ATP bound in the active site of protein kinase A is readily hydrolysed to ADP and free phosphate by X-ray irradiation at room temperature. The phosphate ion observed in the active site causes a dramatic conformational change of the bound peptide inhibitor.
Keywords: protein kinase A, ATPases, ATP hydrolysis
Abstract
Post-translational protein phosphorylation by protein kinase A (PKA) is a ubiquitous signalling mechanism which regulates many cellular processes. A low-temperature X-ray structure of the ternary complex of the PKA catalytic subunit (PKAc) with ATP and a 20-residue peptidic inhibitor (IP20) at the physiological Mg2+ concentration of ∼0.5 mM (LT PKA–MgATP–IP20) revealed a single metal ion in the active site. The lack of a second metal in LT PKA–MgATP–IP20 renders the β- and γ-phosphoryl groups of ATP very flexible, with high thermal B factors. Thus, the second metal is crucial for tight positioning of the terminal phosphoryl group for transfer to a substrate, as demonstrated by comparison of the former structure with that of the LT PKA–Mg2ATP–IP20 complex obtained at high Mg2+ concentration. In addition to its kinase activity, PKAc is also able to slowly catalyze the hydrolysis of ATP using a water molecule as a substrate. It was found that ATP can be readily and completely hydrolyzed to ADP and a free phosphate ion in the crystals of the ternary complex PKA–Mg2ATP–IP20 by X-ray irradiation at room temperature. The cleavage of ATP may be aided by X-ray-generated free hydroxyl radicals, a very reactive chemical species, which move rapidly through the crystal at room temperature. The phosphate anion is clearly visible in the electron-density maps; it remains in the active site but slides about 2 Å from its position in ATP towards Ala21 of IP20, which mimics the phosphorylation site. The phosphate thus pushes the peptidic inhibitor away from the product ADP, while resulting in dramatic conformational changes of the terminal residues 24 and 25 of IP20. X-ray structures of PKAc in complex with the nonhydrolysable ATP analogue AMP-PNP at both room and low temperature demonstrated no temperature effects on the conformation and position of IP20.
1. Introduction
Protein phosphorylation is an important post-translational modification that serves signaling purposes in a myriad of cellular processes (Johnson et al., 1996 ▶). During the chemical process, the γ-phosphoryl group of adenosine triphosphate (ATP) is reversibly transferred to a hydroxyl substituent of a serine, threonine or tyrosine residue of the substrate protein or peptide to generate a phospho-monoester. Phosphorylation is catalyzed by a family of enzymes called protein kinases. Within this large and structurally diverse family, cAMP-dependent protein kinase A (PKA) is the best characterized member and is considered a paradigm for all protein kinases (Taylor et al., 2004 ▶). PKA exists as a tetrameric inactive holoenzyme composed of two regulatory and two catalytic subunits. Following the binding of four cAMP molecules, produced by β-adrenergic stimulation, to the regulatory subunits the tetramer dissociates, releasing the active catalytic subunits PKAc. The catalytic activity of PKAc is then further regulated by conformational changes induced by ATP binding, the coordination of metal cofactors bound to ATP and a heat-stable protein kinase inhibitor (PKI; Walsh et al., 1990 ▶). Although protein kinases may be structurally different, they all share a highly conserved bean-shaped catalytic core of about 220 residues (Fig. 1 ▶ a) with two lobes (small and large) generating a cleft at the base between them for ATP binding and a ledge for protein–substrate docking (Knighton et al., 1991 ▶; Zheng, Knighton et al., 1993 ▶; Zheng, Trafny et al., 1993 ▶). PKAc is a metalloenzyme that requires one or two divalent metal ions to bind in the active site for activity (Adams & Taylor, 1993 ▶). In living cells the physiological metal is magnesium, although in vitro some transition metals also support the phosphotransferase activity of PKAc (Bhatnagar et al., 1983 ▶).
