Abstract
p21-activated kinases (PAKs) are Cdc42 effectors found in metazoans, fungi and protozoa. They are subdivided into PAK1-like (group I) or PAK4-like (group II) kinases. Human PAK4 is widely expressed and its regulatory mechanism is unknown. We show that PAK4 is strongly inhibited by a newly identified auto-inhibitory domain (AID) formed by amino acids 20 to 68, which is evolutionarily related to that of other PAKs. In contrast to group I kinases, PAK4 is constitutively phosphorylated on Ser 474 in the activation loop, but held in an inactive state until Cdc42 binding. Thus, group II PAKs are regulated through conformational changes in the AID rather than A-loop phosphorylation.
Keywords: cytoskeleton, PAK4, Cdc42, phosphorylation
Introduction
There are six mammalian p21-activated kinases (PAKs), which share conserved catalytic and Cdc42–Rac interaction/binding (CRIB) domains, and are classified into two classes. Group I PAKs (PAKs 1–3) are activated by Cdc42 or Rac1 interaction with their CRIB domain [1–3], while the group II PAKs (PAKs 4–6) are supposedly not [4–6]. PAK4, the ubiquitous group II kinase, may be overexpressed in primary cancers and in cancer-derived lines [7]. PAK4 amplifications have been identified in pancreatic cancers [8], and a newly developed PAK inhibitor PF-3758309 is a potent antitumour agent in preclinical models [9]. Loss of PAK4 in mice is embryonic lethal, which can be attributed to multiple developmental defects including cardiac malformation [10]. The kinase is required downstream of Cdc42 in endothelial cell lumen formation [11], and is required for tight junction stability in epithelial cells [12]. Although Cdc42 has never been found to significantly regulate mammalian PAK4 activity, a CRIB mutant of the fly PAK4 Mbt shows elevated activity [13], as does a CRIB-deleted mutant of human PAK4 [4, 14].
In this study we investigate PAK4 regulation in detail and come to three surprising conclusions. First, PAK4 contains a bone fide auto-inhibitory domain (AID) that is conserved from flies to man, and is evolutionarily conserved with the PAK1 AID. Second, the activation loop Ser 474, which is the only residue found to be phosphorylated in the catalytic domain, is constitutively phosphorylated. Third, the binding of Cdc42 serves to activate PAK4, but because kinase activation does not require additional auto-phosphorylation, the kinase retains no covalent memory of this state.
Results and discussion
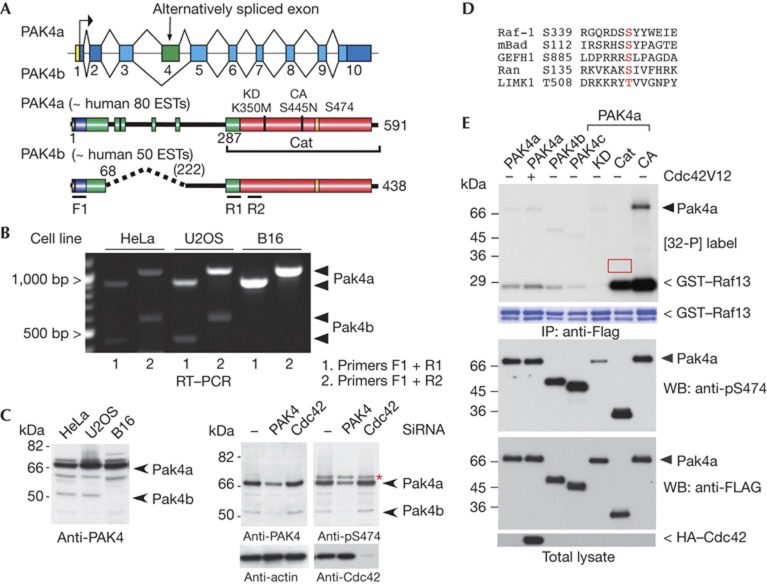
Identification of a new smaller form of human PAK4
The 68-kDa PAK4 was first discovered by homology to the conventional PAK1 kinase [4]. Although much has been revealed regarding the expression and biological roles of PAK4 in vertebrates [15], there are few clues as to its mode of regulation, and the catalytic domain structure is unremarkable [16]. Molecular dynamic simulations based on this X-ray structure suggest that the active form of PAK4 requires phosphorylation of activation loop Ser 474 to stabilize the position of the C-α helix, much like PAK1 [17]. Human PAK4 (591 residues) is a ubiquitous kinase with orthologues in all metazoans, but is absent in plants, fungi and protozoa. We detected an mRNA product encoding a spliced variant lacking 154 residues (exon 4) termed PAK4b (Fig 1A) in HeLa and U2OS cells, but not in mouse B16 cells (Fig 1B). This is consistent with the absence of similar PAK4b mouse sequences in public EST databases. A PAK4-specific antibody raised against the N-terminal region revealed low levels of the smaller PAK4b (Fig 1C, arrows) in HeLa and U2OS, though absent from B16 cells. The PAK4 in these cells was equally reactive towards a more sensitive activation-loop pS474 antibody, which does not react with kinase-inactive PAK4(K350M; Supplementary Fig S1 online). HeLa cells treated with PAK4 short interfering RNA (directed to kinase domain) showed a loss of both PAK4 isoforms. Knockdown of PAK4 but not Cdc42 reduced the corresponding level of pS474 (Fig 1C, arrow). When cells were treated with the staurosporine or the phosphatase inhibitor calyculin (Supplementary Fig S1C online), we saw no change in the pS474 status, suggesting relatively slow turnover of the A-loop modification in vivo.
