SUMMARY
Elongator is required for the synthesis of the mcm5s2 modification found on tRNAs recognizing AA-ending codons. In order to obtain a global picture of the role of Elongator in translation, we used reverse protein arrays to screen the fission yeast proteome for translation defects. Unexpectedly, this revealed that Elongator inactivation mainly affected three specific functional groups including proteins implicated in cell division. The absence of Elongator results in a delay in mitosis onset and cytokinesis defects. We demonstrate that the kinase Cdr2, which is a central regulator of mitosis and cytokinesis, is under translational control by Elongator due to the Lysine codon usage bias of the cdr2 coding sequence. These findings uncover a mechanism by which the codon usage, coupled to tRNA modifications, fundamentally contributes to gene expression and cellular functions.
INTRODUCTION
The coordinated expression of functionally related groups of genes is an universal feature of living cells and can rely on a sequence signature that warrants coexpression of otherwise unlinked genes, as seen for the binding sites of transcription factors. Two recent studies demonstrated that the codon order present within the open reading frame (ORF) also determines the dynamic of expression, in addition to the primary sequence of the encoded polypeptide (Cannarozzi et al., 2010; Tuller et al., 2010). This mechanism relies on tRNA reusage and is particularly significant for groups of genes that are very dynamically regulated. tRNAs from all organisms contain modified nucleosides, and when a uridine is present in the wobble position (U34), this residue is almost universally modified (Agris et al., 2007; Björk, 1995; Suzuki, 2005). Particularly, in eukaryotes, the tRNAs reading codons belonging to split codon boxes, which include the tRNALysUUU, tRNAGluUUC and tRNAGlnUUG, are thiolated (s2) at the 2-carbon and contain a methoxy-carbonyl-methyl modification (mcm5) at the 5-carbon on the uridine (Figures S1A and S1B available online). The complex double modification (mcm5s2U34) is required to offset the translational inefficiency of the AA-ending codons within the corresponding two-codon boxes (Murphy et al., 2004; Yarian et al., 2000). The complete thiolation pathway has been described with Ctu1 catalyzing the final step (Dewez et al., 2008; Leidel et al., 2009; Noma et al., 2009). A screen in budding yeast based on resistance to Zymocin, a toxin that cleaves tRNA only when they bear the modification, identified a list of genes required for the addition of the mcm5 moiety (Huang et al., 2005; Huang et al., 2008). They included the genes coding for the Elongator complex (Elp1p-6p) (Esberg et al., 2006; Huang et al., 2005) although the pathway is not elucidated. Elongator was first isolated based on its ability to coprecipitate with the elongating RNA polymerase II and to acetylate histones H3 (Winkler et al., 2002; Wittschieben et al., 1999). A provocative finding was the fact that the transcription and histone acetylation defects, and the additional phenotypes resulting from the absence of Elongator could be suppressed by the overexpression of two tRNAs (tRNALysUUU and tRNAGlnUUG) in budding yeast, which raised some doubts about the direct involvement of Elongator in other pathways beside tRNA modifications (Esberg et al., 2006). Another twist to the story came from the reports that Elongator is involved in genome demethylation (Okada et al., 2010) and a-tubulin acetylation in metazoans (Creppe et al., 2009) with evidence that this last function is causal in familial dysautonomia (FD), an autosomal recessive disease resulting from defects in neuronal development (Slaugenhaupt and Gusella, 2002).
In the present study, we show that the inactivation of Elongator in fission yeast leads to the loss of the double modification mcm5s2. The resulting phenotypes include mitosis and cytokinesis defects. Using a proteome-wide approach, we have identified target mRNAs that require Elongator for efficient translation. These mRNAs have a codon content signature enriched with the lysine AAA codon and fall into groups of genes implicated, among others, in cell-cycle control. We show that the Cdr2 kinase is a key target of Elongator and that an alternate lysine codon content within the cdr2 coding region uncouples its regulation from Elongator. We propose that Elongator carries out a coordinated control over translation that is essential for cell-cycle progression.
RESULTS
The Inactivation of Elongator Leads to Loss of the Double mcm5s2 Modification and Discrete Phenotypes
To understand the biological importance of the double mcm5s2 modification, we combined a deletion of the ctu1 and elp3 genes in fission yeast which resulted in a viable albeit thermosensitive strain (Figure 1A), contrary to budding yeast where the corresponding mutations are synthetic lethal (Björk et al., 2007). We excluded the possibility that the mcm5 modification was still present in the elp3 mutant by mass spectrometry analyses that also revealed a requirement of the Elongator-dependent mcm5 moiety for efficient thiolation (Figures S1C and S1D), as previously proposed (Noma et al., 2009). The dependency of the thiolation upon the addition of mcm5 was confirmed by PAGE in the presence of APM ([p-(N-acrylamino)-phenyl]mercuric chloride) that highlights the presence of thiolated tRNAs (Figure S1C). These data imply that the Elongator (Δelp3) mutation should be functionally identical to the double Δctu1 Δelp3, which is supported by the identical phenotypes of the Δelp3 and the Δctu1 Δelp3 strains (Figure 1A and data not shown).