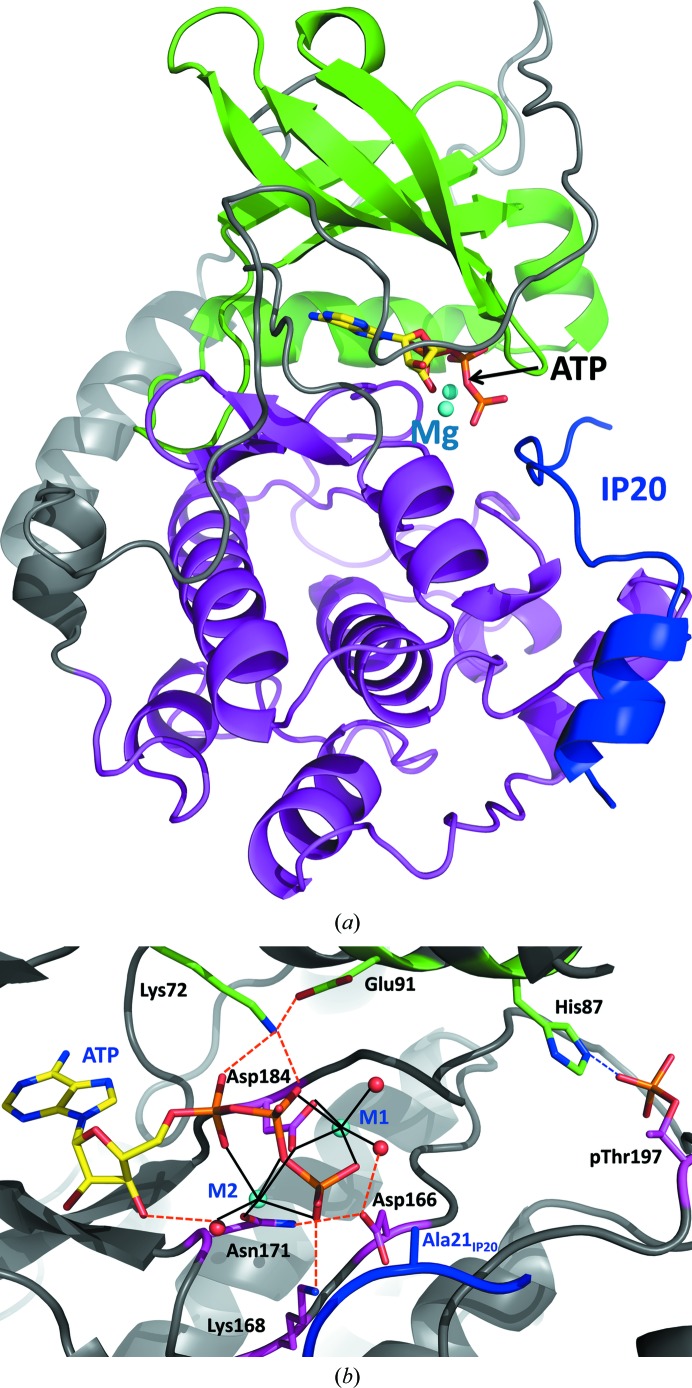
Figure 1.
(a) Overall structure of PKAc in cartoon representation. The small lobe is colored green, the large lobe is colored magenta and IP20 is colored blue. The two Mg2+ ions are represented by cyan spheres, while ATP is shown in stick representation colored by atom type. (b) A close-up view of the active site. Water molecules are represented by red spheres. Metal coordination is shown as black solid lines, whereas hydrogen bonding is represented by orange dashed lines.
There have been extensive efforts to understand the catalytic cycle of PKAc using a variety of experimental and theoretical approaches over the past two decades. The sequence of events and the kinetics of each individual step comprising the overall phosphorylation process have been established; however, the mechanistic details of individual stages are still being debated (Masterson et al., 2010 ▶, 2011 ▶; Montenegro et al., 2011 ▶). Most importantly, global conformational changes of the enzyme determine the rate of turnover, whereas the actual transfer of a phosphoryl group from ATP to OH of Ser/Thr happens on a much faster timescale. X-ray crystallographic studies have identified three conformational states of PKAc: open (apo form), intermediate (with nucleotide bound) and closed (with nucleotide and substrate/inhibitor bound). NMR and molecular-dynamics simulations have shown that during turnover PKAc interconverts between these conformational states, with the nucleotide acting as a dynamic and allosteric activator (Masterson et al., 2010 ▶, 2011 ▶; Montenegro et al., 2011 ▶). The slow global conformational change from open to intermediate induced by nucleotide binding primes the active site for substrate binding, whereas the chemical step only occurs in the closed form of the enzyme, which is stabilized by salt-bridge hydrogen bonding between His87 and phosphorylated Thr197 (Fig. 1 ▶ b; Cox et al., 1994 ▶). The conformational changes associated with binding of substrate ATP and release of product ADP both partially determine the rate of turnover at low (physiological) Mg2+ concentrations of ∼0.5 mM (Shaffer & Adams, 1999a ▶,b ▶). At high Mg2+ concentrations the removal of ADP from the active site after the phosphoryl group has been transferred is the rate-limiting step (Lew et al., 1997 ▶). Additionally, the rate of the chemical step is invariant at any metal concentration.
The phosphotransfer reaction is believed to proceed in a concerted way according to a standard SN2 nucleophilic substitution mechanism, with a pentacoordinated trigonal pyramidal γ P atom in the transition state (Montenegro et al., 2011 ▶; Valiev et al., 2003 ▶; Díaz & Field, 2004 ▶; Cheng et al., 2005 ▶; Madhusudan et al., 1994 ▶; Granot et al., 1980 ▶). The presence of at least one metal ion is crucial for phosphoryl transfer; there is no measurable activity in the absence of metal ions. The binding of the second metal ion to the active site of PKAc is believed to properly position the leaving γ-phosphoryl group while making a tighter complex with the product ADP. Asp184 and Asn171 bind to the metal ions and are important for catalytic function (Gibbs & Zoller, 1991 ▶). The OH of the substrate Ser/Thr either donates its proton to the adjacent conserved Asp166, the carboxylate moiety of which lies within hydrogen-bonding distance, facilitating general base catalysis, or transfers it to the incoming phosphate in the transition state. Although some experimental data and theoretical calculations (Zhou & Adams, 1997 ▶; Blachut-Okrasinska et al., 1999 ▶; Hutter & Helms, 1999 ▶) do not support Asp166 being the catalytic base, a recent QM/MM study by Montenegro et al. (2011 ▶) concluded that Asp166 might act as a ‘proton trap’ late in the reaction process. In fact, the main-chain torsion angles ϕ and ψ of Asp166 (and also of Asp184 coordinating the metal ion) are distorted from their Ramachandran favorable values in PKAc structures in order to position the side chain close to the substrate. Other residues are also important for the phosphotransferase function. Glu91 correctly positions Lys72, which in turn anchors the α- and β-phosphates, facilitating the transfer. Additionally, Lys168 forms a hydrogen bond to the γ-phosphate and acts either to stabilize the negative electrostatic charge in the transition state or to directly transfer one of its protons to the phosphorylated residue (Fig. 1 ▶ b; Montenegro et al., 2011 ▶; Cheng et al., 2005 ▶).
