Abstract
The development of T cells in the thymus involves multiple differentiation and proliferation events during which hematopoietic precursors give rise to T cells responding to antigen stimulation and ready for effector differentiation. This review addresses signaling and transcriptional checkpoints that control the intrathymic journey of T cell precursors. We focus on the divergence of αβ and γδ lineage cells and the elaboration of the αβ T cell repertoire, with special emphasis on the emergence of transcriptional programs that direct lineage decisions.
Introduction
The T cell arm of the immune system is essential for responses against infections. Multiple T cell subsets are distinguished based on the composition of their T cell antigen receptor (TCR) (αβ or γδ), their antigenic specificity and effector potential (Fig. 1). αβ T cells constitute the bulk of T cell populations in lymphoid organs and generally react against peptides presented by MHC-I or MHC-II molecules, whereas γδ T cells are generally not MHC-restricted and seem involved in the surveillance of microbial and non microbial tissue stress1. Unlike other immune cells, T cell develop in the thymus, through a process that can be separated into three broad steps (Fig. 1). The first spans from thymic colonization to T cell commitment, from where the second starts and leads to the divergence of αβ and γδ lineages. The third step sees αβ and γδ lineage cells complete their differentiation and acquire immunological properties, and in some cases effector functions. For most αβ lineage cells, this step is dominated by MHC-induced selection and results in the differentiation of thymocytes (‘single positive’, SP) that express either CD4 or CD8, two molecules that contribute to TCR recognition of MHC-II and MHC-I, respectively; such SP thymocytes are the direct precursors of mature T cells (Fig. 1).
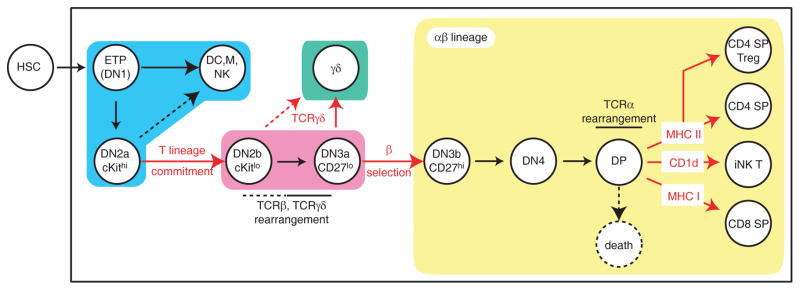
Figure 1. overview of T cell development.
Thymic developmental stages are depicted. Expression of CD4 and CD8 separates CD4−CD8− double negative (DN), CD4+CD8+ double positive (DP) and cells expressing either coreceptor (single positive, SP), whereas the expression of CD44 and CD25 defines four DN subsets: CD44+CD25− [DN1], CD44+CD25+ [DN2], CD44−CD25+ [DN3], and CD44−CD25− [DN4]. The earliest precursors, known as ETP, that enter the thymus from the bone marrow are part of an heterogeneous DN1 subset that includes both subsequent intermediates in the T differentiation pathway and cells belonging to other lineages175. The DN2 and DN3 subsets are themselves divided into two stages based on the expression of the receptor cKit and of CD27, respectively. Critical checkpoints addressed in the text are shown in red. Rounded rectangles group cell subsets according to the key developmental step they belong to: early uncommitted progenitors (blue), T committed progenitors before the separation of αβ and γδ lineages (purple) and committed αβ (yellow) or γδ (green) lineage cells.
This review discusses checkpoints that control the journey of T cell precursors in the thymus. After a brief overview of events that precede T cell commitment, we focus on the divergence of αβ and γδ lineages and the differentiation of αβ T cells. Work from many laboratories over the last few years has put the spotlight on transcriptional ‘circuits’ that control intrathymic checkpoints, and we have placed special emphasis on these emerging transcriptional ‘circuits’ and tried to connect them to intrathymic signals that direct lineage decisions. We refer the reader to recent reviews for important aspects of intrathymic development that are not covered here, including the mechanisms of antigen receptor rearrangement2.
From thymus settling to T cell commitment
While multiple types of progenitors can generate T cells under experimental conditions3–6, recent evidence favors a model whereby the physiological thymic ‘settlers’, referred to as early thymic progenitors (ETP), are uncommitted cells that retain some myeloid but little if any B-lineage potential7,8, although the intrathymic environment normally restrains their myeloid development9. Thymic colonization involves the chemokine receptor CCR9, probably redundantly with CCR7, and PSGL1, a ligand for P-selectin expressed on the thymic epithelium10–12. The loss of multipotency that defines T commitment is a gradual process. It occurs in ‘double negative’ (DN) thymocytes, that do not express CD4 or CD8, a subset itself separated into four sequential phenotypic stages (DN1 to DN4) based on expression of CD44 and CD25, and is not complete before the DN2 stage (Fig. 1).
T lineage commitment involves the sustained repression of alternate gene expression programs characteristic of other lineages. In early thymocytes (up to the DN2 stage), this requires signaling and transcriptional activation by Notch1 upon engagement by its ligand Delta-like 4 (DL4) expressed on the thymic stroma13,14. Such requirement for Notch1 in early precursors is demonstrated by both loss- and gain-of-function analyses15,16. However, even though a constitutively active version of Notch1 can promote extrathymic T cell differentiation, Notch1 is necessary but not sufficient for T cell commitment under physiological circumstances17,18. Other transcription factors, including Runx1, Gata3, and E-box proteins, cooperate with Notch1 to initiate T cell differentiation19. In addition, several factors impinge on Notch1 signaling activity, including the zinc finger transcription factor LRF that restrains Notch signaling in bone marrow progenitors20, and post-translational modifiers of Notch1’s extra-cellular domain21. It is also important to note that Notch1 function is not limited to the exclusion of other developmental fates, as it promotes T lineage-specific gene expression and cell survival, and stimulates cell metabolism (see below)17,22.
Whether and how Notch1 contributes to T cell commitment in DN2 cells is not yet known. In any case, its expression subsides during the late stages of T cell development23,24, and it does not seem involved in maintaining lineage integrity in late thymocytes and mature T cells. Whether a single factor serves such a function in the T lineage, as Pax5 does in B cells25, and its identity, are the focus of ongoing research.
The divergence of αβ and γδ lineages
β-selection and the choice of the αβ lineage
Although incomplete rearrangements at the TCRβ and γ loci occur in pre-commitment thymocytes, the critical rearrangement of variable gene segments occurs in T-committed DN3 cells26. Because of the deletion and untemplated addition of nucleotides during Rag-mediated recombination, most rearrangements are out-of-frame and fail to give rise to genes encoding a functional protein. The first checkpoint that committed thymocytes encounter, at the DN3 stage, precisely verifies proper TCR gene rearrangement.
For αβ precursors, this checkpoint is known as β-selection, and probes in-frame TCRβ gene rearrangement. It requires thymocytes to signal through a pre-TCR consisting of the product of a properly rearranged TCRβ gene, CD3 chains and pre-Tα, that binds TCRβ in the absence of rearranged TCRα27,28. Although the pre-TCR is not detectable at the cell surface, and is not believed to recognize any ligand29, it signals, possibly through oligomerization30, using intra-cellular intermediates similar to those triggered by TCR complexes in mature T cells. At least two additional signals contribute to β-selection: CXCR4, a receptor for the cytokine SDF31,32, and Notch1. Notch1 promotes cell survival and metabolism through activation of the PI-3 kinase and Akt pathway, and contributes to overcome the differentiation block enforced in DN3 thymocytes by E2A proteins (see below)33.