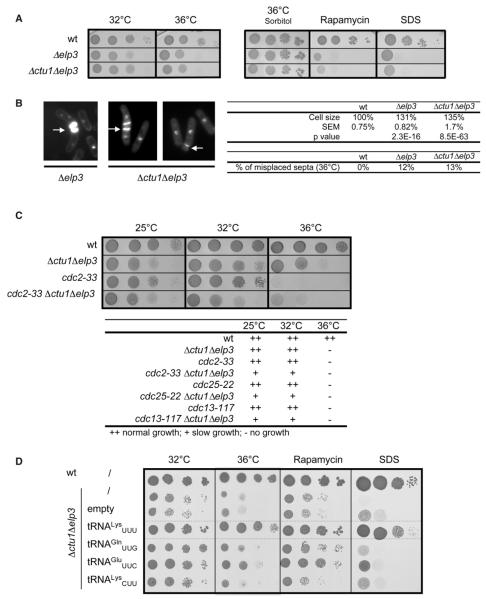
Figure 1. The Absence of the mcm5s2 Modification Leads to Specific Phenotypes Suppressed by Overexpression of the Unmodified tRNALysUUU.
(A) Spot dilution assay of wt, Δelp3 and Δctu1Δelp3 strains grown on rich media at 32°C or 36°C, or supplemented with 1 M sorbitol, 5 ng/ml rapamycin, or 0.005% SDS.
(B) Left panel: DAPI and calcofluor stained Δelp3 or Δctu1Δelp3 cells grown at 36°C. Right panel: measurement of septated cell size (reported to wild-type = 100%; p value by Student’s t test; n > 100) and counting of misplaced septa (n > 100) in the Δelp3 and Δctu1Δelp3 strains.
(C) Spot dilution assay of wt, Δctu1Δelp3, cdc2-33 and Δctu1 Δelp3 cdc2-33 grown on rich media at 25°C, 32°C, or 36°C and a summary of similar interactions with the cdc25-22 and cdc13-117 alleles.
(D) Spot dilution assay of the Δctu1Δelp3 strain expressing elevated levels of tRNALysUUU, tRNAGlnUUG, tRNAGluUUC, or tRNALysCUU on indicated selective minimal media.
See also Figures S1 and S2.
The absence of mcm5s2 double modification resulted in three apparent phenotypes: thermosensitivity, cell elongation, and the presence of multiple, often misplaced septa, which are hallmarks of defects in cell-cycle progression (Figures 1A and 1B). Moreover, the Δctu1 Δelp3 mutant displayed strong negative genetic interactions with mutations in cdc25, cdc13, and cdc2 genes that encode key mitosis inducers (Figure 1C). These genetic interactions are consistent with a role of Elp3 as a positive cellcycle regulator. Broader phenotyping of the Δctu1 Δelp3 and the Δelp3 mutants revealed specificity in drug sensitivity with growth defects obvious only in the presence of the TOR inhibitor rapamycin and detergents (Figure S2). During the course of this assay, we also noted that sorbitol almost completely suppressed the defects at high temperature (Figure 1A).
Consistent with a defect in translation of codons read by the three mcm5s2 modified tRNAs, the overexpression of the unmodified tRNAs rescued all the observed phenotypes with suppression by the tRNALysUUU being the most efficient. The overexpression of tRNALysCUU, which reads the synonymous lysine codon and does not have the mcm5s2 did not rescue the phenotypes (Figure 1D). Similar conclusions could be extended to the deletions of either elp1, elp4 or elp6 (Figure S1E), which supports the requirement of the core fission yeast Elongator complex in tRNA modifications.
The AAA Codon Usage Is Linked to the Expression Level of the Corresponding Genes
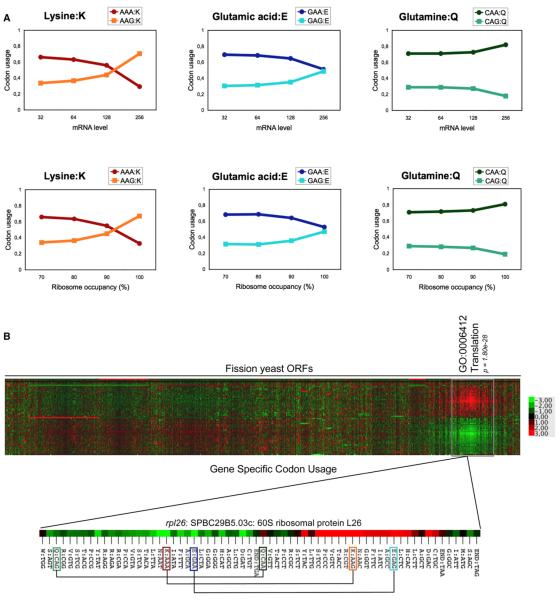
Due to the low G-C content of the fission yeast genome, there is a bias toward the use of the NAA codon in the split box with, for example, 62% of all lysines encoded by AAA and 38% by AAG (Table S1). We took advantage of the availability of large-scale gene expression data sets from fission yeast (Lackner et al., 2007) to study how the codon bias relates to the expression level. Strikingly, at both the transcriptional and the translational levels, highly expressed ORFs reversed the codon usage for lysine with the AAG codon used for nearly 80% of lysines. To a lesser extent, this effect was seen for the GAA-GAG pair but did not occur for the CAA-CAG pair (Figure 2A, Table S1). This indicates that a strong selection pressure is in operation to counteract the low G-C content of the genome and to avoid the use of the AAA codon, and to a lesser extent the GAA codon, when high expression is required. In order to test the prediction that the fission yeast genes with the highest expression level have a biased codon content, we used a Gene-Specific Codon Usage algorithm designed to visually display an integrated measure of codon usage across all the ORFs (Begley et al., 2007). The resulting clustering highlighted a distinct group of genes with a codon usage characterized by overrepresentation of G-ended codons for lysine and glutamic acid. This group is enriched (p < 1.80e-28) for genes encoding highly expressed proteins implicated in translation (Figure 2B).