To advance our understanding of the role of metal cations in the phosphoryl-transfer step in the catalytic process, we have obtained low-temperature (LT) X-ray structures of the ternary complexes of PKAc with ATP and the peptidic inhibitor IP20, which represents residues 5–24 of PKI, at low (∼0.5 mM) and at high (25 mM) Mg2+ concentrations, which are designated LT PKA–MgATP–IP20 (2.0 Å resolution; PDB entry 4dh1) and LT PKA–Mg2ATP–IP20 (2.2 Å resolution; PDB entry 4dh3), respectively. We observed differences in the occupancy of M1, the position of M2 and the flexibility of the ATP phosphates that are significant for the transfer reaction.
Furthermore, we have exploited the ability of X-rays to generate free radicals in water in order to investigate the detailed changes that occur in PKAc during ATP hydrolysis. PKAc possesses intrinsic ATPase activity that is measurable (k cat ≃ 0.01 s−1) but is very weak relative to its ability to phosphorylate protein and peptide substrates. In this chemical reaction the γ-phosphoryl group is cleaved from ATP by an incoming water molecule, resulting in ADP and free phosphate. Damage to proteins during X-ray crystallographic data collection owing to the primary effect of photoelectron generation or the secondary effect of free-radical creation has been known for some time (Blake & Philips, 1962 ▶). One of the dominant processes involved is thought to be the radiolysis of water to free-radical hydroxyl groups, the swift diffusion of which through the crystal can be slowed by data collection at low temperatures. We report a room-temperature (RT) X-ray structure of a ternary complex of PKAc grown in the presence of ATP, IP20 and excess Mg2+ (which we call RT PKA–Mg2ADP·PO4–IP20; PDB entry 4dh5) at a resolution of 2.2 Å. In the RT structure ATP is completely hydrolyzed to ADP and a free phosphate ion, whereas at low temperature in LT PKA–Mg2ATP–IP20 the ATP is not damaged. Furthermore, there are significant conformational differences between the two structures. In order to determine whether these differences are just reflections of different thermal motions or whether they indeed represent two different conformational states of the complex (before and after ATP hydrolysis), we determined two further structures: ternary complexes with IP20, excess Mg2+ and the nonhydrolyzable ATP analog AMP-PNP [adenosine 5′-(β,γ-imido)triphosphate] at LT (LT PKA–Mg2AMPPNP–IP20; PDB entry 4hd7) to 1.8 Å resolution and at RT (RT PKA–Mg2AMPPNP–IP20; PDB entry 4dh8) to 2.3 Å resolution. We observed no significant differences between these structures and LT PKA–Mg2ATP–IP20. The changes in the structures of the ternary complexes at low and high Mg concentration and before and after ATP hydrolysis provide new details on the catalytic activity of PKAc.
2. Materials and methods
2.1. General information
Inhibitory peptide IP20 (TTYADFIASGRTGRRNAIHD; residues 5–24 of the heat-stable PKAc inhibitor PKI) was custom-synthesized by and purchased from GenScript (Piscataway, New Jersey, USA). ATP as the magnesium salt and AMP-PNP as the lithium salt were purchased from Sigma–Aldrich (St Louis, Misssouri, USA). Crystallization reagents were purchased from Hampton Research (Aliso Viejo, California, USA).
2.2. Protein expression and purification
Native wild-type PKAc was expressed and purified as described previously (Yang et al., 2004 ▶). The construction of His6-tagged recombinant mouse PKAc (PKAHis6) has been described elsewhere (Narayana et al., 1997 ▶). PKAHis6 was expressed in Escherichia coli using LB or minimal medium using the same procedure as used for the native enzyme. When minimal medium was used the procedure was altered by allowing expression at 297 K overnight because of slow cell growth. PKAHis6 was purified by affinity chromatography using HisTrap high-performance chromatography columns supplied by GE Healthcare (Piscataway, New Jersey, USA). The enzyme was then buffer-exchanged with 50 mM MES, 20 mM MgCl2, 250 mM NaCl, 2 mM DTT pH 6.5 on a desalting column. The magnesium salt was not used in the solutions intended for the low-Mg2+-concentration complex. Isoforms of PKAHis6 were not separated, without any noticeable effect on crystallization.