Thymocytes that ‘pass’ β-selection enter the last proliferative burst they will encounter in the thymus. They initiate CD4 and CD8 expression (becoming ‘double positive’, DP) and TCRα gene rearrangement, resulting in the surface expression of TCRαβ complexes, whereas they cease to express receptors characteristic of hematopoietic cells, marking their ‘coming of age’ as immune cells. They also become unresponsive to cytokine signals, and specifically to IL-7 expressed by the thymic stroma and critical to early T cell development34, due to their down-regulation of IL-7Rα and their expression of the inhibitor of cytokine signaling SOCS-135.
αβ vs. γδ lineage choice
Developing γδ cells proof-read their TCR rearrangement at a DN3 checkpoint similar to β-selection, although differing in two important respects. First, unlike for αβ cells, there is no ‘pre-TCRγδ’ and the DN3 checkpoint assesses signaling by complete TCRγδ complexes. Second, γδ lineage differentiation does not depend on Notch signaling36. How DN3 cells distinguish pre-TCR from TCRγδ signals, and make the corresponding developmental decisions, has attracted much attention37. In a simple perspective, TCRγδ signals would ‘instruct’ DN3 thymocytes to become γδ T cells, whereas pre-TCR signals would cause differentiation into DP cells. While the importance for β-selection of a proline-rich motif in the pre-Tα cytosolic tail agrees with this view27,28, the ‘instructive’ perspective is challenged by the fact that each type of receptor can in certain circumstances direct development of the other lineage. Expression of αβ transgenic TCRs can ‘redirect’ cells into the γδ lineage38, whereas disruption of the TCRβ gene, which prevents pre-TCR expression, results in the generation of small populations of γδ thymocytes that express CD4 and CD8, the hallmark of αβ precursors39.
These findings have thrown the ‘instructive’ perspective into disfavor, and the current view is that γδ TCR signals are ‘stronger’ than pre-TCR signals, and that such ‘strong’ signals promote γδ lineage choice. Indeed, impairing TCRγδ signaling, through the expression of signaling-defective CD3ζ chains, causes the differentiation of TCRγδ-bearing DP thymocytes40. It is not yet clear why TCRγδ signals would be stronger than pre-TCR signals; notably, both receptors differ from TCRαβ as they do not require CD3δε to signal41,42. Two complementary hypotheses are that greater signaling by TCRγδ results from its higher expression compared to pre-TCR, and from engagement by intrathymic ligands. Indeed, thymocytes expressing a transgenic TCRγδ receptor develop into γδ T cells in mice expressing the receptor ligand, but adopt an αβ fate when the ligand is absent43. There is additional evidence that γδ T cell development requires engagement by an intrathymic TCR ligand44, and it seems reasonable to assume this to be the rule, even though the identity of such ligands remains in most cases unknown.
Whether specific environmental signals direct thymocytes towards the γδ fate before the DN3 checkpoint (e.g. by affecting TCR gene rearrangement) has been a contentious issue37,45. The strong IL-7 requirement for TCRγ but not TCRβ gene rearrangement26, and the remarkable emergence of waves of γδ T cells with mono- or oligo-clonal receptors carrying invariant rearrangements during embryonic development, are suggestive of ‘induced’ events. The complete loss of αβ but not γδ T cells caused by inactivation of the transcription factor Bcl11b is also consistent with the idea of an early divergence, as the block in αβ thymocyte development occurs at the DN2 stage before the completion of TCRβ rearrangement46. However, these findings are also consistent with distinct requirements for the survival or amplification of αβ vs. γδ T cells. Indeed, recent single cell analyses support the idea that γδ lineage commitment is solely the result of TCR signaling ‘strength’ at the DN3 checkpoint47. As recently highlighted48, the heterogeneity of γδ T cells challenges the idea that they form a single lineage, and it is conceivable that distinct rules apply to distinct γδ subsets.
Transcriptional control
How TCR or pre-TCR signals impinge on the transcriptional circuitry in DN3 thymocytes is coming to light49. E-box binding proteins of the E2A family have emerged as a key target of these signals. This activity, contributed in developing T cells by the two molecules E47 and HEB, and that we will refer to as ‘E2A’, blocks development at the DN3 stage50. Cells use two complementary mechanisms to overcome this block: reduced expression of the genes encoding E47 or HEB, or increased expression of Id-family molecules, that inhibit E2A protein function51. A member of this family, Id3, is preferentially up-regulated in γδ lineage cells52. Although Id3 is also a target of pre-TCR signals, it has been proposed that the extent of Id3 up-regulation depends on signal strength, and distinguishes between pre-TCR and TCRγδ signals52. High Id3 expression in γδ cells would suffice to overcome the ‘E2A block’, whereas lower Id3 levels in pre-TCR signaled cells could do so only in cooperation with Notch1-induced down-regulation of E2A expression36,43,52–54. This model, which provides an elegant rationale for the differential Notch requirement of αβ vs. γδ lineage cell differentiation53, predicts that Id3 disruption would impair γδ T cells development. While that is the case in fetal thymi, Id3 disruption unevenly affects adults γδ thymocyte subsets, and actually enhances the pool of γδ thymocytes displaying effector function52,55. Thus, additional studies will be needed to fully comprehend the role of Id3 in γδ lineage differentiation52,55,56.
While Id3 emerges as a possible ‘sensor’ converting signal strength into distinct transcriptional outcomes50, how it is linked to ‘effector’ transcription factors that are selectively required for the differentiation of DN3 cells into αβ or γδ lineages remains to be determined. Three effector activities specifically or preferentially contribute to the generation of DP thymocytes from ‘β-selected’ cells. Runx1 is necessary for the proliferative burst that follows β-selection, but not for the generation of γδ T cells57. RORγt, encoded by the Rorc gene, is a hallmark of DP thymocytes in the thymus (in addition to its other functions in the immune system) and notably promotes their survival by up-regulating expression of the anti-apoptotic protein Bcl-XL58. However, there is evidence that Rorc expression depends on E2A and inhibits thymocyte proliferation59, suggesting that it may not directly be induced by pre-TCR signals, which reduce E2A activity and promote proliferation, or that its induction by pre-TCR signals is delayed by the concurrent up-regulation of the transcription factor Egr359. The HMG protein TCF1 (encoded by the Tcf7 gene, and partly redundant with the related factor LEF) is needed for the generation of DP thymocytes, whereas the role of its ‘conventional’ partner β-catenin is not yet fully delineated60–64.
Reciprocally, Sox13, another HMG transcription factor, inhibits the generation of αβ T cells and is important for the development of at least some γδ T cells65, in part by antagonizing the function of TCF1, even though it remains unclear whether Sox13 contributes to γδ lineage ‘choice’ per se. Members of the Egr transcription factor family (Egr1, Egr2 and Egr3) are triggered by both pre-TCR and TCRγδ signals; high expression of these factors promotes γδ T cell development, possibly by repressing RORγt expression59 and as intermediates for Id3 up-regulation43,52. Whether such factors, and others preferentially expressed in γδ T cells66, contribute to γδ lineage differentiation per se or promote the survival or expansion of γδ T cell subsets remains to be determined.