Figure 2. Highly Expressed Fission Yeast Genes Use a Skewed Codon Content Excluding the AAA and GAA Codons.
(A) Codon bias between G- and A-ending codons for lysine, glutamic acid, and glutamine in genes as a function of mRNA level (upper panel) or ribosome occupancy (lower panel). The data sets used are from a previous study (Lackner et al., 2007).
(B) Hierarchical clustering and heat map analysis of the gene-specific codon usage for each codon (rows) in each S. pombe ORF (column). Clustering was performed based on Z-scores shown in Table S1. Deviations from genome average are shown for each codon according to the color-code key. One main cluster is highlighted and is enriched for the “Translation” GO category that includes highly expressed genes. The codon usage of the rpl26 gene (rotated) exemplifies a skewed codon content for lysine and glutamic acid.
The Targets of Elongator Are Enriched in AAA and Fall into Four Groups of Functionally Related Genes
Considering the specific defects resulting from the absence of the mcm5s2 modification, we postulated that mRNA-specific rather than transcriptome-wide translation defects would account for the observed phenotypes. To test this hypothesis, we combined the fission yeast integrated ORFeome strain library with Reverse Protein Macro-arrays (Matsuyama et al., 2006) to determine the relative expression of all fission yeast proteins in strains lacking either ctu1 or elp3. Expression levels were determined by normalization against a-tubulin whose level is not affected by the double modification (Figure S3). The 4910 tagged ORFs are expressed from the same promoter, terminator and chromatin locus in the various genetic backgrounds. Cluster analysis revealed an overall similarity in the protein expression profiles in the absence of either ctu1 or elp3 (Figures 3A and 3B), with a moderate global effect on the proteome: the average expression level of the proteome is 96% in the absence of ctu1 and 87% in the absence of elp3 (Table S2). In the Δelp3 strain, where the double modification is abolished, the expression level of 494 proteins was decreased (fold change > = 2). A gene ontology analysis revealed a significant enrichment of four distinct functional groups encompassing the regulation of the cell cycle, cytokinesis, response to nutrients, and chromatin modifications at the centromere (Figure 3B, Table S2). Further analyses confirmed that the protein expression level within these groups was statistically more affected than the whole proteome in the absence of elp3 (Figure 3C). Analysis of the codon bias within these groups showed that they were overrepresented for the A-ended codon in the case of lysine (AAA) and also, in the case of the “chromatin silencing by small RNA” category, for glutamic acid (GAA) whereas the glutamine codon usage was not statistically different from the genome (Figures 3D–3F). This experiment reveals that unlike highly expressed genes, which mostly rely on the AAG codon, some defined functional groups are further enriched for the AAA codon, which renders their expression highly dependent upon the double modification mcm5s2 and the Elongator complex. Importantly, the functional groups identified could readily be linked to phenotypes resulting from the absence of the modification. However, the fourth group, encompassing proteins related to chromatin modifications at centromeric repeats, was unexpected. To investigate the biological relevance of the enrichment of this group for the AAA codon, we analyzed the silencing of an ura4 reporter inserted in the outer repeat of the centromere of chromosome 1 (Grewal, 2010). As shown in Figure S4, the absence of elp3 led to desilencing of ura4 and elevated expression of the mRNA, most likely due to a defect in the RNAi-dependent heterochromatin formation. This data suggests that Elongator is required for efficient translation of mRNAs encoding factors regulating chromatin silencing at centromeres, as supported by the fact that well characterized RNAi regulators from this group, including Chp1, Ago1, and Rdp1, show decreased protein level in either the ctu1 or the elp3 strains (Figure S4). The overexpression of the unmodified tRNALysUUU suppressed the silencing defect of elp3 but had no effect on the swi6 mutant (data not shown). Moreover, gene expression profiling of an elp3 mutant did not reveal any decrease in the steady state mRNA expression of the genes comprised in the functional group (Nugent et al., 2010). Taken together, these data show that the proteome expression profile of an Elongator mutant is sufficient to predict a phenotype resulting from the inactivation of the complex.
Figure 3. Proteome-wide Analysis of Strains Lacking tRNA Modifications by Reverse Protein Arrays.
(A) Hierarchical clustering and heat map analysis of the proteome expression level in the Δctu1 and Δelp3 strains compared to wild-type. The data are presented as log2 mutant/wild-type ratio of immunoblotting signals and are color-coded as indicated in the key.
(B) Left panel: protein expression profiling in the Δelp3 mutant. Scatter plot of protein expression changes between mutant and the wild-type: signals above the red line are increased in the mutant relative to wild-type, whereas signals below the green line are decreased with a 2-fold cutoff. The proteins whose expression is significantly decreased are reported in Table S2. Right panel: Enriched GO groups are listed with their accession number, name and associated p value(see Experimental Procedures). (GO:0009267, n = 70; GO:0031929, n = 56; GO:0031048, n = 14; GO:0030702, n = 56; GO:0030466, n = 37; GO:0031047, n = 24; GO:0010389, n = 16; GO:0051325, n = 106; GO:0010972, n = 36; GO:0071341, n = 6).
(C) Average expression level of the proteome in the Δelp3 strain compared to wt set to 100%. Average expression level within four GO functional groups significantly more affected than the average in the absence of the double modification (*p < 0.05; **p < 0.01, Student’s t test). Expression level of the Cdr2 protein in the Δelp3 strain.