2.3. Crystallization and data collection
For crystallization, all PKAc batches were concentrated to 10 mg ml−1. The ternary complexes with ATP (or AMP-PNP) and IP20 were made before crystallization was set up by mixing the enzyme, nucleoside and peptide inhibitor in a molar ratio of 1:10:10. Crystals grew as long sticks using well solutions consisting of 50 mM MES pH 6.5, 50 mM Mg2Cl, 5 mM DTT, 12–15% PEG 4000 at 277 K. The magnesium salt was not used in the crystallization mother liquor when crystals of the low-Mg2+ complex were grown.
2.4. Structure determination and refinement
X-ray crystallographic data were collected at room temperature for RT PKA-Mg2ADP·PO4–IP20 and RT PKA–Mg2AMPPNP–IP20 and from cooled samples at 100 K for LT PKA–MgATP–IP20, LT PKA–Mg2ATP–IP20 and LT PKA–Mg2AMPPNP–IP20. The data sets were collected on Rigaku FR-E or Rigaku MicroMax-007 HF generators equipped with Osmic VariMax optics. Diffraction images were obtained using an R-AXIS IV++ detector or a Bruker CCD 1000 detector. Diffraction data were integrated and scaled using the CrystalClear/d*TREK (Pflugrath, 1999 ▶) and HKL-3000 software suites (Minor et al., 2006 ▶) and the structures were refined using CNS (Brünger et al., 1998 ▶; Brunger, 2007 ▶). A summary of the crystallographic data is given in Table 1 ▶. All of the structures were of isoform 2 and contained three phosphorylated residues, namely Ser139, Thr197 and Ser338, as observed in PDB entry 1atp (Zheng, Trafny et al., 1993 ▶), which was used as a starting model to solve all of the structures described here. The structures were built and manipulated with Coot (Emsley et al., 2010 ▶), while the figures were generated using the PyMOL molecular-graphics system (v.1.4; Schrödinger LLC).
Table 1. X-ray crystallographic data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell. Data were collected from one crystal for each structure.
| LT PKA–MgATP–IP20 | LT PKA–Mg2ATP–IP20 | RT PKA–Mg2ADP·PO4–IP20 | LT PKA–Mg2AMPPNP–IP20 | RT PKA–Mg2AMPPNP–IP20 | |
|---|---|---|---|---|---|
| Data collection | |||||
| Temperature (K) | 100 | 100 | 293 | 100 | 293 |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit-cell parameters | |||||
| a (Å) | 58.43 | 58.90 | 58.90 | 57.54 | 59.21 |
| b (Å) | 79.92 | 79.80 | 79.63 | 79.65 | 80.03 |
| c (Å) | 98.61 | 98.51 | 99.49 | 98.01 | 99.20 |
| Resolution (Å) | 20.00–2.00 (2.07–2.00) | 28.95–2.20 (2.28–2.20) | 19.86–2.20 (2.28–2.20) | 20.00–1.80 (1.86–1.80) | 20.00–2.30 (2.38–2.30) |
| R merge | 0.089 (0.438) | 0.118 (0.412) | 0.132 (0.449) | 0.044 (0.256) | 0.107 (0.488) |
| 〈I/σ(I)〉 | 9.8 (2.5) | 5.5 (1.9) | 4.8 (1.9) | 38.5 (8.2) | 15.7 (3.3) |
| Completeness (%) | 91.8 (87.1) | 93.3 (91.1) | 95.1 (94.7) | 99.6 (99.0) | 96.3 (94.3) |