Figure 1.
PAK4 activity is independent of Ser 474 phosphorylation. (A) Schematic of the PAK4 gene in chromosome 19q13.2 and the exon organization of human PAK4. The PAK4b splice isoform lacks exon 4: identified domains include the nuclear localization signal (NLS, black), Cdc42–Rac interaction/binding (CRIB, blue) and catalytic domain (red). Other conserved but undefined regions in group II PAKs are in green. The number of human EST (expressed sequence tags) in the NCBI database is indicated. (B) Reverse transcriptase–PCR analysis (RT–PCR, 30 cycles) of mRNA isolated from HeLa, U2OS and mouse B16 cell lines. The position of the PAK4a and PAK4b cDNA products are marked. (C) Western blots (WBs) of cell lysates derived cell lines (40 μg) as indicated were probed with affinity-purified antibodies raised against PAK4(1–280) or a pSer 474 13mer peptide. The PAK4 and Cdc42 targeted short interfering RNAs (siRNAs) have been described previously [12]. The asterisk marks a 72-kDa nonspecific band. (D) Alignment of sequences surrounding known PAK4 substrate sites. (E) Flag–PAK4 constructs were immunoprecipitated (IP) and assayed in vitro with [32-P]ATP with the Raf1 S338/9 peptide substrate (see Methods). Autoradiograph on the top panel is a 2-h exposure. The A-loop phosphorylation was probed using CS#3241. The red box highlights that PAK4–Cat does not undergo auto-phosphorylation. CA, constitutively active, PAK4(S445N); Cat, PAK4(286–591); GST, glutathione S-transferase; HA, haemagglutinin; KD, kinase dead, PAK4(K350M); PAK, p21-activated kinase; PAK4c lacks residues 121–285, under accession number AAH02921.1.
The PAK4 Ser 474 is constitutively phosphorylated
Coexpressed Cdc42V12 marginally increased the activity of immunoprecipitated Flag–PAK4a (Fig 1E lane 2). Both Flag–PAK4a and Flag–PAK4b expressed in mammalian cells show very low activity (lanes 1 and 3) compared with catalytic domain PAK4–Cat or the full-length activated PAK4(S445N) [18]. The Raf1 substrate peptide used in these assays conforms to an ideal PAK4 target [19], and has similar sequence to Bad (Fig 1D), which is phosphorylated to a similar extent in vitro (Supplementary Fig S3B online). The fact that the PAK4 N-terminal strongly suppresses kinase activity was not appreciated [7]. In spite of their very different activities, we noted that the PAK4 activation-loop residue (Ser474) was phosphorylated to a similar level among these constructs, excepting catalytically inactive PAK4a(K350M). The PAK4–Cat is fully phosphorylated at Ser 474 when isolated from mammalian cells, as is bacterially expressed PAK4–Cat [16], and as a result does not show auto-phosphorylation in vitro (Fig 1E red box). The fact that PAK4 Ser 474 A-loop phosphorylation does not correlate with kinase activity contrasts starkly with the behaviour of PAK1 [20]. Most protein kinases rely on changes to A-loop phosphorylation for activation [21], and it is currently thought that PAK4 pS474 levels are a marker of kinase activity [9, 22]. Our data instead indicate that an auto-inhibitory region of PAK4 controls kinase activity, not by regulating A-loop phosphorylation, but by allosterically modulating the constitutively phosphorylated kinase. A different human PAK4 form lacking residues 121–285 (designated PAK4c) is also auto-inhibited (Fig 1E). Taken together, the low activity of PAK4b and PAK4c suggested that PAK4(1–68) contains an AID that acts on the PAK4–Cat. Given the similarity of the PAK1 and PAK4 catalytic domains one might anticipate that their AIDs are sequence related.