(D–F) Codon usage for lysine (AAA-AAG), glutamic acid (GAA-GAG), and glutamine (CAA-CAG) within the same groups shown in (C), and the Cdr2 protein. The ORFeome codon usage is indicated by the black line. Note that the 0%–20% and 40%–100% of the y axis are not shown to highlight variations from the ORFeome codon usage. (*p < 0.05; **p < 0.01; ***p < 0.001, Student’s t test).
See also Figure S3.
Reversing the AAA Codon Content to AAG Is Sufficient to Uncouple the cdr2 mRNA from Elongator Regulation
With respect to the cell-cycle defects caused by the absence of elp3, we noticed that the Cdr2 kinase was among the more strongly downregulated proteins within the cell-cycle and cytokinesis functional groups. Cdr2 is a key player in cell-cycle progression at the G2-M transition and in cytokinesis (Breeding et al., 1998; Kanoh and Russell, 1998; Martin and BerthelotGrosjean, 2009; Moseley et al., 2009; Young and Fantes, 1984), and its downregulation could therefore link two prominent facets of the elp3 deletion phenotypes. Interestingly, all phenotypes known to result from the deletion of cdr2 were shared by the Δelp3 strain (Table S3), and similarly to Δelp3, sorbitol efficiently suppressed the thermosensitive phenotypes of Δcdr2 (Breeding et al., 1998), although the molecular mechanism of this suppression remained unclear. Quantitative western blotting confirmed a strong reduction in the level of hemagglutinin-tagged Cdr2 in the absence of the double modification irrespective of whether the HA tag was appended to the N or C terminus, while the steady state mRNA level was unaffected (Figure 4A). The overexpression of the unmodified tRNALysUUU restored Cdr2 expression to levels close to those in wild-type and suppressed the cell-cycle defect of the elp3 mutant (Figures 1A and 4C, data not shown). In contrast, the tRNALysCUU, which reads the synonymous codon, or tRNAGluUUC and tRNAGlnUUG had no effect (Figures 4B and 4C). Based on these data presented above, we hypothesized that the overrepresentation of the AAA codon within the cdr2 coding sequence was causal in the translational downregulation. Indeed, replacing all AAA codons in cdr2 by the synonymous AAG rendered Cdr2 expression insensitive to the absence of the double modification (Figure 4C). These data confirm that cdr2 is a target of Elongator. However, the Δctu1 Δelp3 mutation was not epistatic to the deletion of cdr2 with the triple mutant showing stronger defects than either of the single mutants (Figure 4D). This is expected as besides Cdr2, the translation of additional mRNAs within the same functional groups related to cell-cycle progression is regulated by the mcm5s2 modification, as suggested by the reverse protein array (Figure 4E) and codon content analyses (Figure 3).
Figure 4. The Cdr2 Kinase Is Regulated Translationally by Elongator-Dependent tRNALysUUU Modification.
(A) Quantitative analysis of cdr2 mRNA and Cdr2-HA or HA-Cdr2 protein abundance in a wt or Δctu1Δelp3 strains tagged at chromosomal locus. Error bars represent the SEM of biological triplicates.
(B) Western blot analysis of plasmid born Cdr2-HA in wt or Δctu1Δelp3 strains expressing elevated level of tRNALysUUU, tRNAGlnUUG, tRNAGluUUC, or tRNALysCUU. Tubulin was used as a loading control and quantification was performed with a LI-COR scanner (see Experimental Procedures). The value from the wild-type strain transformed with an empty vector was set to 1.
(C) Western blot analysis of plasmid born Cdr2-HA, or Cdr2-HAno-AAA in wt or Δctu1Δelp3 strains. Tubulin was used as a loading control and quantification was performed with a LI-COR scanner (see Experimental Procedures). The value from the wild-type strain was set to 1.
(D) Measure of septated cell size (reported to wild-type = 100%; SEM = standard error of the mean; n > 100; p value obtained from Student’s t test) and counting of misplaced septa (n > 100) in wt, Δcdr2, Δctu1Δelp3, and Δctu1Δelp3Δcdr2 grown at 36°C.
(E) Hierarchical clustering and heat map analysis of the protein expression level of the gene ontology group “Regulation of the G2-M transition of the mitotic cell cycle” (GO 0010389) in the Δctu1 and Δelp3 strain compared to wild-type. The data are presented as log2 mutant/wild-type ratio of hybridization signals and are color-coded as in Figure 3A.
See also Figure S4.
Elongator has been implicated in various processes (Svejstrup, 2007) but our data reveal that its genuine biological function in fission yeast is to promote the translation of mRNA required for cell-cycle progression and potentially other functional groups.
DISCUSSION
The inherent redundancy of the genetic code imposes the existence of synonymous codons, and a universal feature of gene expression is that synonymous codons are not used at the same frequency, a phenomenon referred to as the codon usage bias. Our work reveals that the codon usage bias, coupled to tRNA modification by Elongator, is used as a mechanism to coordinate the expression of groups of genes involved in specific pathways, including cell-cycle progression.