| Multiplicity | 4.8 (4.0) | 3.4 (3.3) | 2.9 (2.8) | 6.9 (6.7) | 5.5 (5.6) |
| Refinement | |||||
| Resolution (Å) | 20–2.00 | 20–2.20 | 20–2.20 | 20–1.80 | 20–2.30 |
| No. of reflections | 29134 (2719) | 22475 (2161) | 23181 (2264) | 42275 (4112) | 20784 (2004) |
| R work/R free | 0.199/0.234 | 0.203/0.233 | 0.187/0.222 | 0.179/0.194 | 0.155/0.189 |
| No. of water molecules | 432 | 321 | 211 | 454 | 224 |
| B factors (Å2) | |||||
| Protein | 27.6 | 32.3 | 37.0 | 22.1 | 30.9 |
| Ligand + Mg | 39.5 | 33.0 | 40.8 | 17.4 | 20.0 |
| IP20 | 29.1 | 33.3 | 37.7 | 22.2 | 30.3 |
| Water | 39.8 | 40.6 | 49.8 | 34.8 | 43.9 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.005 | 0.006 | 0.006 | 0.004 | 0.005 |
| Bond angles (Å) | 1.226 | 1.267 | 1.210 | 1.186 | 1.189 |
3. Results and discussion
3.1. Mg2+ concentration controls ATP-binding affinity, its mobility and its correct positioning for phosphoryl transfer
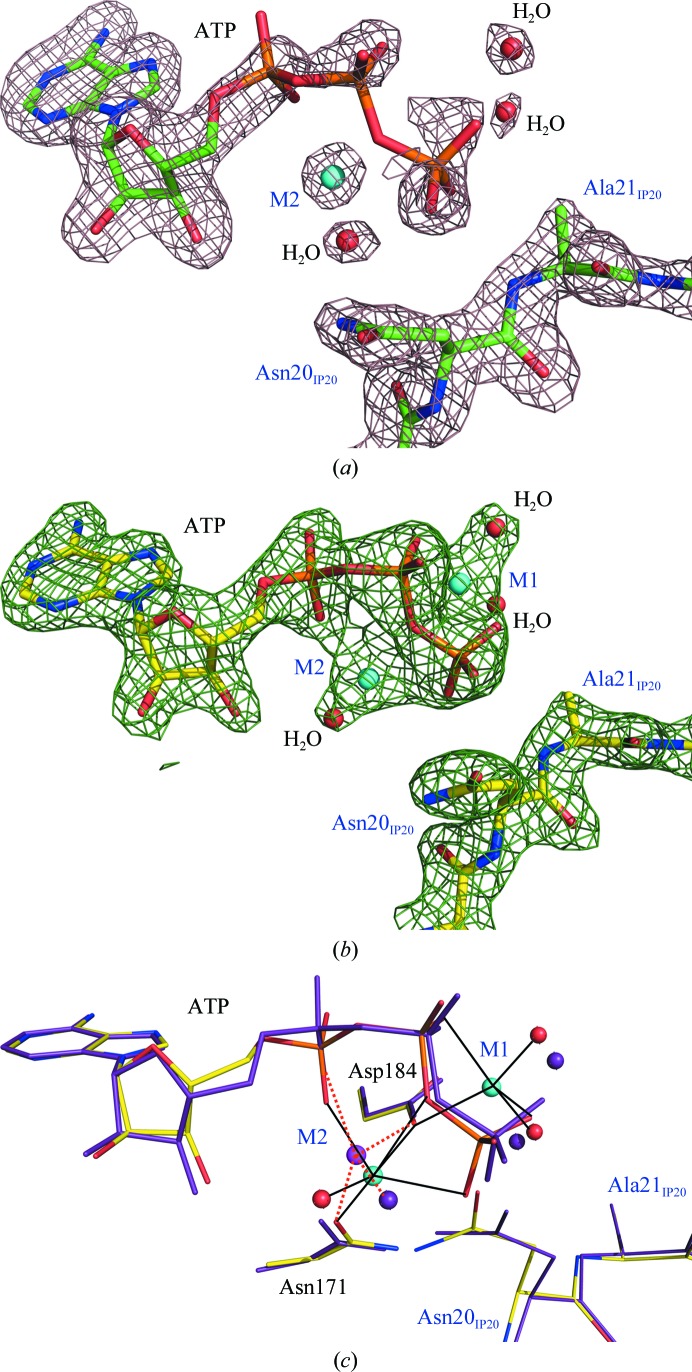
It has previously been proposed, based on 2.7 Å resolution structures of PKAc ternary complexes (Zheng, Knighton et al., 1993 ▶), that at the low physiologically relevant Mg2+ concentration of ∼0.5 mM it binds at an essential high-affinity site M1, coordinating the β- and γ-phosphoryl groups of ATP and two water molecules and chelating a conserved Asp184. It has further been suggested that this metal probably enters the PKAc active site already bound to an ATP molecule (Cini et al., 1983 ▶). At higher Mg2+ concentrations a second metal site, M2, can subsequently be occupied, with the metal ion bound to the α- and γ-phosphates, Asn171 and Asp184 (Fig. 1 ▶ b). However, our higher resolution (2.0 Å) structure LT PKA–MgATP–IP20 demonstrates that when Mg2+ is present at low concentrations during purification and in the crystallization conditions only as part of MgATP a single metal ion is found at M2 rather than at M1 (Fig. 2 ▶ a). Furthermore, M2 slides slightly (0.7 Å) from its position in the high-Mg2+ structure LT PKA–Mg2ATP–IP20 towards the α-phosphoryl group of ATP and now only binds to an α-phosphate oxygen, a conserved water molecule, Asn171 and Asp184. Thus, it no longer chelates ATP by binding to the α- and γ-phosphoryl O atoms as it does in LT PKA–Mg2ATP–IP20 (Fig. 2 ▶ b). The metal ion in LT PKA–MgATP–IP20 has a severely distorted coordination sphere that is altered from the normal octahedral coordination that is found in LT PKA–Mg2ATP–IP20 (Fig. 2 ▶ c). Moreover, the conserved water moves by 1.7 Å towards the terminal phosphate in LT PKA–MgATP–IP20 and is located within hydrogen-bonding distances of its O atoms, while it is about 4 Å away from the γ-phosphoryl group in LT PKA–Mg2ATP–IP20.
Figure 2.