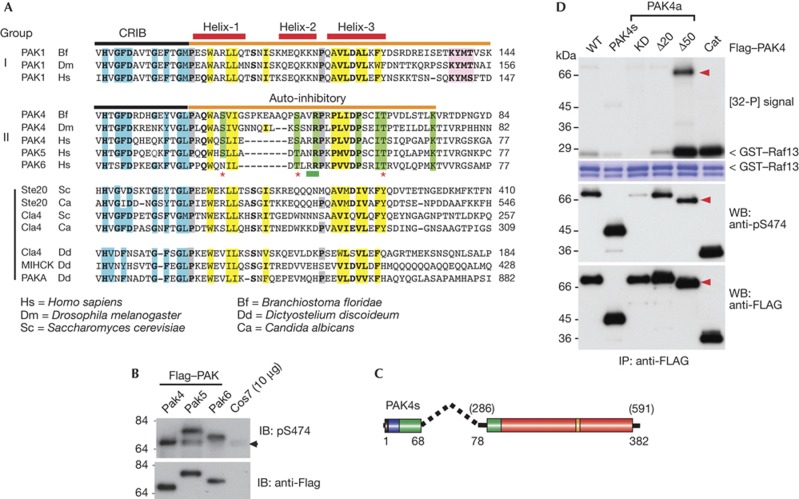
All PAKs have sequence-related AIDs
Mammalian PAK1 possesses a functional AID adjacent to and overlapping the CRIB motif [2]. Structural studies of auto-inhibited human PAK1 indicate that the AID (Fig 2A orange bar) inhibits in trans at the dimerization interface PAK1 [1, 23]. Both PAK1 and PAK4 CRIB sequences form an almost identical fold when bound to Cdc42 (pdb: 1E0A and 2OV2). In these structures the N termini of the PAK4 and PAK1 CRIBs bind, as a β-sheet, to Cdc42 strand β2, and similar CRIB residues bind to switches I and II of Cdc42.GTP. The putative helix 1 of the PAK4 AID (marked on Fig 2A) is indeed an α-helix when bound to Cdc42. The PAK1 AID structure shows a core of two α-helices that pack against each other (as we illustrate in Supplementary Fig S2A online) with conserved hydrophobic residues largely buried (Fig 2A in yellow). The second long helix of the PAK1/2 AID is a target of the Escherichia coli toxin espG [24] and Fragile-X proteins [25]. By alignment of group II PAKs from humans with PAK4 from diverse metazoans, one can see that PAK4 contains motifs related to the PAK1 AID (Fig 2A). Consistent with full-length group II kinases being fully phosphorylated in the activation loop (PRRKpSLVG), which is different from the PAK1 sequence (SKRSpTMVG), we find that human PAK5 and PAK6 (PKRKpSLVG) are equally reactive using this anti-pS474 antibody (Fig 2B). The ∼70 kDa nonspecific band observed with most cell lines (Fig 1C) is not likely to be PAK6, which consistently runs higher on SDS–PAGE.
Figure 2.
Identification of a conserved PAK4 auto-inhibitory domain (AID). (A) An alignment of the AID (orange bar) comparing group I PAKs with those from metazoan PAK4 and fungal and protozoan PAKs. The three α-helices in the PAK1 AID are marked (red bars). The A-loop binding motif (KYMS/T, pink box) is not found in fungal or protozoan PAKs. Proline residues preceding helixes 1 and 3 are marked in grey. Conserved residues involved in Cdc42 binding are in blue. Hydrophobic AID residues are highlighted in yellow, while those conserved only among group II PAKs are in green. The position of the PAK4 AID mutations tested in this study are indicated by asterisk marks (potential phosphorylation sites) or underlined in green (R48, R49). (B) Flag–tagged PAK4/5 and 6 expressed in COS7 were analysed for activation loop phosphorylation. (C) Schematic of a minimal synthetic construct PAK4(s) comprising residues PAK4(1–68) joined to PAK4(286–591) with a 10-residue linker. (D) The various Flag–PAK4 were used for in vitro kinase assays performed as described in Fig 1. The A-loop phosphorylation was probed using CS#3241. Cat, PAK4(286–591); CRIB, Cdc42–Rac interaction/binding; GST, glutathione S-transferase; IP, immunoprecipitated; KD, kinase dead, PAK, p21-activated kinase; PAK4a(K350M); WB, western blot; WT, wild type; Δ20, PAK4a(21–591) and Δ50, PAK4a(51–591).