The molecular origin of this mechanism relies on the fact that unmodified tRNAs for glutamine, glutamic acid, and lysine have a decreased ribosome binding as supported by both their inability to bind appropriately programmed ribosomes (Ashraf et al., 1999) and efficient suppression by overexpression of the unmodified molecule (Dewez et al., 2008; Esberg et al., 2006). A detailed analysis of the phenomena concluded that pyrimidine-rich anticodon loops lack effective stacking interactions and low enthalpy resulting from the A-U base interaction (Krüger et al., 1998; Murphy et al., 2004; Yarian et al., 2000). In vitro binding data suggest that the equilibrium binding affinity is reduced by less than 2-fold in the absence of the double modification (Yarian et al., 2000). Our proteome-wide analysis is in agreement with these data as the effect of elp3 deletion is globally modest (Figure 3). Misreading and inability to read the near cognate codons were also proposed to underlie the defects observed in the absence of the modifications (Agris et al., 2007; Murphy et al., 2004). However, the overexpression of the unmodified tRNA would be expected to aggravate rather than rescue the phenotypes in case of misreading, and the existence in yeast of tRNA species that do not require modifications to read the near cognate codon discourage these two alternative possibilities. In that context, the tRNALysUUU codon is expected to present the highest dependency upon the modification, which is supported by several lines of evidence. First, using independent genome wide data sets, we show that a strong selection operating against the genome GC content is exerted to avoid the use of this codon when high expression is required (Figure 2). Second the overexpression of the unmodified tRNALysUUU was the most efficient to suppress the phenotypes resulting from the absence of Elongator, and the codon content analysis we have performed (Figure 3) mostly highlighted an enrichment in the AAA codon as the key determinant of the regulation by Elongator. Finally, we demonstrated that the replacement of all AAA codons within the cdr2 open reading frame was sufficient to restore efficient translation (Figure 4).
We propose that a key aspect of the codon-dependent control of gene expression we present is the fact that it applies to groups of functionally related genes rather than single genes. Specifically in the fission yeast, our work explains at the molecular level how Elongator influences the cell-cycle control over mitosis and cytokinesis mostly, but not entirely, through regulating the level of the Cdr2 kinase, which provides a conceptual frame for the early puzzling connection between cell-cycle progression and tRNA metabolism in fission yeast (Grossenbacher et al., 1986; Heyer et al., 1984; Nurse and Thuriaux, 1984; Young and Fantes, 1984).
The coordinated expression of groups of functionally related genes makes biological sense, but an important issue is to determine if modification-dependent translational controls are integrated with environmental cues. Although environmentally induced RNA modifications were reported (Wu et al., 2011), it was difficult until recently to provide a large-scale view of tRNA modifications dynamics in different growth conditions. A quantitative system approach has recently shown that tRNA modifications, including the double mcm5s2, are regulated by cellular response pathways (Chan et al., 2010). In addition, Elongator activity was reported to be controlled by a complex phosphorylation balance (Jablonowski et al., 2009; Mehlgarten et al., 2009).
A peculiarity of the Elongator complex is its involvement in higher eukaryotes in apparently unrelated processes including RNA polymerase II transcription, tubulin acetylation and genome demethylation (Creppe et al., 2009; Okada et al., 2010; Otero et al., 1999). A fission yeast Elongator mutant also presents apparently unrelated phenotypes, and, taken together, our data support that the biologically relevant function of Elongator in fission yeast is in tRNA modification. The demonstration of a conservation of this activity in distantly related plant (Mehlgarten et al., 2010) and nematode (Chen et al., 2009) supports an evolutionary conserved role in tRNA modifications, which could unify the pleiotropic defects resulting from Elongator inactivation in higher eukaryotes. However, the predominance of the AAG over the AAA codon to encode lysine in mammals, consequent to high G-C genome content, may have attenuated the primary translation regulation by Elongator in aid of the emergence of additional, maybe critical substrates in metazoans.
EXPERIMENTAL PROCEDURES
General Methods
Fission yeast growth, microscopy, gene tagging and mating were performed as described (Bamps et al., 2004; Fersht et al., 2007; Hermand and Nurse, 2007). Reverse protein arrays were processed, scanned, and analyzed as described (Matsuyama et al., 2006). The purification of tRNA and the analysis of tRNA hydrolysates by APM-PAGE northern blot were previously described in detail (Dewez et al., 2008). A more efficient method for synthesizing APM [p-(N-acrylamino)-phenyl]mercuric chloride was used (Lemau de Talancé et al., 2011).
Bioinformatics and Statistical Analyses
The reverse protein arrays were scanned with a LI-COR scanner. The expression levels of each protein were calculated by western blotting using an anti-HIS antibody (MBL) compared to those of the a-tubulin internal control detected by the B-512 antibody (Sigma). Each signal was analyzed by the Odyssey Imaging System as described (Matsuyama et al., 2006). Clustering and heat map analyses were performed using Cluster and Treeview on M data (where M = log2(D)-log2(wt)). A hierarchical clustering of the data was performed using the centroid linkage method. Gene annotations were taken from the Schizosaccharomyces pombe DB at the Sanger Institute. The Gene Ontology groups were defined using the AMIGO (http://amigo.geneontology.org/cgi-bin/amigo/term_enrichment), GO-miner (http://discover.nci.nih.gov/gominer/index.jsp), and GOEAST (http://omicslab.genetics.ac.cn/GOEAST/) software toolkit according to the developer instructions (Zeeberg et al., 2005). The large-scale data set used in Table S2 were obtained from Lackner et al. (2007). Statistical analyses were performed using a t test (GraphPad Prism 4). The Codon usage presented in Table S1 was obtained from the Codon Usage Database (http://www.kazusa.or.jp/codon/) and computed with the EMBOSS cusp (http://emboss.sourceforge.net/apps/cvs/emboss/apps/cusp.html). The Z-score presented in Table S1, that indicates how many standard deviations a codon frequency is above or below the mean, was calculated as the actual codon frequency minus the average codon frequency, the resulting value was divided by the standard deviation obtained from the 5082 genes presented in Table S1. The Gene-Specific Codon Usage shown in Figure 2 was obtained as previously described (Begley et al., 2007).