(a) Electron density for the active-site components ATP, Mg2+, water molecules and IP20 in LT PKA–MgATP–IP20 contoured at the 1.0σ level (2.0σ for the metal). (b) Electron density for the active-site components ATP, Mg2+, water molecules and IP20 in LT PKA–Mg2ATP–IP20 contoured at the 1.6σ level. (c) Superposition of low-Mg2+ (purple; metal in magenta) and high-Mg2+ (colored by atom type) structures; the coordination of the metal ions is shown by orange dashed and black solid lines, respectively.
Not unexpectedly, ATP is found to be quite mobile in the low-Mg2+ structure. The phosphoryl groups of the nucleoside have high B factors of the order of 60–80 Å2 that increase in value going from the α-phosphate to the γ-phosphate. The B factors of the phosphoryl groups in LT PKA–Mg2ATP–IP20 are significantly smaller, at ∼30–40 Å2. Clearly, the phosphates have lost a second anchor, the metal ion at M1, and are somewhat free to move. The absence of M1 leads to an increase in the mobility of the phosphoryl groups, whereas the adenine moiety remains tightly bound in its cleft by hydrogen bonds and hydrophobic interactions. This observation may explain why the affinity of ATP for PKAc increases at metal concentrations above 0.5 mM and demonstrates that M1 is important for rigid positioning of the γ-phosphate for the transfer reaction. Interestingly, occupation of the M1 site is not necessary for binding of IP20. In both structures Ala21 of the inhibitor is located at the same position with similar B factors (Fig. 2 ▶ c).
3.2. Structural changes to PKAc and IP20 are associated with in situ ATP hydrolysis
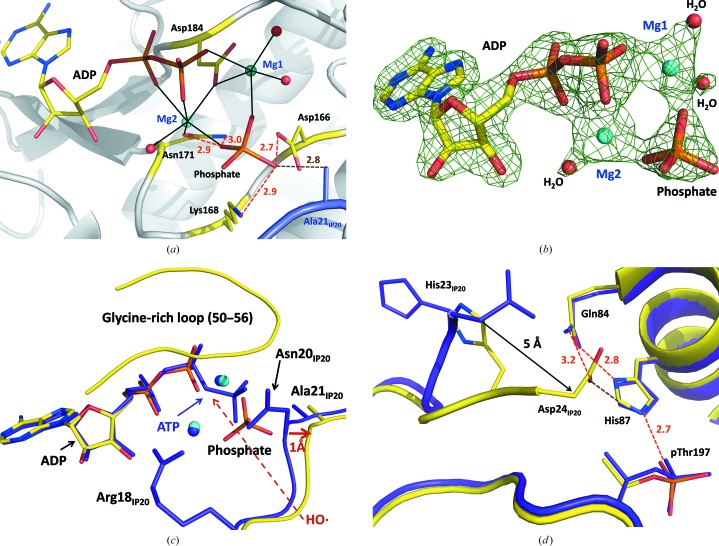
We observe complete hydrolysis of the ATP molecule in the RT X-ray structure of the high-Mg2+ ternary complex of PKAc with ATP and IP20 (Fig. 3 ▶ a). The products, ADP and a free phosphate ion (in RT PKA–Mg2ADP·PO4–IP20), have 100% occupancy; there is no electron density that would indicate the presence of nonhydrolyzed ATP (Fig. 3 ▶ b). Close inspection of the LT PKA–Mg2ATP–IP20 structure suggests that the O atom bridging the β and γ P atoms of ATP would be reachable by a diffusing HO· through a channel flanked by the Arg18 and Asn20 side chains of IP20 and the glycine-rich loop (residues 50–56 of PKAc; Fig. 3 ▶ c). Similar, but only partial, hydrolysis of ATP had been detected in our previous high-resolution structure of the Y204A mutant at 100 K (Yang et al., 2004 ▶). At the time, we proposed that the hydrolysis might have occurred owing to an increased ATPase activity of the mutant compared with the wild-type enzyme. However, based on our current observations of full ATP hydrolysis at room temperature and no ATP hydrolysis at low temperature, it is possible that the partial ATP hydrolysis in our previous low-temperature structure of the PKAc mutant could have resulted from the high X-ray radiation dose necessary for atomic resolution data collection at a synchrotron source. Complete hydrolysis of ATP to ADP has also been detected in the binary ATP–citrate lyase complex, possibly caused by the synchrotron X-ray radiation, although in this case the cleaved free phosphate anion was not visible in the electron density (Sun et al., 2011 ▶). RT PKA–Mg2ADP·PO4–IP20 is therefore unique in representing a transient stage in the reaction after hydrolysis but before the free phosphate has been transferred to the protein substrate.
Figure 3.
(a) Active site of PKAc in the room-temperature structure RT PKA–Mg2ADP·PO4–IP20. Mg2+ coordination with the enzyme residues and water molecules is shown as black solid lines. (b) Electron density for ADP, phosphate, Mg2+ and water molecules in RT PKA–Mg2ADP·PO4–IP20 contoured at the 1.4σ level. (c, d) Superposition of the room- and low-temperature structures RT PKA–Mg2ADP·PO4–IP20 (yellow C atoms, cyan Mg2+ ions) and LT PKA–Mg2ATP–IP20 (purple C atoms and Mg2+ ions), respectively. Hydrogen bonds are shown as orange dashed lines. C—H⋯O interactions are shown as brown dashed lines. All distances are in Å.