The group I metazoan PAKs have an KYMS/T motif not found in protozoan PAKs (Fig 2A, pink box), which controls kinase activity by inserting between the activation loop and the α-C helix, as illustrated in the model shown in Supplementary Fig S2A online [1]. The KYMS/T motif, which is also not present in PAK4, prevents auto-phosphorylation of PAK1 Thr423 by holding the A-loop well away from its normal position. The absence of such a motif in PAK4 (Fig 2A) could allow constitutive A-loop auto-phosphorylation. Conserved putative AID residues that are only found in group II PAKs (Fig 2A) are marked in green. N-terminal deletion of PAK4a residues 1–20 (part of the CRIB region) had no effect on kinase activity. However, PAK4a lacking residues 1–50 is extremely active (Fig 2D) and also undergoes auto-phosphorylation, while PAK4–Cat(286–591) does not because A-loop Ser 474 is fully phosphorylated. Thus, PAK4 residues 20–50 contain residues essential for the functioning of the PAK4 AID: this region is conserved among all the group II PAKs, and related to AID sequences found in other PAKs (Fig 2A).
Residues 1–68 are sufficient to inhibit PAK4
Is the putative PAK4 AID sufficient of itself to inhibit the kinase domain, or do residues 69–285 have a supporting role? We generated a short synthetic PAK4(s) by joining PAK4(1–68) with a 10-residue linker to the catalytic domain (286–591) as illustrated in Fig 2C (total 384 residues). This synthetic PAK4s was fully auto-inhibited as compared to PAK4a (Fig 2D, lane 2). This experiment in combination with the deletion studies demonstrates that residues 20–68 are necessary and sufficient for PAK4 inhibition. As PAK5 and PAK6 contain the same related CRIB/AID (Fig 2A), we suggest that these kinases have the same basic mechanisms of regulation. Thus, PAK4 contains an AID positioned similarly to the PAK1 AID, but it differs in its mechanism of action because activation loop of PAK4 is constitutively phosphorylated. Nonetheless, active Δ50-PAK4a undergoes auto-phosphorylation on N-terminal sites (arrowheads).
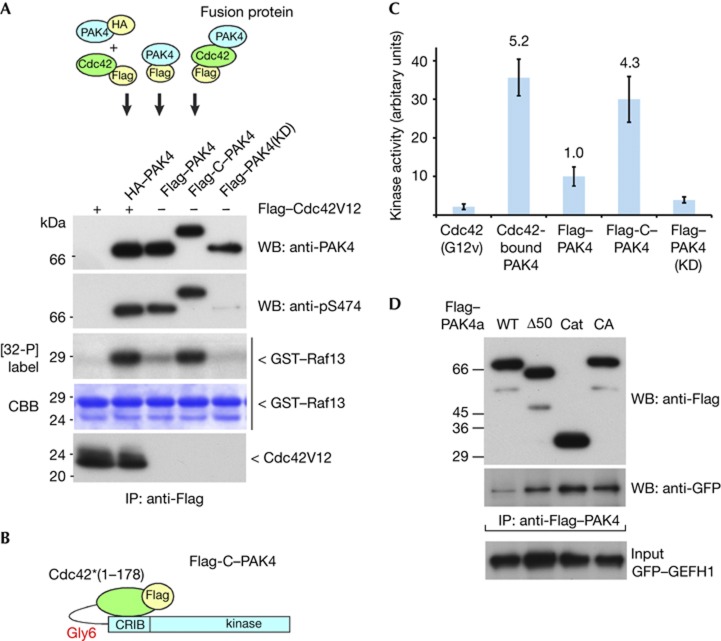
PAK4 is directly activated by Cdc42.GTP in vivo
It was originally noted that Cdc42 coexpression fails to increase PAK4 activity when subsequently tested in vitro [4]. Consideration of the sequence homology between PAKs suggested to us that Cdc42 binding does indeed regulate PAK4. To test this we immunoprecipitated haemagglutinin (HA)–PAK4 indirectly though its association with Flag–Cdc42 (G12V; as illustrated in Fig 3A). Averaged across multiple experiments HA–PAK4 bound to the Flag–Cdc42 was ∼5 times more active than Flag–PAK4 (Fig 3C). The Cdc42-bound kinase does undergoes significant auto-phosphorylation (Supplementary Fig S3A online) as for the PAK4(S445N) mutant [18]. We also generated a chimaeric Cdc42–Gly6–PAK4 (as illustrated in Fig 3B), allowing stoichiometric intramolecular activation but without Cdc42 membrane interaction (the CAAX motif being missing). This construct was ∼4-fold more active than wild-type Flag–PAK4. Finally, we show that 6His–PAK4a purified from E. coli, which is phosphorylated on Ser 474, is essentially inactive (Supplementary Fig S3B online). That recombinant full-length PAK4(S474E) is not active (lane 5) also indicating the phospho-mimetic is similarly held in an inactive state. Taken together we conclude that group II PAKs are bone fide ‘p21-activated kinases’ regulated by an AID.