The elaboration of the αβ T cell repertoire
Positive and negative selection of αβ lineage cells
The rest of this review discusses the differentiation of αβ lineage cells, that emerge from β-selection as DP thymocytes and are the precursors of conventional CD4 and CD8 cells. Three key events mark the developmental progression of these cells: (i) positive selection, the rescue from programmed cell death of DP thymocytes whose TCRαβ productively interacts with self MHC peptide complexes (MHCp) expressed by the thymic epithelium (or with other MHC or MHC-like molecules)67 (ii) negative selection, the elimination of self-reactive cells, and (iii) acquisition of functional competence, notably marked by the termination of either CD4 or CD8 expression (‘lineage differentiation’) and its matching to MHC specificity.
The need for positive selection is a direct consequence of the random nature of TCR rearrangement and of the high diversity of MHC alleles. As a result, most DP thymocytes in a given individual fail to productively interact with MHCp and die by ‘neglect’ within a few days, even though evolutionary pressure has resulted in a ‘germline-encoded’ MHC reactivity of TCR variable regions, thereby increasing the yield of positive selection68,69. CD4 and CD8 molecules provide an additional guard against the selection of non-MHC reactive cells, as they sequester the tyrosine kinase Lck required to initiate TCR signaling, thereby restricting its activity to TCRs engaged by MHCp, which, unlike non MHC-ligands, co-engage CD4 or CD870.
The avidity of T cells for MHC-bound self peptides, that underpins positive selection, is a correlate of their reactivity against MHC-bound foreign peptides. Thus, mechanisms have evolved to prevent the development of T cells with overt reactivity against MHC-bound self peptides, or to redirect such cells towards immune suppression. Thymocytes carrying receptors with the highest avidity for MHCp undergo TCR-induced programmed cell death (negative selection), a process essential for central tolerance71. It notably involves the exposure of thymocytes to tissue-specific antigens ectopically expressed by medullary epithelial cells in a manner dependent on the transcription factor Aire72. In addition, there is evidence that Foxp3 expressing thymic-derived Treg cells, another essential facet of central tolerance, express receptors with higher avidity for self than conventional MHC II-restricted cells73,74. We will not address further these issues, despite their obvious importance for the elaboration of the T cell repertoire, as they have been recently discussed71. Instead, the rest of this section will focus on how positively selected thymocytes ‘sense’ low avidity MHCp, and on the transcriptional circuitries that convert TCR signals into lineage decisions.
Setting the signaling threshold
Thymocytes that undergo positive selection, including those that are not subsequently eliminated by negative selection, undergo phenotypic changes that resemble those of mature T cells upon stimulation by their cognate antigen. While these changes in thymocytes do not lead to cell proliferation and effector differentiation, this raises the question of why MHCp that promote positive selection do not cause a similar ‘abortive’ activation program in mature T cells. One obvious possibility is that the set of peptides generated by thymic epithelial cells, that present positively selecting MHCp, differs from that of professional bone marrow derived APCs, which post-thymic T cells interrogate in their quest for antigen. There is evidence that this is the case, and that may contribute to the unique reactivity of thymocytes (and to reduce the footprint of negative selection over the positively selected repertoire)75–77. However, DP thymocytes are intrinsically more sensitive than mature T cells to low-avidity MHCp78–80, raising the question of what lowers the threshold to TCR engagement in DP thymocytes.
In addition to CD5, which is thought to ‘tune’ the sensitivity of mature and immature T cells to TCR signals81, recent studies have highlighted the potential role of MicroRNA (miR) 181a in DP thymocytes82. MiR181a is highly expressed in immature thymocytes, and rapidly down-regulated after positive selection, and both gain and loss-of-function analyses supports its role in raising thymocyte sensitivity to TCR engagements. MiR repress gene expression by targeting messenger RNA (mRNA) for degradation or translational repression: while the full set of targets of miR181a remains to be identified, it impairs expression of phosphatases that inhibit TCR signaling, and notably that de-phosphorylate Erk kinases. Erk kinases have been proposed to have increased responsiveness to TCR engagement in DP thymocytes, as a result of calcineurin-mediated signaling in cells undergoing β-selection83; it is possible that this is due to miR181a, although its expression is not increased by β-selection82. It will be important to confirm the role of miR181a genetically, and to examine how disruption of miRNA generation affects positive selection84–86. Nonetheless, miRNAs are required for the generation of Treg cells87 and the terminal differentiation of CD8 cells86,87, suggesting that the repertoire of miRNA functions in the thymus is just being discovered88.
Control of gene expression in DP thymocytes
Identifying the gene expression programs that ‘orchestrate’ the differentiation of DP thymocytes into mature T cells has proven a difficult endeavor. This is due in part to our incomplete understanding of the underlying effector mechanisms. Thus, although rescue from cell death defines positive selection, how TCR signals prevent thymocyte apoptosis is not yet well understood. TCR signaling increases expression of the anti-apoptotic molecule Bcl-2, but Bcl-2 is not required for T cell development89,90, presumably because of functional redundancy with other members of the Bcl-2 family, including Mcl191,92. Another complicating factor is the multiplicity of events induced by TCR signaling in thymocytes, in addition to positive selection per se. These include (i) the expression of the chemokine receptor CCR7, that promotes the migration of thymocytes from the cortex to the medulla93–95, (ii) the acquisition of mature T cell gene expression programs, notably the expression of IL-7Rα96, and (iii) the differentiation into the CD4 or the CD8 lineage97,98. While it could be conceived that specific signal transduction pathways trigger these events in DP thymocytes, there is little or no evidence that this is the case. Rather, the signaling cascades immediately downstream of TCR appear similar to those in mature T cells, including the Erk kinase and the calcineurin-NFAT pathways that are both required for positive selection99–101.
Evidence is emerging that these cascades initially target a set of ‘sensor’ transcription factors that, most likely indirectly101, control the expression of effectors molecules involved in the generation of mature αβ T cells, including IL-7Rα and CCR7. As in DN3 thymocytes, E2A is a primary target of these sensors, reflecting its keystone role in the transcriptional circuitry of DP cells (Fig. 2A). E2A activity promotes TCR gene rearrangement and expression of genes characteristic of immature thymocytes (including CXCR4)102, and contributes to repress genes characteristic of mature thymocytes, including Foxo1, Klf2, IL-7Rα and CCR7103. The upregulation of Id3, through a cascade involving the Erk pathway, its direct target Elk4, and Egr proteins104,105, is an early consequence of TCR signaling (Fig. 2A); the ensuing reduction in E2A activity appears as a key step in the transition from pre-selection DP to post-selection SP thymocyte. Of special interest are mechanisms that restrain Foxo1 and Klf2 expression in DP thymocytes, given the role that these factors play in T cell survival and homeostasis. How E2A contributes to such repression103, and whether positive factors induced by TCR signals contribute to Foxo1 up-regulation, remain to be determined (Fig. 2B).
Figure 2. transcriptional circuitries in differentiating αβ T cells.