Quantitative Analysis of Gene and Protein Expression
qRT-PCR was performed as described previously (Coudreuse et al., 2010). For quantitative western blotting, 5 3 107 cells were treated by the alkaline protein extraction method (Matsuo et al., 2006), separated by SDS-PAGE and transferred on nitrocellulose membranes Immunoblotting using anti-HA and anti-tubulin, used to normalize, were performed and each signal was analyzed using LI-COR scanner following the instructions of the manufacturer.
Supplementary Material
Figure S1. The mcm5s2U34 Modified Nucleotide Structure and Synthesis, Related to Figure 1
Green: s2 modification ; red: mcm5 modification
(A) The genetic code. Modified wobble base are indicated and the double mcm5s2U is highlighted (mcm5 in red, s2 34 in green).
(B) Structure of the doubly modified uridine (left 3D, right 2D) with the same color code as in A. The unmodified molecule is shown in the insert.
(C) Purified tRNAs from the indicated strains were separated in the presence or absence of APM ([p-(N-acrylamino)-phenyl]mercuric chloride) that band shifts thiolated molecules. A northern blot was performed with the indicated labeled probes. The tRNAMetCUT, recognizing the methionine codon is not thiolated and used as a control.
(D) LC-MS analysis and quantification of an hydrolysate of small RNAs from the indicated strains. The ratio of the single or double mcm5 and s2 modifications in the indicated strains is indicated. While mcm5U is increased in the ctu1 mutant, no similar increase in s2U is detected in the absence of elp3, which indicates that the mcm5 modification is required for efficient thiolation. The values obtained for the wild-type strain were set to 1. The actual values are also indicated in brackets. See Experimental Procedures for details.
(E) Spot dilution assay of the Δelp1, Δelp3, Δelp4 and Δelp6 strains (from the Bioneer S. pombe deletion library) expressing elevated levels of tRNALysUUU or tRNALysCUU on indicated selective minimal media.
Figure S2. The Absence of the mcm5s2 Modification Results in Specific Phenotypes, Related to Figure 1
(A) Phenotyping of the Δctu1, Δelp3 or Δctu1Δelp3 mutant strains by spot dilution assay for growth in the presence of the listed compounds.
(B) Relevant growth defects from a shown for the Δctu1 Δelp3 mutant.
Figure S3. Proteome-wide Analysis by Reverse Protein Macroarrays, Related to Figure 3
(A) Left panel: schematic representation of the ORFeome strains used. Right panel: a representative membrane after immunoblotting. Tagged proteins were quantified with the anti-His (red) and the internal control a-tubulin was simultaneously quantified with an anti-tubulin (green) in the wt, Δctu1 or Δelp3 strains together with a negative control.
(B) The reverse protein macro-array analysis was performed in six steps. 1/ ORFeome on YE in a 384 spot grid; 2/ Streaking of each 96 strains of an ORFeome plate on YE; 3/ Streaking of each 96 strains of an ORFeome plate on EMM during 22h to induce the expression of nmt1 promoter; 4/ Harvest of each 96 strains in 96-well plate; 5/ Whole cell lysate by a guanidine buffer; 6/ Spotting of the protein extracts on nitrocellulose membrane and dot blot.
(C) Schematic representation of the detection used for quantification of the his-tagged proteins and normalization by the internal control.
(D) Plots showing the correlation between the expression levels in three representative replicate experiments of the wt, Δctu1 or Δelp3. The top-right part shows the correlation coefficients for each pair-wise combination, while the bottom-left part shows the corresponding scatter plots.
Figure S4. RNAi-Dependent Heterochromatin Silencing Is Defective in the Absence of mcm5s2 tRNA Modification, Related to Figure 4
(A) Schematic representation of the centromere of Chr.1 with the insertion of the ura4 reporter gene in the outer repeat sequences (otr1R). The presence of heterochromatin in the region leads to silencing of ura4 and consequent 5-FOA resistance in the wt.
(B) Spot dilution assay on YES medium in the presence or absence of 5-FOA. The wt and Δelp3 strains harboring, or not, a copy of ura4 inserted at the peri- centromeric otr region (otr1R::ura4) were spotted on the indicated medium. The Δswi6 that lacks centromeric heterochromatin in the outer repeats was used as control for loss of silencing and consequent 5-FOA sensitivity.
(C) Quantitative RT-PCR measurement of outer repeat expressed ura4 mRNA level in the indicated strains, normalized to act1 (mean of otr1::ura4 mRNA expression; SEM = standard error of the mean; n = replicate number; p value obtained by Student’s t test).
(D) Hierarchical clustering and heat map analysis of the protein expression level of the Gene Ontology group “Chromatin Silencing by small RNA” (GO 0031048) in the Δctu1 and Δelp3 strain compared to wild-type. The data are presented as log2 mutant/wild-type ratio of hybridization signals and are color-coded as in Figure 3A.