Figure 3.
Cdc42.GTP directly activates PAK4. (A) A schematic of the pull-down protocol from COS7 cell lysates is shown. The PAK4 proteins were recovered as indicated, and assayed for activity towards glutathione S-transferase (GST)–Raf13 (20 μg) in the presence of 10 μM [γ32-P] ATP. The pS474 was probed using CS#3241. (B) Schematic of the active Cdc42–Gly6–PAK4 chimaera (Flag-C–PAK4). (C) The relative in vitro kinase activity of purified PAK4 tested in three independent experiments was calculated after subtracting PAK4 KD (that is, background value) and setting the Flag–PAK4 value to 1. The bars represent the standard error for each measurement. (D) The efficiency of green fluorescent protein–GEFH1 binding to various Flag–PAK4 constructs as indicated was tested by coimmunoprecipitation. CA, constitutively active; Cat, PAK4(286–591); CRIB, Cdc42–Rac interaction/binding; GFP, green fluorescent protein; GST, glutathione S-transferase; HA, haemagglutinin; IP, immunoprecipitated; KD, kinase dead; PAK, p21-activated kinase; WB, western blot; WT, wild type; Δ50=PAK4a(51–591).
PAK4 AID kinase contacts are different to those in PAK1
The PAK1 AID functions as an inhibitory ‘switch’, in which Cdc42 or Rac1 binding induces conformational changes to release some contacts with catalytic domain [1]. Subsequent PAK1 auto-phosphorylation of Thr 423 and Ser 144 sustains activity even in the absence of bound Cdc42 [20]. Critical residues of the PAK1 C-lobe αG-helix contact the AID (illustrated in Supplementary Fig S2A online) and when mutated prevent AID interaction and lead to kinase activation [23]. The PAK4 αG-helix [16] was therefore mutated at equivalent residues to those that are involved with AID contact in PAK1 (Supplementary Fig S2B online): mutation of PAK4 L521, K522 and K525 (Supplementary Fig S2C online) did not increase the activity of the full-length PAK4 (Supplementary Fig S2D online), as would be expected if the PAK4 αG-helix bound the AID. We then tested if the PAK4 AID blocks substrate access to GEF–H1, a validated PAK4 target [26]. The amount of GEF–H1 bound to PAK4 correlated with the activity status of the kinase (Fig 3D). Thus, the PAK4 AID defined in this study likely controls substrate access. Proof of this model will require structural analysis of the auto-inhibited kinase.