In pre-selection DP thymocytes (left), E2A is thought to promote expression of Rorc and Rag genes, thereby ensuring TCR gene receptor expression and TCRα locus accessibility. In addition, E2A restrains expression of IL-7Rα, Bcl-2 and CCR7, although it is not clear whether such effects are direct or indirect (e.g. through effects on Foxo1 expression). Positive selection signals (right) reduce E2A activity by increasing Id3 expression, indirectly through Erk-dependent up-regulation of Egr proteins. Positively selected thymocytes have ceased expression of DP-stage genes and up-regulated Bcl-2, CCR7 and IL-7Rα, presumably as a result of the termination of E2A activity and of the induction of activators that may include Foxo1. Arrows or block signs do not necessarily indicate direct effects.
CD4-CD8 lineage differentiation
In addition to being rescued from cell death (positive selection per se) TCR signaled thymocytes undergo functional differentiation into mature T cells. One key aspect of this process is the differentiation into the CD4 or CD8 lineage. This includes the termination of expression of either coreceptor, and the initiation of gene expression programs characteristic of helper (CD4) or cytotoxic (CD8) cells, two events that have long been recognized as being mechanistically coupled106,107. Because a functional immune system requires that this lineage ‘decision’ be matched to MHC specificity, so that MHC II-restricted thymocytes become helper CD4 cells, and MHC I-restricted thymocytes cytotoxic CD8 cells, its mechanisms have attracted much attention over the last two decades. The last few years have seen significant progress in the elucidation of the transcriptional circuits that promote CD4 or CD8 differentiation, that we will discuss first.
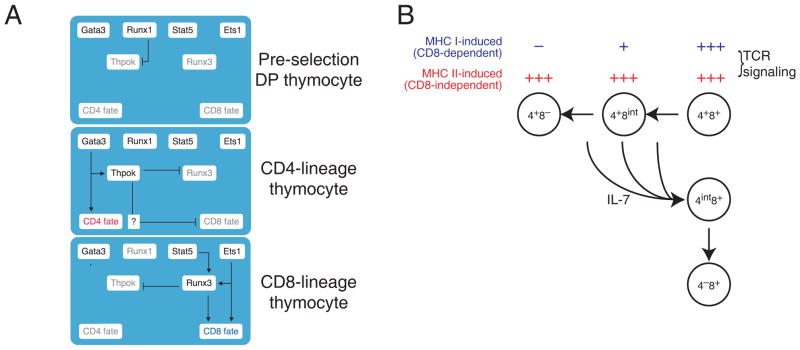
Two transcription factors, Thpok and Runx3, specifically expressed in CD4 and CD8 differentiating thymocytes, respectively, are important for this process108–111. Thpok is required for CD4 commitment and acts at least in part by repressing expression of CD8 lineage genes, including Runx3110,112–114. Runx3 is important for the silencing of Cd4 in CD8 cells108,109, and the complete disruption of Runx activity (that is of Runx3 and of the partly redundant factor Runx1) prevents CD8 cell development57,113,115. While this effect is due in part to unrestrained Thpok expression in Runx-deficient thymocytes115, cells lacking both Runx and Thpok activities fail to become CD8, indicating a specific role of Runx proteins in CD8-lineage differentiation113. These findings have led to the proposal that a dual negative regulatory loop involving Thpok and Runx3, which mutually prevent expression of each other, results in lineage commitment (Fig. 3A).
Figure 3. CD4-CD8 lineage differentiation.
(A) Components of the transcriptional circuitry that promotes CD4-CD8 differentiation are schematically depicted and interconnected at three stages of T cell development. In preselection cells, Runx1-nucleated activities repress Thpok expression. In CD4-differentiating cells, Runx1-mediated Thpok expression is relieved, although Runx1 is still expressed in CD4 cells in which it binds the Thpok gene. Gata3 promotes both Thpok expression and additional developmental events required for CD4 cell differentiation. Thpok prevents Runx3 up-regulation and CD8 differentiation. In CD8-differentiating cells, Thpok repression is maintained, presumably through Runx3. Ets1 promotes Runx3 expression, and binds the Runx3 locus, whereas Stat5 has been reported to relay IL-7 signaling to Runx3131. Grey lettering indicates factors not expressed at a particular stage. Other factors (including Tox) are omitted for clarity. Arrows or block signs do not imply direct effects.
(B) The ‘kinetic signaling’ model of lineage differentiation posits that intrathymic TCR signaling, regardless of MHC specificity, represses Cd8 expression, causing thymocytes to adopt a CD4+CD8int surface phenotype. TCR signaling in MHC II-restricted thymocytes is not affected by CD8 down-regulation, and its persistence eventually seals CD4 commitment. In contrast, TCR signaling in MHC I-restricted thymocytes is impaired by CD8 down-regulation, and its cessation causes ‘coreceptor reversal’ i.e. the cessation of Cd4 expression and the resumption of Cd8 expression.
A critical question is how thymocytes initiate expression of Runx3 and Thpok, neither of which is expressed in preselection DP cells. There is little information yet available for Runx3. The transcription factor Ets1 binds Runx3 and promotes its expression in CD8-lineage cells, but Ets1 is not CD8-lineage specific and its function in CD8-lineage commitment remains to be clarified116. Our understanding of Thpok gene regulation is more advanced. Thpok expression is limited to CD4-lineage cells by an upstream regulatory element with both positive and negative (‘silencing’) functions115,117. The binding of Runx molecules to this element is required, although not sufficient, for its silencing activity in DP thymocytes115,117, and ongoing efforts search for additional factors that contribute to prevent Thpok expression in DP and MHC I-restricted thymocytes, including the zinc finger protein Mazr118. The impaired CD4-differentiation of thymocytes deficient for E2A and HEB could suggest that this activity is required for Thpok expression103. However E2A activity is important for Cd4 expression in DP thymocytes103. Thus, it is possible that the reduced CD4 expression in E2A-deficient DP cells, rather than an intrinsic E2A requirement for Thpok expression, accounts for the impaired CD4-differentiation of E2A-HEB deficient thymocytes103.
While Thpok and Runx are necessary for lineage commitment, they are not sufficient. Notably, both the transcription factor Gata3 and the HMG protein Tox are required for CD4 cell differentiation and Thpok expression112,119,120. Although Gata3 binding to the Thpok locus112 and its preferential expression in MHC II-signaled thymocytes121 make it a good candidate as an ‘inducer’ of Thpok expression, whether Gata3 acts by relieving Runx-dependent Thpok repression remains to be elucidated. Of note, a Thpok transgene fails to rescue Gata3-deficiency in CD4-differentiating cells, indicating a role for Gata3 beyond its ability to promote Thpok expression112.
How intrathymic signals set this transcriptional circuitry in motion so that it matches lineage differentiation with MHC specificity has attracted much interest122–124. Although it was initially envisioned that specific signals induced by MHC-I or MHC-II molecules, and transduced through CD4 and CD8 could ‘instruct’ lineage differentiation125,126, recent evidence favors the possibility that the kinetics of TCR signals play a decisive role in lineage ‘choice’98. Genetic analyses supports the concept that ‘longer’ TCR signals are required for CD4- than for CD8-lineage commitment98,127,128, specifically that CD4-lineage commitment requires TCR signals to persist until the cessation of CD8 expression, whereas no reciprocal requirement exists for CD8-lineage commitment. The ‘kinetic signaling’ model of lineage choice (Fig. 3B) proposes that asymmetric changes in CD4 and CD8 expression induced by TCR signaling in DP thymocytes cause MHC II-induced TCR signals to persist longer than those induced by MHC-I128–130, and ongoing research is directed at determining how TCR signals affect Thpok or Runx3 expression. In addition, the transcription factor Stat5, a key messenger of IL-7 signals, is important for CD8 but not CD4 T cell development and, unexpectedly acts in a manner redundant with the related protein Stat6131; these findings underscore the unique role of IL-7 in the generation of CD8 cells131,132.