In RT PKA–Mg2ADP·PO4–IP20, ADP and the free phosphate ion maintain their interactions with the Mg ions, chelating them at M1 and M2, even though the free phosphate has moved by ∼2 Å towards IP20 from its position as part of ATP. ADP does not significantly change its orientation relative to its position as part of ATP in LT PKA–Mg2ATP–IP20 (Fig. 3 ▶ c). The free phosphate is also held in the active site by hydrogen-bonding interactions with the side chains of Asp166, Lys168 and Asn171 (Fig. 3 ▶ a). Hydrogen bonding to Asn171 is not observed for the γ-phosphoryl group of ATP in LT PKA–Mg2ATP–IP20, as it is over 4 Å away from the amide group of the side chain of Asn171. Conversely, the cleaved phosphate ion loses hydrogen bonding to the main-chain amide of Ser53 in the glycine-rich loop that stabilized the orientation of the terminal phosphoryl of ATP. Intriguingly, an O atom of the free phosphate that points towards Ala21 of IP20 gains a new short contact to the carboxylic group of the catalytic Asp166, while the γ-phosphoryl of ATP lacks such an interaction in the LT structure. The O⋯O distance suggests hydrogen-bond formation, which would require that either the phosphate ion or Asp166 is protonated. The close proximity of the free phosphate to IP20 results in the movement of residues 20–22 of the inihibitor by ∼1 Å away from the active site. This sliding motion of IP20 near the active site may induce the dramatic change in the conformation of its C-terminus relative to that of LT PKA–Mg2ATP–IP20 (Fig. 3 ▶ d). Asp24 of IP20 rotates toward the protein surface, thus moving ∼5 Å closer to the peptide substrate-binding groove of PKAc. In this new position it forms additional bonds to the enzyme. Asp24 of IP20 forms a hydrogen bond to the side chain of Gln84 and makes an unconventional C—H⋯O hydrogen bond with His87 C∊1; these interactions are absent from the LT structure (Fig. 3 ▶ d).
3.3. Structural changes are driven by the chemical reaction and not by temperature
The structural changes in the position and the conformation of the terminal residues 20–24 of IP20 observed between RT PKA–Mg2ADP·PO4–IP20 and LT PKA–Mg2ATP–IP20 may be driven either by the temperature differences between the two data sets or the chemical reaction in the PKAc active site. In order to differentiate between these two possibilities, we determined X-ray structures of similar complexes containing the substrate analogue AMP-PNP at both room and low temperature. The only chemical difference between ATP and AMP-PNP is that the β,γ-bridging O atom of the former is replaced by an NH group in the latter, making AMP-PNP less prone to hydrolysis.
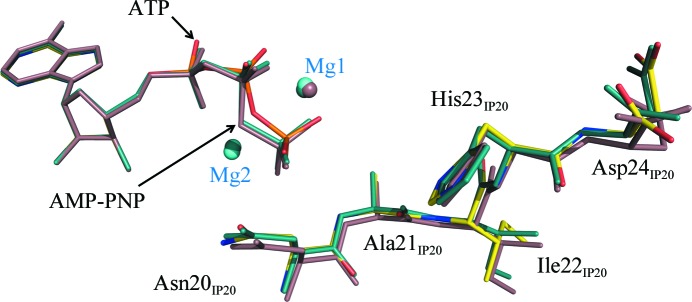
When the LT PKA–Mg2AMPPNP–IP20 and RT PKA–Mg2AMPPNP–IP20 structures were superimposed with that of LT PKA–Mg2ATP–IP20, the r.m.s. deviation was just 0.2 Å. The difference in the IP20 conformations between the three structures is insignificant, with the C-termini of the peptide inhibitors occupying similar positions (Fig. 4 ▶). Thus, crystal cooling does not lead to significant structural changes in the ternary complexes of PKAc. We therefore propose that the position of the free phosphate in the active site of RT PKA–Mg2ADP·PO4–IP20, 2 Å away from the γ-phosphoryl group of ATP and closer to IP20, imposes an electrostatic effect on the surrounding residues owing to its large negative charge, which may trigger a cascade of conformational changes in IP20 that culminate in a large swing of the C-terminus of the inhibitor.
Figure 4.
Superposition of LT PKA–Mg2ATP–IP20 (yellow C atoms), LT PKA–Mg2AMPPNP–IP20 (dark cyan C atoms) and RT PKA–Mg2AMPPNP–IP20 (light brown C atoms).
In summary, we have determined the X-ray crystallographic structures of various ternary complexes of PKAc with IP20 and either ATP or its analogue AMP-PNP in the presence of different amounts of Mg2+ and at both RT and LT. The results revealed that the concentration of Mg2+ controls the binding affinity of ATP, its mobility and its positioning for phosphoryl transfer. We exploited the ability of X-ray irradiation to produce diffusible free radicals in order to hydrolyze ATP and trap a transient stage after hydrolysis but before the transfer of the released phosphate. The results revealed significant conformation changes of both PKAc and IP20 associated with the chemical reaction. In future studies, we intend to pursue neutron protein crystallographic studies of PKAc complexes in order to directly determine the protonation states of active-site residues and ATP and the identities of solvent molecules bound to the metal centers.