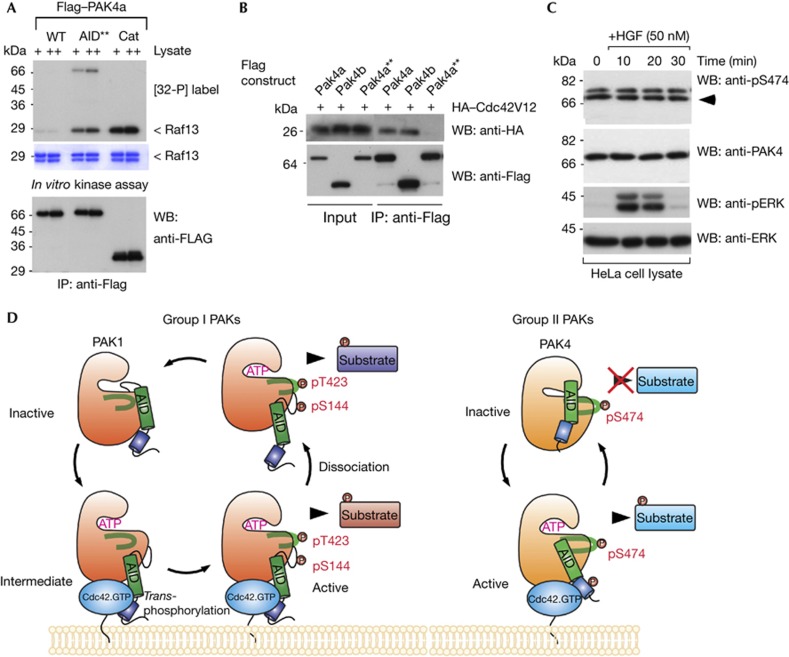
Activation does not require AID autophosphorylation
Does Cdc42 promote activation through auto-phosphorylation of the PAK4 AID? This could provide the means of disinhibiting PAK4, as already noted for PAK1 via Ser 144 [20]. Three conserved PAK4 serine/threonine residues in the AID (S41, S46 and T60) were mutated to phospho-mimetics and tested for their in vitro activity. Only PAK4a(T60E) exhibited above basal activity (Supplementary Fig S4A online), although still essentially inactive compared with ΔN50–PAK4. Thus, covalent AID phosphorylation probably does not drive PAK4 activation, which can explain why immunoprecipitated PAK4 shows similar activity when coexpressed with Cdc42 (Fig 1E). We then mutated two conserved basic AID residues (R48/49; Fig 2A green bar), which we suspected as being involved in contacting the kinase domain. This full-length PAK4a mutant designated PAK4a–AID** (see Fig 4A legend for details) was active. When tested for their ability to bind Cdc42(G12V) in cells, we noted that both PAK4a and PAK4b bind equally well to the GTPase (Fig 4B), however the PAK4a–AID** did not. Given that the R48/49 residues lie well outside the conventional CRIB region (that is, 10–35), we conclude that the CRIB and AID domains are folded into a single unit, much like with PAK1 [1]. Mutations in the AID can therefore perturb the structure of the CRIB. This is consistent with the observation that a Drosophila Mbt (HH/LL) CRIB mutant yields a more active kinase [13], exactly as seen with PAK1 [2].
Figure 4.
An auto-inhibitory domain (AID) active mutant has reduced binding to Cdc42. (A) Constructs of wild-type or mutant PAK4a as indicated were expressed in Cos7 cells, immunoprecipitated and assayed for activity in vitro. WT, wild type; AID**, RR48/49AE, T60E; Cat, PAK4(286–591). (B) PAK4a, PAK4b and the active PAK4a–AID** were immunoprecipitated and tested for the presence of coexpressed HA–Cdc42V12. (C) HeLa cells were serum-starved (16 h), treated with HGF and harvested at the times shown. Triton X-100 soluble cell lysates (30 μg/lane) were probed with new antibodies directed to PAK4, and PAK4 pS474, or ERK1/2 and pERK1/2 (Cell Signaling). (D) This model summarizes our findings regarding the alternate activation mechanisms for group I and II PAKs. Cdc42 binding in both cases results in relief from auto-inhibition, but through different underlying mechanisms. In the case of PAK1, this requires new phosphorylation of Ser 144 and Thr 423. For simplicity we do not show the PAK1 as a dimer. In contrast, PAK4 is constitutively phosphorylated on Ser 474, and requires Cdc42.GTP to induce and sustain an active kinase conformation that allows substrate interaction. HA, haemagglutinin; HGF, hepatocyte growth factor; IB, immunoblot; IP, immunoprecipitated; PAK, p21-activated kinase; WB, western blot; WT, wild type.
PAK4 is constitutively phosphorylated at Ser474
Several studies have used A-loop Ser 474 phosphorylation as a marker of PAK4 activity [22, 27, 28]. However, as illustrated in Supplementary Fig S1 online, commercial antibodies used in these studies can react more strongly with irrelevant bands. Also generic basic substrates such as histone H4 have significant levels of nonspecific phosphorylation by contaminant kinases. By contrast the glutathione S-transferase–Raf13 substrate is quite PAK selective. Our anti-pS474 antibodies (characterized in Supplementary Fig S1 online) were used to reinvestigate endogenous PAK4 after hepatocyte growth factor treatment, where PAK4 activity has a role in cell motility in response to this growth factor [14, 27]. There was no change in pS474 signal under these conditions, although robust ERK activation was confirmed (Fig 4C). Similarly immunoprecipitated PAK4 derived from LNCaP cells treated with serum or sorbitol (Supplementary Fig S4C online), which activates Cdc42 and p38 MAPK [29], showed unaltered pS474 levels. Finally, we show that immunoprecipitated PAK4 showed equivalent pS474 modification to recombinant 6His–PAK4 purified from E. coli (Supplementary Fig S4B online), which has stoichiometric phosphate on Ser 474 as confirmed by mass spectrometry (data not shown).