Terminal maturation
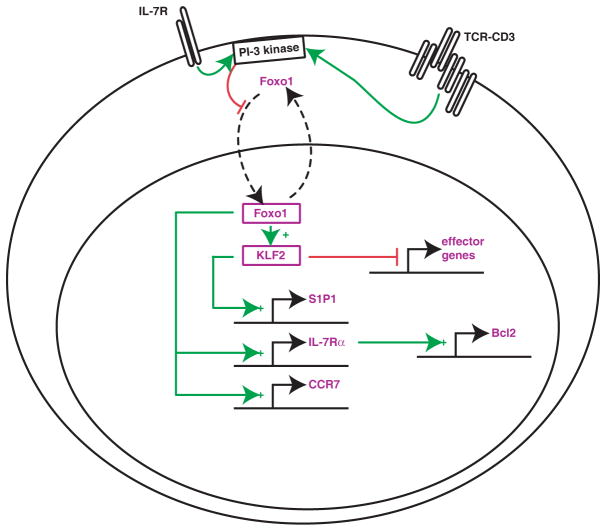
In addition to CD4-CD8 differentiation, the functional differentiation of thymocytes includes aspects that appear common to both lineages, notably controling thymic egress and cell quiescence. The zinc finger transcription factor Klf2 is essential to both phenomena, as it promotes expression of surface molecules involved in T cell trafficking, including L-selectin and the receptor for sphingosine 1 phosphate that is required for mature thymocytes to enter the bloodstream133,134 and restrains effector cytokine production in naïve T cells135. Foxo1 is also essential for in the survival and homeostasis of mature T cells, notably because it promotes expression of IL-7Rα 136,137 and of Klf2137,138 (Fig. 4). While redundancy among Foxo factors has so far prevented an exhaustive study of their role in the thymus139, it is possible that they act similarly in mature thymocytes, and preliminary evidence suggests that this is the case133,134. The post-translational control of Foxo1 activity adds a critical layer of regulation139. In mature T cells, IL-7 signaling inhibits Foxo1 nuclear translocation and therefore its activity in a PI3-kinase dependent manner (Fig. 4). While it is possible that the same happens in thymocytes, TCR signals also activate PI3 kinase140, suggesting a scenario whereby TCR signals would act in a dual fashion to control Foxo1 and its targets: they would promote Foxo1 expression, but need to subside to allow it to stimulate gene expression. While hypothetical, such a mechanism would prevent Foxo1 activity in, and therefore thymic egress of, self-reactive thymocytes undergoing continued TCR signaling in the medulla.
Figure 4. An hypothetical transcriptional network in mature thymocytes and T cells.
Based on analyses in mature T cells, a transcriptional circuitry is proposed in mature thymocytes, that enables expression of IL-7Rα, CCR7 and Klf12, which itself controls thymic egress by increasing expression of the receptor for sphingosine 1-phophate (S1P1), T cell trafficking and quiescence. Foxo1 activity is inhibited by PI-3 kinase-dependent phosphorylation, that promotes its sequestration in the cytosol, contributing to the self-limiting IL-7Rα expression characteristic of mature T cells176. It may also act as a ‘licensing’ factor in the thymus to prevent the release of self reactive thymocytes due to their persistent TCR signaling.
Invariant iNK T cell differentiation
While the vast majority of αβ T cells are restricted by MHC-I or MHC-II molecules, small subsets are selected on other MHC or MHC-like molecules. The most abundant of these, invariant NK T cells (iNK T) recognize lipid-bound MHC I-like CD1d molecules141,142. These cells carry TCRs made of a nearly invariant TCRα chain (Vα14Jα18 in mice) associated to a small set of TCRβ partners (Vβ8.2, Vβ7 or Vβ2). Although NK T cells account for a minor fraction of mature thymocyte and T cell populations in the spleen or LN, they accumulate in non lymphoid tissues, including the gut and liver where they form a large fraction of resident T cells and are stimulated by macrophages143,144. Like all αβ T cells, NK T cells are generated in the thymus from DP thymocytes145. Unexpectedly, although they are selected by MHC I-like CD1d, they fail to express CD8 and often retain CD4 expression (although a subset will eventually lose CD4 to become DN).
The selection of iNK T cells differs from that of conventional αβ T cells in three key respects142,146. First, CD1d selecting molecules are expressed on DP thymocytes, not on the cortical epithelium. Second, there is an additional requirement for signaling through homotypic interactions of SLAM family receptors, requiring activity of the SAP adaptor and the Fyn tyrosine kinase. Although the potential redundancy between SLAM family members and their close genetic linkage complicates genetic analyses, SLAMF1 and SLAMF6-deficient thymocytes demonstrate partial defects in iNKT cells implying a role in the development and function of these cells147. Third, these cells expand in the thymus, presumably as a consequence of TCR and SAP-mediated signaling and subsequently acquire effector properties, including expression of NK receptors and production of cytokines such as IL-4 and IFNγ.
The unique properties of iNK T cells are dependent on the transcription factor PLZF148,149. Unlike CD1d or SLAM-SAP signals, PLZF is not required for iNK T cell development, as iNK T cells can be detected in PLZF-deficient mice; however, such cells are in vastly reduced numbers, they fail to exhibit the high-level expression of NK receptors and cytokines that is typical of their wild-type counterparts, and, similar to conventional T cells, they display a naïve phenotype and home to lymph nodes and spleen.
Future challenges
Signals and circuits in developing thymocytes
The preceding pages have highlighted the considerable progress made over the last few years in our understanding of the signals and circuits that control thymocyte development. These years have also seen the emergence of new questions, including the role of pre-senilins, a class of enzymes notably involved in Notch signaling, in positive selection150, and the discovery of Themis151–155, the prototype a newly identified class of molecules. Themis is required for positive selection and the generation of a normal T cell repertoire, and that requirement appears quite specific as Themis-deficient mice do not display other overt phenotypic defects. While there is no consensus yet on whether Themis directly promotes TCR signal transduction, or serves downstream in the cytosol or nucleus, its disruption impairs TCR signaling during the DP to SP transition. In fact, the developmental arrest of Themis-deficient thymocytes resembles that of thymocytes engineered to cease TCR signaling during the DP to SP transition156, suggesting that Themis functions in the kinetic integration of TCR signals over time, or in tuning thymocyte sensitivity to low-avidity ligands during positive selection.
Molecular bases of commitment
A recurring theme in the preceding pages is that circuitries operating at binary checkpoints promote the expression of ‘commitment’ factors that seal lineage fate, with Runx3 and Thpok at the CD4-CD8 checkpoint illustrating this thinking. While the identification of such factors, most notably of a putative T commitment factor, is a major objective of current research, a broader question is how they maintain lineage ‘integrity’, i.e. prevent the re-emergence of alternate gene expression programs in committed cells. CD4-CD8 differentiation offers a striking example, where the circuitry that decides commitment in the thymus is ‘recycled’ during CD4 cell effector differentiation157. While Thpok is important to prevent re-expression of CD8-lineage genes in post-thymic CD4 cells158, epigenetic mechanisms also contribute to maintaining mature T cell CD4 or CD8 identity, including the silencing of either coreceptor gene158,159. Much is expected from current investigations that aim at determining what underpins epigenetic control, including histone methylation, nuclear localization, and at delineating the respective role of epigenetic and direct transcription control, especially in light of recent progress in cell ‘reprogramming’160.
