which allows for the transcriptionally driven dedifferentiation of SCs. We also observed that, similar to what has been reported for SC postnatal development (Bremer et al., 2010; Pereira et al., 2010; Yun et al., 2010), the expression of most SC miRNAs is upregulated once again shortly after axonal regrowth. Because most SC miRNAs target positive regulators of SC dedifferentiation/proliferation that must be repressed for proper SC differentiation to occur (Le et al., 2005), this serves to facilitate redifferentiation and stabilization of the mature SC phenotype. This observation is consistent with studies showing that miRNAs are generally expressed at lower levels in immature or dedifferentiated tissues than in differentiated ones (Lu et al., 2005; Wienholds et al., 2005) and with the idea that miRNAs are largely involved in promoting, inducing, and stabilizing differentiated phenotypes (Wienholds and Plasterk, 2005). It is possible that manipulation of SC miRNAs could be used therapeutically to facilitate SC differentiation after injury or reverse the effects of neuropathy.
The therapeutic modulation of SC miRNAs, however, requires the identification of specific miRNAs playing critical roles in the establishment and maintenance of SC differentiation as well as in the modulation of the SC injury response. We identify miR-34a as one such candidate. We found that miR-34 was able to critically regulate SC cell cycle exit by directly interacting with Ccnd1 transcripts and impinging on Notch signaling, a central modulator of SC dedifferentiation after injury (Woodhoo et al., 2009). Other miRNAs are likely to play similar roles; for example, let-7 family members enhance differentiation and inhibit stem cell self-renewance (Melton et al., 2010). Moreover, this family of miRNAs participates in regenerative processes in zebrafish (Ramachandran et al., 2010). Intriguingly, our microarray analysis found that several members of the let-7 family were differentially regulated in SCs after peripheral nerve injury.
While our work shows that most SC miRNAs interact with targets involved in SC dedifferentiation/proliferation, we also identified miR-140 as a potential regulator of Egr2. Egr2 is an essential transcription factor for myelinogenesis and is often mutated in patients with peripheral myelinopathy (Svaren and Meijer, 2008). Accordingly, we found that overexpression of miR-140 in SCs impaired their ability to form myelin in vitro. Interestingly, the expression pattern of miR-140 and Egr2 indicates the presence of an incoherent FFL (Hornstein and Shomron, 2006; Tsang et al., 2007) in which both transcript and miRNA are coexpressed at the same time in SCs. This suggests that miR-140 may act to maintain Egr2 expression within precise functional levels. This is particularly interesting because myelin formation and maintenance is very sensitive to gene dosage effects. In fact, both abnormally low and high levels of specific myelin proteins can cause peripheral neuropathy in humans (Meyer zu Hörste et al., 2006). Regulation of expression thresholds could therefore be a second mechanism, together with reinforcement of transcriptional silencing, by which SC miRNAs modulate the injury response of these glia and promote regeneration of peripheral nerves.
In summary, through miRNA expression profiling after nerve damage and examination of the effects of impaired miRNA processing on the SC injury response, the present study establishes a critical role for miRNAs in the modulation of the SC regenerative response in peripheral nerves. We demonstrate that SC miRNAs ensure rapid and robust transitions between the distinct differentiation states necessary to support nerve regeneration by reinforcing the transcriptional silencing of dedifferentiation- and proliferation-related genes. Modulation of SC miRNAs could therefore offer a new therapeutic avenue to enhance nerve regeneration and improve the treatment of neurodegenerative diseases characterized by axonopathy.
Footnotes
This work was also supported by NIH Neuroscience Blueprint Center Core Grant P30 NS057105 to Washington University, the HOPE Center for Neurological Disorders, NIH Grants NS040745 (J.M.) and AG13730 (J.M.). L.-W.-C. is supported by a NIH Pathway to Independence Award LM010824. We thank Amy Strickland and Nina Panchenko for experimental assistance; Matthias Merkenschlager for the Dicer conditional knock-out mice; members of the Milbrandt laboratory for their comments on the manuscript and helpful discussions. We thank the Genome Technology Access Center (GTAC) in the Department of Genetics for help with genomic analysis. We also thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO, for the use of the Biomedical Informatics Core (BMC), which provided the in silico analysis service. Both GTAC and BMC are partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by Institute of Clinical and Translational Sciences/Clinical and Translational Science Award UL1RR024992 from the National Center for Research Resources (a component of the NIH), and NIH Roadmap for Medical Research.