In summary, we show for the first time that the prototype group II kinase PAK4 is strongly auto-inhibited by an AID, and is activated by the binding of Cdc42 as illustrated in Fig 4D. The presence of a number of residues of the PAK4 AID in common with PAK1 suggest that the metazoan-specific PAK4 could have coevolved from a protozoan Ste20p-like kinase. Unlike PAK1, which is inhibited in trans [23], we find that PAK4 behaves as a monomer by gel filtration analysis (data not shown). Why has PAK4 dispensed with A-loop Ser 474 phosphorylation as a mechanism of kinase regulation? One idea is that rapid and reversible response to changes to Cdc42.GTP levels are transduced by tight spatial and temporal coupling with the PAK4 kinase. Active group I PAKs by contrast may function at sites not accessible to the membrane-bound GTPase, for example, in the nucleus. The group II PAKs, which are constitutively phosphorylated in the A-loop, are thus regulated through conformational changes in the AID. This is not unlike the situation with PKAc, which becomes phosphorylated on Thr 197 soon after synthesis in the cell, and responds primarily to changes in the conformation of the regulatory subunits [30]. This paper also highlights the pitfall of assuming that immunoprecipitated protein kinases retain a ‘memory’ of their cellular activity state.
Methods
Antibodies. Commercial anti-PAK4 antibodies (Cell Signaling, CS #3242) and anti-PAK4 pS474 (CS #3241) were used as indicated in the legends. Other panels used antibodies raised and purified against human PAK4(1–350). Anti-PAK4 pS474 was generated in rabbit using a synthetic 13mer phospho-peptide surrounding the pSer474 site, and purified under standard conditions. Rabbit anti-ERK (Cell Signaling) and pERK (Santa Cruz) were used to confirm HGF treatment.
Cloning and mutagenesis. Constructs were introduced into pXJ40-based vectors [31] contain N-terminal Flag-, HA- or green fluorescent protein-fusion tags. Truncation mutants were generated by PCR and point substitutions were generated by QuikChange (Stratagene).
In vitro kinase assays. Mammalian constructs were immunoprecipitated from COS7 cells using 20 μl of mouse M2 anti-Flag agarose beads (Sigma-Aldrich). Kinase assays (30 min at 30 °C) were performed in 50 μl buffer (with 5 mM MgCl2, 2 μCi [32-P] γATP and 10 μM ATP) including 20 μg glutathione S-transferase–Raf1(332–344). Other methods are provided in supplementary information online.
Supplementary Material
Acknowledgments
Part of this work was supported by the GSK-IMCB Research Fund.
Author contributions: Y.B. and E.M. designed the experiments that were performed by Y.B., Y.-W.N., W.S. and F.T.P.L. The manuscript was written by Y.B. and E.M.
Footnotes
The authors declare that they have no conflict of interest.