The biology of quiescence
Aside from these mechanistic issues, the past years have brought new tools to tackle unanswered ‘biological’ questions. One key issue is what controls quiescence in thymocytes and T cells. Following the discovery that PLZF is critical for the acquisition of effector properties by iNK T cells, it was soon realized that its expression is not limited to that lineage. Indeed, subsets of γδ T cells express PLZF, and transgenic expression of PLZF seems to confer an ‘innate-like’ effector-type phenotype to developing thymocytes55,161,162. As recurrent themes are emerging, notably that such cells often react against low-diversity foreign antigens or self determinants released by stress tissues163, and that their development involves homotypic SLAM-SAP signals and cytokine signaling164,165, it will be important to examine whether this extends to other effector subsets in the thymus. The potential impact of such ‘innate-like’ populations is illustrated by the recent discovery that their increased number in mice with impaired activity of Tec-family tyrosine kinases164,165 promotes in trans the effector differentiation of conventional CD8 thymocytes (Kristin Hogquist, personal communication). The other side of the question is to identify the specific factors that maintain quiescence in ‘conventional’ thymocytes and mature T cells, with Klf2, Foxp1, a transcription factor distantly related to Foxo1, and Slfn2, a protein of unknown biochemical function, appearing as likely candidates135,137,166,167.
Integrating at the organ level: cross talk in the thymus
The homotypic SLAM interactions in iNK T cell development illustrate the importance of the ‘cross-talk’ between thymocytes in intrathymic development. While it has long been known that such cross talk, or that between thymocytes and stroma, affects thymus biology168,169, recent studies have started to dissect the signals involved66,170. Interactions between SP thymocytes and medullary epithelial cells have received particular attention: newly generated SP thymocytes spend several days in the medulla before exiting the thymus170, where their exposure to tissue antigens expressed in medullary epithelial cells prevents the generation of self-reactive T cells76. Feed-back signaling from SP thymocytes is important for this effect, as it induces the expression of the autoimmune regulator Aire in medullary epithelial cells, through interactions between thymocyte-expressed RANK (receptor activator of NF-κB), and its ligand RANKL on epithelial cells, in a manner partly redundant with CD40-CD40L interactions171–173. The ongoing investigations of these and other cell-cell interactions build the foundation of an ‘organ biology’ approach to T cell development.
In summary, T cell development has generated immense interest for more than two decades, both because of the key immunological questions it raises, and as model system for cell differentiation processes, from programmed cell death to epigenetic silencing. The time now appears ripe for new harvests on these two fronts. New genetic and investigative tools tackle outstanding immunological questions, including the emergence of the distinct T lineages and the ligands that promote selection, whereas analyses of signaling and transcriptional mechanisms define new paradigms for gene expression and cell differentiation. In addition, emerging data on human T cell development highlight similarities and differences with mouse experimental systems174. These are exciting times for thymus biology !
Acknowledgments
We thank Jon Ashwell, Avinash Bhandoola, Anne Gégonne, Paul Love and Al Singer for their comments on the manuscript. Research work in the authors’ laboratory is supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH.
References
- 1.Hayday AC. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity. 2009;31:184–196. doi: 10.1016/j.immuni.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Matthews AG, Oettinger MA. RAG: a recombinase diversified. Nat Immunol. 2009;10:817–821. doi: 10.1038/ni.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Chi AW, Bell JJ, Zlotoff DA, Bhandoola A. Untangling the T branch of the hematopoiesis tree. Curr Opin Immunol. 2009;21:121–126. doi: 10.1016/j.coi.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feyerabend TB, et al. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 2009;30:67–79. doi: 10.1016/j.immuni.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Kawamoto H, Katsura Y. A new paradigm for hematopoietic cell lineages: revision of the classical concept of the myeloid-lymphoid dichotomy. Trends Immunol. 2009;30:193–200. doi: 10.1016/j.it.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Wada H, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 8.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 9.Schlenner SM, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, et al. Coordination between CCR7- and CCR9-mediated chemokine signals in prevascular fetal thymus colonization. Blood. 2006;108:2531–2539. doi: 10.1182/blood-2006-05-024190. [DOI] [PubMed] [Google Scholar]
- 11.Rossi FM, et al. Recruitment of adult thymic progenitors is regulated by P-selectin and its ligand PSGL-1. Nat Immunol. 2005;6:626–634. doi: 10.1038/ni1203. [DOI] [PubMed] [Google Scholar]
- 12.Zlotoff DA, et al. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2009 doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hozumi K, et al. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med. 2008;205:2507–2513. doi: 10.1084/jem.20080134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch U, et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 16.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 17.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 18.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 19.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda T, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visan I, et al. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat Immunol. 2006;7:634–643. doi: 10.1038/ni1345. [DOI] [PubMed] [Google Scholar]
- 22.Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Yashiro-Ohtani Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiorini E, et al. Dynamic regulation of notch 1 and notch 2 surface expression during T cell development and activation revealed by novel monoclonal antibodies. J Immunol. 2009;183:7212–7222. doi: 10.4049/jimmunol.0902432. [DOI] [PubMed] [Google Scholar]
- 25.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 26.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 28.von Boehmer H. Unique features of the pre-T-cell receptor alpha-chain: not just a surrogate. Nat Rev Immunol. 2005;5:571–577. doi: 10.1038/nri1636. [DOI] [PubMed] [Google Scholar]
- 29.Irving BA, Alt FW, Killeen N. Thymocyte development in the absence of pre-T cell receptor extracellular immunoglobulin domains. Science. 1998;280:905–908. doi: 10.1126/science.280.5365.905. [DOI] [PubMed] [Google Scholar]
- 30.Yamasaki S, et al. Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat Immunol. 2006;7:67–75. doi: 10.1038/ni1290. [DOI] [PubMed] [Google Scholar]
- 31.Janas ML, et al. Thymic development beyond beta-selection requires phosphatidylinositol 3-kinase activation by CXCR4. J Exp Med. 2010;207:247–61. S1–2. doi: 10.1084/jem.20091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trampont PC, et al. CXCR4 acts as a costimulator during thymic beta-selection. Nat Immunol. 2009 doi: 10.1038/ni.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maillard I, et al. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fry TJ, Mackall CL. The many faces of IL-7: from lymphopoiesis to peripheral T cell maintenance. J Immunol. 2005;174:6571–6576. doi: 10.4049/jimmunol.174.11.6571. [DOI] [PubMed] [Google Scholar]
- 35.Chong MM, et al. Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation. Immunity. 2003;18:475–487. doi: 10.1016/s1074-7613(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 36.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Hayes SM, Love PE. A retrospective on the requirements for gammadelta T-cell development. Immunol Rev. 2007;215:8–14. doi: 10.1111/j.1600-065X.2006.00476.x. [DOI] [PubMed] [Google Scholar]
- 38.Bruno L, Fehling HJ, von Boehmer H. The alpha beta T cell receptor can replace the gamma delta receptor in the development of gamma delta lineage cells. Immunity. 1996;5:343–352. doi: 10.1016/s1074-7613(00)80260-5. [DOI] [PubMed] [Google Scholar]