References
- Araki T, Nagarajan R, Milbrandt J. Identification of genes induced in peripheral nerve after injury. expression profiling and novel gene discovery. J Biol Chem. 2001;276:34131–34141. doi: 10.1074/jbc.M104271200. [DOI] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Atanasoski S, Shumas S, Dickson C, Scherer SS, Suter U. Differential cyclin D1 requirements of proliferating Schwann cells during development and after injury. Mol Cell Neurosci. 2001;18:581–592. doi: 10.1006/mcne.2001.1055. [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bosse F, Hasenpusch-Theil K, Küry P, Müller HW. Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J Neurochem. 2006;96:1441–1457. doi: 10.1111/j.1471-4159.2005.03635.x. [DOI] [PubMed] [Google Scholar]
- Bremer J, O'Connor T, Tiberi C, Rehrauer H, Weis J, Aguzzi A. Ablation of dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS One. 2010;5:e12450. doi: 10.1371/journal.pone.0012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QR, Yu LR, Tsang P, Wei JS, Song YK, Cheuk A, Chung JY, Hewitt SM, Veenstra TD, Khan J. Systematic proteome analysis identifies transcription factor YY1 as a direct target of miR-34a. J Proteome Res. 2011;10:479–487. doi: 10.1021/pr1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. Axon degeneration mechanisms: commonality amid diversity. Nat Rev Neurosci. 2005;6:889–898. doi: 10.1038/nrn1788. [DOI] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuna S, Raimondo S, Ronchi G, Di Scipio F, Tos P, Czaja K, Fornaro M. Chapter 3: histology of the peripheral nerve and changes occurring during nerve regeneration. Int Rev Neurobiol. 2009;87:27–46. doi: 10.1016/S0074-7742(09)87003-7. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Kim JY, Dupree J, Tewari A, Melendez-Vasquez C, Svaren J, Casaccia P. YY1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat Neurosci. 2010;13:1472–1480. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Hunter DA, Moradzadeh A, Whitlock EL, Brenner MJ, Myckatyn TM, Wei CH, Tung TH, Mackinnon SE. Binary imaging analysis for comprehensive quantitative histomorphometry of peripheral nerve. J Neurosci Methods. 2007;166:116–124. doi: 10.1016/j.jneumeth.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia. 2008;56:1552–1565. doi: 10.1002/glia.20761. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R, Morgan L. Role of cyclic AMP and proliferation controls in Schwann cell differentiation. Ann N Y Acad Sci. 1991;633:78–89. doi: 10.1111/j.1749-6632.1991.tb15597.x. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Kim HA, Ratner N, Roberts TM, Stiles CD. Schwann cell proliferative responses to cAMP and Nf1 are mediated by cyclin D1. J Neurosci. 2001;21:1110–1116. doi: 10.1523/JNEUROSCI.21-04-01110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RH. Nerve trunks and spinal roots. In: Dyck PJ, Thomas PK, editors. Peripheral neuropathy. Philadelphia: Elsevier Saunders; 2005. pp. 35–42. [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le N, Nagarajan R, Wang JY, Araki T, Schmidt RE, Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of schwann cell differentiation and myelination. Proc Natl Acad Sci U S A. 2005;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT. MicroRNA-responsive ‘sensor’ transgenes uncover hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer zu Hörste G, Prukop T, Nave KA, Sereda MW. Myelin disorders: causes and perspectives of charcot-marie-tooth neuropathy. J Mol Neurosci. 2006;28:77–88. doi: 10.1385/jmn:28:1:77. [DOI] [PubMed] [Google Scholar]
- Monje PV, Rendon S, Athauda G, Bates M, Wood PM, Bunge MB. Non-antagonistic relationship between mitogenic factors and cAMP in adult Schwann cell re-differentiation. Glia. 2009;57:947–961. doi: 10.1002/glia.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. doi: 10.1016/s0896-6273(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Nagarajan R, Le N, Mahoney H, Araki T, Milbrandt J. Deciphering peripheral nerve myelination by using schwann cell expression profiling. Proc Natl Acad Sci U S A. 2002;99:8998–9003. doi: 10.1073/pnas.132080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, Lee KF, Yeung WS. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31:1037–1044. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- Pereira JA, Baumann R, Norrmén C, Somandin C, Miehe M, Jacob C, Lühmann T, Hall-Bozic H, Mantei N, Meijer D, Suter U. Dicer in schwann cells is required for myelination and axonal integrity. J Neurosci. 2010;30:6763–6775. doi: 10.1523/JNEUROSCI.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates muller glia dedifferentiation and retinal regeneration through a lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu EJ, Yang M, Gustin JA, Chang LW, Freimuth RR, Nagarajan R, Milbrandt J. Analysis of peripheral nerve expression profiles identifies a novel myelin glycoprotein, MP11. J Neurosci. 2008;28:7563–7573. doi: 10.1523/JNEUROSCI.1659-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Araki T, Milbrandt J. Stimulation of nicotinamide adenine dinucleotide biosynthetic pathways delays axonal degeneration after axotomy. J Neurosci. 2006;26:8484–8491. doi: 10.1523/JNEUROSCI.2320-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Vohra BPS, Lund FE, Milbrandt J. Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J Neurosci. 2009;29:5525–5535. doi: 10.1523/JNEUROSCI.5469-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56:1541–1551. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci U S A. 2008;105:18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier JD, Lau P, Hudson L, Murashov AK, Renne R, Notterpek L. Peripheral myelin protein 22 is regulated post-transcriptionally by miRNA-29a. Glia. 2009;57:1265–1279. doi: 10.1002/glia.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier JD, Semple-Rowland S, Madorsky I, Papin JE, Notterpek L. Reduction of dicer impairs schwann cell differentiation and myelination. J Neurosci Res. 2010;88:2558–2568. doi: 10.1002/jnr.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wood P, Moya F, Eldridge C, Owens G, Ranscht B, Schachner M, Bunge M, Bunge R. Studies of the initiation of myelination by Schwann cells. Ann N Y Acad Sci. 1990;605:1–14. doi: 10.1111/j.1749-6632.1990.tb42376.x. [DOI] [PubMed] [Google Scholar]
- Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D'Antonio M, Parkinson DB, Wilton DK, Al-Shawi R, Simons P, Shen J, Guillemot F, Radtke F, Meijer D, Feltri ML, Wrabetz L, Mirsky R, Jessen KR. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009;12:839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B, Anderegg A, Menichella D, Wrabetz L, Feltri ML, Awatramani R. MicroRNA-deficient schwann cells display congenital hypomyelination. J Neurosci. 2010;30:7722–7728. doi: 10.1523/JNEUROSCI.0876-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]