References
- Lei M, Lu W, Meng W, Parrini MC, Eck MJ, Mayer BJ, Harrison SC (2000) Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102: 387–397 [DOI] [PubMed] [Google Scholar]
- Zhao ZS, Manser E, Chen XQ, Chong C, Leung T, Lim L (1998) A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol 18: 2153–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z-S, Manser E (2005) PAK and other Rho-associated kinases—effectors with surprisingly diverse mechanisms of regulation. Biochem J 386: 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo A, Qu J, Cammarano MS, Dan C, Fritsch A, Baud V, Belisle B, Minden A (1998) PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J 17: 6527–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan C, Nath N, Liberto M, Minden A (2002) PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. Mol Cell Biol 22: 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Li X, Sharma M, Zarnegar M, Lim B, Sun Z (2001) Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J Biol Chem 276: 15345–15353 [DOI] [PubMed] [Google Scholar]
- Arias-Romero LE, Chernoff J (2008) A tale of two Paks. Biol Cell 100: 97–108 [DOI] [PubMed] [Google Scholar]
- Chen S et al. (2008) Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther 7: 1793–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BW et al. (2010) Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci USA 107: 9446–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasova T, Minden A (2011) Role for p21-activated kinase PAK4 in development of the mammalian heart. Transgenic Res [Epub ahead of print] doi:10.1007/s11248-011-9578-7 [DOI] [PubMed] [Google Scholar]
- Koh W, Mahan RD, Davis GE (2008) Cdc42- and Rac1-mediated endothelial lumen formation requires Pak2, Pak4 and Par3, and PKC-dependent signaling. J Cell Sci 121: 989–1001 [DOI] [PubMed] [Google Scholar]
- Wallace SW, Durgan J, Jin D, Hall A (2010) Cdc42 regulates apical junction formation in human bronchial epithelial cells through PAK4 and Par6B. Mol Biol Cell 21: 2996–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger D, Raabe T (2003) Mbt, a Drosophila PAK protein, combines with Cdc42 to regulate photoreceptor cell morphogenesis. Development 130: 427–437 [DOI] [PubMed] [Google Scholar]
- Wells CM, Abo A, Ridley AJ (2002) PAK4 is activated via PI3K in HGF-stimulated epithelial cells. J Cell Sci 115: 3947–3956 [DOI] [PubMed] [Google Scholar]
- Wells CM, Jones GE (2010) The emerging importance of group II PAKs. Biochem J 425: 465–473 [DOI] [PubMed] [Google Scholar]
- Eswaran J, Lee WH, Debreczeni JE, Filippakopoulos P, Turnbull A, Fedorov O, Deacon SW, Peterson JR, Knapp S (2007) Crystal structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs. Structure 15: 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Y-W, Raghunathan D, Chan PM, Baskaran Y, Smith DJ, Lee C-H, Verma C, Manser E (2010) Why an A-loop phospho-mimetic fails to activate PAK1: understanding an inaccessible kinase state by molecular dynamics simulations. Structure 18: 879–890 [DOI] [PubMed] [Google Scholar]
- Gnesutta N, Qu J, Minden A (2001) The serine/threonine kinase PAK4 prevents caspase activation and protects cells from apoptosis. J Biol Chem 276: 14414–14419 [DOI] [PubMed] [Google Scholar]
- Rennefahrt UEE, Deacon SW, Parker SA, Devarajan K, Beeser A, Chernoff J, Knapp S, Turk BE, Peterson JR (2007) Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J Biol Chem 282: 15667–15678 [DOI] [PubMed] [Google Scholar]
- Chong C, Tan L, Lim L, Manser E (2001) The mechanism of PAK activation. Autophosphorylation events in both regulatory and kinase domains control activity. J Biol Chem 276: 17347–17353 [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Siu MKY et al. (2010) p21-activated kinase 4 regulates ovarian cancer cell proliferation, migration, and invasion and contributes to poor prognosis in patients. Proc Natl Acad Sci USA 107: 18622–18627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrini MC, Lei M, Harrison SC, Mayer BJ (2002) Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol Cell 9: 73–83 [DOI] [PubMed] [Google Scholar]
- Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, Tomchick DR, Alto NM (2011) The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature 469: 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Say E, Tay H-G, Zhao Z-S, Baskaran Y, Li R, Lim L, Manser E (2010) A functional requirement for PAK1 binding to the KH(2) domain of the fragile X protein-related FXR1. Mol Cell 38: 236–249 [DOI] [PubMed] [Google Scholar]
- Callow MG, Zozulya S, Gishizky ML, Jallal B, Smeal T (2005) PAK4 mediates morphological changes through the regulation of GEF-H1. J Cell Sci 118: 1861–1872 [DOI] [PubMed] [Google Scholar]
- Paliouras GN, Naujokas MA, Park M (2009) Pak4, a novel Gab1 binding partner, modulates cell migration and invasion by the Met receptor. Mol Cell Biol 29: 3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lock JG, Olofsson H, Kowalewski JM, Teller S, Liu Y, Zhang H, Strömblad S (2010) Integrin-mediated cell attachment induces a PAK4-dependent feedback loop regulating cell adhesion through modified integrin αv β5 clustering and turnover. Mol Biol Cell 21: 3317–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PM, Lim L, Manser E (2008) PAK is regulated by PI3K, PIX, CDC42, and PP2Cα and mediates focal adhesion turnover in the hyperosmotic stress-induced p38 pathway. J Biol Chem 283: 24949–24961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Buechler JA, Yonemoto W (1990) cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem 59: 971–1005 [DOI] [PubMed] [Google Scholar]
- Manser E, Huang HY, Loo TH, Chen XQ, Dong JM, Leung T, Lim L (1997) Expression of constitutively active aPAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol 17: 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.