ACKNOWLEDGMENTS
We thank P. Russell, K. Gould, J.-P. Javerzat, A. Byström, J. Kohli, and P. Nurse for reagents; A. Chariot and A. Byström for fruitful discussions and critical reading of the manuscript; Y. Kawamura, H. van Bakel, and W. Van Delm for biostatistical analyses; the GEMO laboratory for discussions. This work was supported by an EMBO STF grant (58-2008), grant FRFC 2.4510.10, Credit aux chercheurs 1.5.013.09, and grant MIS F.4523.11 to D.H. J.S. and D.A.W. are supported by grant 0920229 from the US National Science Foundation. F.B. is an FRIA research fellow, and D.H. is an FNRS research associate. This work is dedicated to the memory of Pierre Thuriaux.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, four figures, and three tables and can be found with this article online atdoi:10.1016/j.celrep.2012.04.001.
LICENSING INFORMATION This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
REFERENCES
- Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Ashraf SS, Sochacka E, Cain R, Guenther R, Malkiewicz A, Agris PF. Single atom modification (O/S) of tRNA confers ribosome binding. RNA. 1999;5:188–194. doi: 10.1017/s1355838299981529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamps S, Westerling T, Pihlak A, Tafforeau L, Vandenhaute J, Mäkelä TP, Hermand D. Mcs2 and a novel CAK subunit Pmh1 associate with Skp1 in fission yeast. Biochem. Biophys. Res. Commun. 2004;325:1424–1432. doi: 10.1016/j.bbrc.2004.10.190. [DOI] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR. Biosynthesis and function of modified nucleosides. In: D. S, RajBhandary U, editors. tRNA: Structure, Biosynthesis, and Foundation. American Society for Microbiology; Washington, DC: 1995. pp. 165–205. [Google Scholar]
- Björk GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeding CS, Hudson J, Balasubramanian MK, Hemmingsen SM, Young PG, Gould KL. The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell. 1998;9:3399–3415. doi: 10.1091/mbc.9.12.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y. A role for codon order in translation dynamics. Cell. 2010;141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Tuck S, Byström AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5:e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudreuse D, van Bakel H, Dewez M, Soutourina J, Parnell T, Vandenhaute J, Cairns B, Werner M, Hermand D. A gene-specific requirement of RNA polymerase II CTD phosphorylation for sexual differentiation in S. pombe. Curr. Biol. 2010;20:1053–1064. doi: 10.1016/j.cub.2010.04.054. [DOI] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, Hermand D. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. USA. 2008;105:5459–5464. doi: 10.1073/pnas.0709404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Byström AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Fersht N, Hermand D, Hayles J, Nurse P. Cdc18/CDC6 activates the Rad3-dependent checkpoint in the fission yeast. Nucleic Acids Res. 2007;35:5323–5337. doi: 10.1093/nar/gkm527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossenbacher AM, Stadelmann B, Heyer WD, Thuriaux P, Kohli J, Smith C, Agris PF, Kuo KC, Gehrke C. Antisuppressor mutations and sulfur-carrying nucleosides in transfer RNAs of Schizosaccharomyces pombe. J. Biol. Chem. 1986;261:16351–16355. [PubMed] [Google Scholar]
- Hermand D, Nurse P. Cdc18 enforces the S-phase checkpoint by anchoring the Rad3-Rad26 complex to chromatin. Mol. Cell. 2007;6:553–563. doi: 10.1016/j.molcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Thuriaux P, Kohli J, Ebert P, Kersten H, Gehrke C, Kuo KC, Agris PF. An antisuppressor mutation of Schizosaccharomyces pombe affects the post-transcriptional modification of the “wobble” base in the anticodon of tRNAs. J. Biol. Chem. 1984;259:2856–2862. [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Lu J, Byström AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski D, Taübert JE, Bär C, Stark MJ, Schaffrath R. Distinct subsets of Sit4 holophosphatases are required for inhibition of Saccharomyces cerevisiae growth by rapamycin and zymocin. Eukaryot. Cell. 2009;8:1637–1647. doi: 10.1128/EC.00205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Russell P. The protein kinase Cdr2, related to Nim1/ Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol. Biol. Cell. 1998;9:3321–3334. doi: 10.1091/mbc.9.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger MK, Pedersen S, Hagervall TG, Sørensen MA. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 1998;284:621–631. doi: 10.1006/jmbi.1998.2196. [DOI] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bähler J. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol. Cell. 2007;26:145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- Lemau de Talancé V, Bauer F, Hermand D, Vincent SP. A simple synthesis of APM ([p-(N-acrylamino)-phenyl]mercuric chloride), a useful tool for the analysis of thiolated biomolecules. Bioorg. Med. Chem. Lett. 2011;21:7265–7267. doi: 10.1016/j.bmcl.2011.10.051. [DOI] [PubMed] [Google Scholar]
- Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Asakawa K, Toda T, Katayama S. A rapid method for protein extraction from fission yeast. Biosci. Biotechnol. Biochem. 2006;70:1992–1994. doi: 10.1271/bbb.60087. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Breunig KD, Stark MJ, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol. Microbiol. 2009;73:869–881. doi: 10.1111/j.1365-2958.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jäger G, Gong Z, Byström AS, Schaffrath R, Breunig KD. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010;76:1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- Murphy FV, 4th, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent RL, Johnsson A, Fleharty B, Gogol M, Xue-Franzén Y, Seidel C, Wright AP, Forsburg SL. Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation. BMC Genomics. 2010;11:59. doi: 10.1186/1471-2164-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P. Temperature sensitive allosuppressor mutants of the fission yeast S. pombe influence cell cycle control over mitosis. Mol. Gen. Genet. 1984;196:332–338. [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Gusella JF. Familial dysautonomia. Curr. Opin. Genet. Dev. 2002;12:307–311. doi: 10.1016/s0959-437x(02)00303-9. [DOI] [PubMed] [Google Scholar]
- Suzuki T. Biosynthesis and function of wobble modifications. In: Grosjean H, editor. Finetuning of RNA functions by modification and editing. Springer-Verlag; Heidelberg: 2005. [Google Scholar]
- Svejstrup JQ. Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA. 2002;99:3517–3522. doi: 10.1073/pnas.022042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseu-douridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011;30:79–89. doi: 10.1038/emboj.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A, Agris PF. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry. 2000;39:13390–13395. doi: 10.1021/bi001302g. [DOI] [PubMed] [Google Scholar]
- Young PG, Fantes PA. Changed division response mutants function as allosuppressors. In: Friedman PSSJ, editor. Growth, Cancer and the Cell Cycle. Humana Press; Clifton, N-Y: 1984. pp. 221–228. [Google Scholar]
- Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The mcm5s2U34 Modified Nucleotide Structure and Synthesis, Related to Figure 1
Green: s2 modification ; red: mcm5 modification
(A) The genetic code. Modified wobble base are indicated and the double mcm5s2U is highlighted (mcm5 in red, s2 34 in green).