- 39.Mombaerts P, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 40.Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 41.Dave VP, et al. CD3 delta deficiency arrests development of the alpha beta but not the gamma delta T cell lineage. EMBO J. 1997;16:1360–1370. doi: 10.1093/emboj/16.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes SM, Love PE. Stoichiometry of the murine gammadelta T cell receptor. J Exp Med. 2006;203:47–52. doi: 10.1084/jem.20051886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haks MC, et al. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Boyden LM, et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet. 2008;40:656–662. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayday AC, Pennington DJ. Key factors in the organized chaos of early T cell development. Nat Immunol. 2007;8:137–144. doi: 10.1038/ni1436. [DOI] [PubMed] [Google Scholar]
- 46.Wakabayashi Y, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 47.Kreslavsky T, Garbe AI, Krueger A, von Boehmer H. T cell receptor-instructed alphabeta versus gammadelta lineage commitment revealed by single-cell analysis. J Exp Med. 2008;205:1173–1186. doi: 10.1084/jem.20072425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kreslavsky T, Gleimer M, von Boehmer H. alphabeta versus gammadelta lineage choice at the first TCR-controlled checkpoint. Curr Opin Immunol. 2010 doi: 10.1016/j.coi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taghon T, Rothenberg EV. Molecular mechanisms that control mouse and human TCR-alphabeta and TCR-gammadelta T cell development. Semin Immunopathol. 2008;30:383–398. doi: 10.1007/s00281-008-0134-3. [DOI] [PubMed] [Google Scholar]
- 50.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 52.Lauritsen JP, et al. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garbe AI, Krueger A, Gounari F, Zuniga-Pflucker JC, von Boehmer H. Differential synergy of Notch and T cell receptor signaling determines alphabeta versus gammadelta lineage fate. J Exp Med. 2006;203:1579–1590. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 55.Alonzo ES, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gammadelta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol. 2009;182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo J, et al. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 59.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–826. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 60.Ohteki T, Wilson A, Verbeek S, MacDonald HR, Clevers H. Selectively impaired development of intestinal T cell receptor gamma delta+ cells and liver CD4+ NK1+ T cell receptor alpha beta+ cells in T cell factor-1-deficient mice. Eur J Immunol. 1996;26:351–355. doi: 10.1002/eji.1830260213. [DOI] [PubMed] [Google Scholar]
- 61.Ioannidis V, Beermann F, Clevers H, Held W. The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat Immunol. 2001;2:691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 62.Jeannet G, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 63.Koch U, et al. Simultaneous loss of beta- and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood. 2008;111:160–164. doi: 10.1182/blood-2007-07-099754. [DOI] [PubMed] [Google Scholar]
- 64.Yu Q, Sharma A, Sen JM. TCF1 and beta-catenin regulate T cell development and function. Immunol Res. 2010 doi: 10.1007/s12026-009-8137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Melichar HJ, et al. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 66.Silva-Santos B, Pennington DJ, Hayday AC. Lymphotoxin-mediated regulation of gammadelta cell differentiation by alphabeta T cell progenitors. Science. 2005;307:925–928. doi: 10.1126/science.1103978. [DOI] [PubMed] [Google Scholar]
- 67.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 68.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 69.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 71.von Boehmer H, Melchers F. Checkpoints in lymphocyte development and autoimmune disease. Nat Immunol. 2010;11:14–20. doi: 10.1038/ni.1794. [DOI] [PubMed] [Google Scholar]
- 72.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 73.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bautista JL, et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakagawa T, et al. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280:450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- 76.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 77.Nitta T, et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32:29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 78.Davey GM, et al. Preselection thymocytes are more sensitive to T cell receptor stimulation than mature T cells. J Exp Med. 1998;188:1867–1874. doi: 10.1084/jem.188.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lucas B, Stefanova I, Yasutomo K, Dautigny N, Germain RN. Divergent changes in the sensitivity of maturing T cells to structurally related ligands underlies formation of a useful T cell repertoire. Immunity. 1999;10:367–376. doi: 10.1016/s1074-7613(00)80036-9. [DOI] [PubMed] [Google Scholar]
- 80.Daniels MA, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 81.Azzam HS, et al. Fine tuning of TCR signaling by CD5. J Immunol. 2001;166:5464–5472. doi: 10.4049/jimmunol.166.9.5464. [DOI] [PubMed] [Google Scholar]
- 82.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 83.Gallo EM, et al. Calcineurin sets the bandwidth for discrimination of signals during thymocyte development. Nature. 2007;450:731–735. doi: 10.1038/nature06305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 89.Nakayama K, et al. Disappearance of the lymphoid system in Bcl-2 homozygous mutant chimeric mice. Science. 1993;261:1584–1588. doi: 10.1126/science.8372353. [DOI] [PubMed] [Google Scholar]
- 90.Veis DJ, Sorenson CM, Shutter JR, Korsmeyer SJ. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell. 1993;75:229–240. doi: 10.1016/0092-8674(93)80065-m. [DOI] [PubMed] [Google Scholar]
- 91.Dzhagalov I, Dunkle A, He YW. The anti-apoptotic Bcl-2 family member Mcl-1 promotes T lymphocyte survival at multiple stages. J Immunol. 2008;181:521–528. doi: 10.4049/jimmunol.181.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Opferman JT, et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 93.Ueno T, et al. CCR7 Signals Are Essential for Cortex-Medulla Migration of Developing Thymocytes. J Exp Med. 2004;200:493–505. doi: 10.1084/jem.20040643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kwan J, Killeen N. CCR7 directs the migration of thymocytes into the thymic medulla. J Immunol. 2004;172:3999–4007. doi: 10.4049/jimmunol.172.7.3999. [DOI] [PubMed] [Google Scholar]
- 95.Nitta T, Nitta S, Lei Y, Lipp M, Takahama Y. CCR7-mediated migration of developing thymocytes to the medulla is essential for negative selection to tissue-restricted antigens. Proc Natl Acad Sci U S A. 2009;106:17129–17133. doi: 10.1073/pnas.0906956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sudo T, et al. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc Natl Acad Sci U S A. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang L, Bosselut R. CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. J Immunol. 2009;183:2903–2910. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neilson JR, Winslow MM, Hur EM, Crabtree GR. Calcineurin B1 is essential for positive but not negative selection during thymocyte development. Immunity. 2004;20:255–266. doi: 10.1016/s1074-7613(04)00052-4. [DOI] [PubMed] [Google Scholar]
- 100.Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–443. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 101.Wang L, Xiong Y, Bosselut R. Tenuous paths in unexplored territory: From T cell receptor signaling to effector gene expression during thymocyte selection. Seminars in Immunology. 2010 doi: 10.1016/j.smim.2010.04.013. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jones ME, Zhuang Y. Regulation of V(D)J recombination by E-protein transcription factors. Adv Exp Med Biol. 2009;650:148–156. doi: 10.1007/978-1-4419-0296-2_12. [DOI] [PubMed] [Google Scholar]
- 103.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bain G, et al. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 105.Costello PS, Nicolas RH, Watanabe Y, Rosewell I, Treisman R. Ternary complex factor SAP-1 is required for Erk-mediated thymocyte positive selection. Nat Immunol. 2004;5:289–298. doi: 10.1038/ni1038. [DOI] [PubMed] [Google Scholar]
- 106.Corbella P, et al. Functional commitment to helper T cell lineage precedes positive selection and is independent of T cell receptor MHC specificity. Immunity. 1994;1:269–276. doi: 10.1016/1074-7613(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 107.Matechak EO, Killeen N, Hedrick SM, Fowlkes BJ. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 1996;4:337–347. doi: 10.1016/s1074-7613(00)80247-2. [DOI] [PubMed] [Google Scholar]
- 108.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 109.Woolf E, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 111.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 112.Wang L, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muroi S, et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 115.Setoguchi R, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 116.Zamisch M, et al. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med. 2009;206:2685–2699. doi: 10.1084/jem.20092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.He X, et al. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 118.Sakaguchi S, et al. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pai SY, et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 120.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 122.von Boehmer H. CD4/CD8 lineage commitment: back to instruction? J Exp Med. 1996;183:713–715. doi: 10.1084/jem.183.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singer A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr Opin Immunol. 2002;14:207–215. doi: 10.1016/s0952-7915(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 124.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 125.Janeway CAJ. T-cell development. Accessories or coreceptors? Nature. 1988;335:208–210. doi: 10.1038/335208a0. [DOI] [PubMed] [Google Scholar]
- 126.Robey E. Introduction: commitment to CD4 and CD8 lineages--stochastic or instructive? Semin Immunol. 1994;6:207–208. doi: 10.1006/smim.1994.1026. [DOI] [PubMed] [Google Scholar]
- 127.Liu X, Bosselut R. Duration of TCR signaling controls CD4-CD8 lineage differentiation in vivo. Nat Immunol. 2004;5:280–288. doi: 10.1038/ni1040. [DOI] [PubMed] [Google Scholar]
- 128.Sarafova SD, et al. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 2005;23:75–87. doi: 10.1016/j.immuni.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 129.Brugnera E, et al. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. doi: 10.1016/s1074-7613(00)00008-x. [DOI] [PubMed] [Google Scholar]
- 130.Sarafova SD, et al. Upregulation of CD4 expression during MHC class II-specific positive selection is essential for error-free lineage choice. Immunity. 2009;31:480–490. doi: 10.1016/j.immuni.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park JH, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yu Q, Erman B, Bhandoola A, Sharrow SO, Singer A. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J Exp Med. 2003;197:475–487. doi: 10.1084/jem.20021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 134.Shiow LR, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 135.Weinreich MA, et al. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fabre S, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181:2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 139.Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol. 2009;10:1057–1063. doi: 10.1038/ni.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Finlay D, Cantrell D. Phosphoinositide 3-kinase and the mammalian target of rapamycin pathways control T cell migration. Ann N Y Acad Sci. 2010;1183:149–157. doi: 10.1111/j.1749-6632.2009.05134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 143.Lee WY, et al. An intravascular immune response to Borrelia burgdorferi involves Kupffer cells and iNKT cells. Nat Immunol. 2010;11:295–302. doi: 10.1038/ni.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barral P, et al. CD169(+) macrophages present lipid antigens to mediate early activation of iNKT cells in lymph nodes. Nat Immunol. 2010;11:303–312. doi: 10.1038/ni.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Egawa T, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 146.Borowski C, Bendelac A. Signaling for NKT cell development: the SAP-FynT connection. J Exp Med. 2005;201:833–836. doi: 10.1084/jem.20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Griewank K, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Laky K, Fowlkes BJ. Presenilins regulate alphabeta T cell development by modulating TCR signaling. J Exp Med. 2007;204:2115–2129. doi: 10.1084/jem.20070550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johnson AL, et al. Themis is a member of a new metazoan gene family and is required for the completion of thymocyte positive selection. Nat Immunol. 2009;10:831–839. doi: 10.1038/ni.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lesourne R, et al. Themis, a T cell-specific protein important for late thymocyte development. Nat Immunol. 2009;10:840–847. doi: 10.1038/ni.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fu G, et al. Themis controls thymocyte selection through regulation of T cell antigen receptor-mediated signaling. Nat Immunol. 2009;10:848–856. doi: 10.1038/ni.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kakugawa K, et al. A novel gene essential for the development of single positive thymocytes. Mol Cell Biol. 2009;29:5128–5135. doi: 10.1128/MCB.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Patrick MS, et al. Gasp, a Grb2-associating protein, is critical for positive selection of thymocytes. Proc Natl Acad Sci U S A. 2009;106:16345–16350. doi: 10.1073/pnas.0908593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu X, et al. Restricting Zap70 expression to CD4+CD8+ thymocytes reveals a T cell receptor-dependent proofreading mechanism controlling the completion of positive selection. J Exp Med. 2003;197:363–373. doi: 10.1084/jem.20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang L, et al. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 2008;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zou YR, et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 160.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 161.Kreslavsky T, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Raberger J, et al. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci U S A. 2008;105:17919–17924. doi: 10.1073/pnas.0805733105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Strid J, Tigelaar RE, Hayday AC. Skin immune surveillance by T cells--a new order? Semin Immunol. 2009;21:110–120. doi: 10.1016/j.smim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 164.Schwartzberg PL, Mueller KL, Qi H, Cannons JL. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol. 2009;9:39–46. doi: 10.1038/nri2456. [DOI] [PubMed] [Google Scholar]
- 165.Prince AL, Yin CC, Enos ME, Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate conventional versus innate T-cell development. Immunol Rev. 2009;228:115–131. doi: 10.1111/j.1600-065X.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Feng X, et al. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood. 2009 doi: 10.1182/blood-2009-07-232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Berger M, et al. An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat Immunol. 2010;11:335–343. doi: 10.1038/ni.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shores EW, Van EW, Singer A. Disorganization and restoration of thymic medullary epithelial cells in T cell receptor-negative scid mice: evidence that receptor-bearing lymphocytes influence maturation of the thymic microenvironment. Eur J Immunol. 1991;21:1657–1661. doi: 10.1002/eji.1830210711. [DOI] [PubMed] [Google Scholar]
- 169.Canelles M, Park ML, Schwartz OM, Fowlkes BJ. The influence of the thymic environment on the CD4-versus-CD8 T lineage decision. Nat Immunol. 2003;4:756–764. doi: 10.1038/ni953. [DOI] [PubMed] [Google Scholar]
- 170.McCaughtry TM, Wilken MS, Hogquist KA. Thymic emigration revisited. J Exp Med. 2007;204:2513–2520. doi: 10.1084/jem.20070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Irla M, et al. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity. 2008;29:451–463. doi: 10.1016/j.immuni.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 172.Hikosaka Y, et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 2008;29:438–450. doi: 10.1016/j.immuni.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 173.Akiyama T, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 2008;29:423–437. doi: 10.1016/j.immuni.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 174.Van de Walle I, et al. An early decrease in Notch activation is required for human TCR-alphabeta lineage differentiation at the expense of TCR-gammadelta T cells. Blood. 2009;113:2988–2998. doi: 10.1182/blood-2008-06-164871. [DOI] [PubMed] [Google Scholar]
- 175.Porritt HE, et al. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 176.Park JH, et al. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21:289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]