(B) Structure of the doubly modified uridine (left 3D, right 2D) with the same color code as in A. The unmodified molecule is shown in the insert.
(C) Purified tRNAs from the indicated strains were separated in the presence or absence of APM ([p-(N-acrylamino)-phenyl]mercuric chloride) that band shifts thiolated molecules. A northern blot was performed with the indicated labeled probes. The tRNAMetCUT, recognizing the methionine codon is not thiolated and used as a control.
(D) LC-MS analysis and quantification of an hydrolysate of small RNAs from the indicated strains. The ratio of the single or double mcm5 and s2 modifications in the indicated strains is indicated. While mcm5U is increased in the ctu1 mutant, no similar increase in s2U is detected in the absence of elp3, which indicates that the mcm5 modification is required for efficient thiolation. The values obtained for the wild-type strain were set to 1. The actual values are also indicated in brackets. See Experimental Procedures for details.
(E) Spot dilution assay of the Δelp1, Δelp3, Δelp4 and Δelp6 strains (from the Bioneer S. pombe deletion library) expressing elevated levels of tRNALysUUU or tRNALysCUU on indicated selective minimal media.
Figure S2. The Absence of the mcm5s2 Modification Results in Specific Phenotypes, Related to Figure 1
(A) Phenotyping of the Δctu1, Δelp3 or Δctu1Δelp3 mutant strains by spot dilution assay for growth in the presence of the listed compounds.
(B) Relevant growth defects from a shown for the Δctu1 Δelp3 mutant.
Figure S3. Proteome-wide Analysis by Reverse Protein Macroarrays, Related to Figure 3
(A) Left panel: schematic representation of the ORFeome strains used. Right panel: a representative membrane after immunoblotting. Tagged proteins were quantified with the anti-His (red) and the internal control a-tubulin was simultaneously quantified with an anti-tubulin (green) in the wt, Δctu1 or Δelp3 strains together with a negative control.
(B) The reverse protein macro-array analysis was performed in six steps. 1/ ORFeome on YE in a 384 spot grid; 2/ Streaking of each 96 strains of an ORFeome plate on YE; 3/ Streaking of each 96 strains of an ORFeome plate on EMM during 22h to induce the expression of nmt1 promoter; 4/ Harvest of each 96 strains in 96-well plate; 5/ Whole cell lysate by a guanidine buffer; 6/ Spotting of the protein extracts on nitrocellulose membrane and dot blot.
(C) Schematic representation of the detection used for quantification of the his-tagged proteins and normalization by the internal control.
(D) Plots showing the correlation between the expression levels in three representative replicate experiments of the wt, Δctu1 or Δelp3. The top-right part shows the correlation coefficients for each pair-wise combination, while the bottom-left part shows the corresponding scatter plots.
Figure S4. RNAi-Dependent Heterochromatin Silencing Is Defective in the Absence of mcm5s2 tRNA Modification, Related to Figure 4
(A) Schematic representation of the centromere of Chr.1 with the insertion of the ura4 reporter gene in the outer repeat sequences (otr1R). The presence of heterochromatin in the region leads to silencing of ura4 and consequent 5-FOA resistance in the wt.
(B) Spot dilution assay on YES medium in the presence or absence of 5-FOA. The wt and Δelp3 strains harboring, or not, a copy of ura4 inserted at the peri- centromeric otr region (otr1R::ura4) were spotted on the indicated medium. The Δswi6 that lacks centromeric heterochromatin in the outer repeats was used as control for loss of silencing and consequent 5-FOA sensitivity.
(C) Quantitative RT-PCR measurement of outer repeat expressed ura4 mRNA level in the indicated strains, normalized to act1 (mean of otr1::ura4 mRNA expression; SEM = standard error of the mean; n = replicate number; p value obtained by Student’s t test).
(D) Hierarchical clustering and heat map analysis of the protein expression level of the Gene Ontology group “Chromatin Silencing by small RNA” (GO 0031048) in the Δctu1 and Δelp3 strain compared to wild-type. The data are presented as log2 mutant/wild-type ratio of hybridization signals and are color-coded as in Figure 3A.