Supplementary Material
PDB reference: LT PKA–MgATP–IP20, 4dh1
PDB reference: LT PKA–Mg2ATP–IP20, 4dh3
PDB reference: RT PKA–Mg2ADP·PO4–IP20, 4dh5
PDB reference: LT PKA–Mg2AMPPNP–IP20, 4dh7
PDB reference: RT PKA–Mg2AMPPNP–IP20, 4dh8
Acknowledgments
ST, PL, HJ and AYK were partly supported by a UCOP grant. AYK, HJ, MJW and SZF were partly supported by a DOE-OBER grant to the neutron Protein Crystallography Station at LANSCE. PL was partly supported by an NIH–NIGMS-funded consortium (1R01GM071939-01) between ORNL and LBNL to develop computational tools for neutron protein crystallography. BLH was supported by NSF 446218.
References
- Adams, J. A. & Taylor, S. S. (1993). Protein Sci. 2, 2177–2186. [DOI] [PMC free article] [PubMed]
- Bhatnagar, D., Roskoski, R., Rosendahl, M. S. & Leonard, N. J. (1983). Biochemistry, 22, 6310–6317. [DOI] [PubMed]
- Blachut-Okrasinska, E., Lesyng, B., Briggs, J. M., McCammon, J. A. & Antosiewicz, J. M. (1999). Eur. J. Biophys. 28, 457–467. [DOI] [PubMed]
- Blake, C. C. F. & Philips, D. C. (1962). Biological Effects of Ionizing Radiation at the Molecular Level, pp. 183–191. Vienna: International Atomic Energy Agency.
- Brunger, A. T. (2007). Nature Protoc. 2, 2728–2733. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Cheng, Y., Zhang, Y. & McCammon, J. A. (2005). J. Am. Chem. Soc. 127, 1553–1562. [DOI] [PubMed]
- Cini, R., Sabat, M., Sundaralingam, M., Burla, M. C., Nunzi, A., Polidori, G. & Zanazzi, P. F. (1983). J. Biomol. Struct. Dyn. 1, 633–637. [DOI] [PubMed]
- Cox, S., Radzio-Andzelm, E. & Taylor, S. S. (1994). Curr. Opin. Struct. Biol. 4, 893–901. [DOI] [PubMed]
- Díaz, N. & Field, M. J. (2004). J. Am. Chem. Soc. 126, 529–542. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gibbs, C. S. & Zoller, M. J. (1991). J. Biol. Chem. 266, 8923–8931. [PubMed]
- Granot, J., Mildvan, A. S., Bramson, H. N. & Kaiser, E. T. (1980). Biochemistry, 19, 3537–3543. [DOI] [PubMed]
- Hutter, M. C. & Helms, V. (1999). Protein Sci. 8, 2728–2733. [DOI] [PMC free article] [PubMed]
- Johnson, L. N., Noble, M. E. & Owen, D. J. (1996). Cell, 85, 149–158. [DOI] [PubMed]
- Knighton, D. R., Zheng, J. H., Ten Eyck, L. F., Xuong, N.-H., Taylor, S. S. & Sowadski, J. M. (1991). Science, 253, 414–420. [DOI] [PubMed]
- Lew, J., Taylor, S. S. & Adams, J. A. (1997). Biochemistry, 36, 6717–6724. [DOI] [PubMed]
- Madhusudan, Trafny, E. A., Xuong, N.-H., Adams, J. A., Ten Eyck, L. F., Taylor, S. S. & Sowadski, J. M. (1994). Protein Sci. 3, 176–187. [DOI] [PMC free article] [PubMed]
- Masterson, L. R., Cheng, C., Yu, T., Tonelli, M., Kornev, A., Taylor, S. S. & Veglia, G. (2010). Nature Chem. Biol. 6, 821–828. [DOI] [PMC free article] [PubMed]
- Masterson, L. R., Shi, L., Metcalfe, E., Gao, J., Taylor, S. S. & Veglia, G. (2011). Proc. Natl Acad. Sci. USA, 108, 6969–6974. [DOI] [PMC free article] [PubMed]
- Montenegro, M., Garcia-Viloca, M., Lluch, J. M. & González-Lafont, A. (2011). Phys. Chem. Chem. Phys. 13, 530–539. [DOI] [PubMed]
- Narayana, N., Cox, S., Shaltiel, S., Taylor, S. S. & Xuong, N. (1997). Biochemistry, 36, 4438–4448. [DOI] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Shaffer, J. & Adams, J. A. (1999a). Biochemistry, 38, 5572–5581. [DOI] [PubMed]
- Shaffer, J. & Adams, J. A. (1999b). Biochemistry, 38, 12072–12079. [DOI] [PubMed]
- Sun, T., Hayakawa, K. & Fraser, M. E. (2011). Acta Cryst. F67, 1168–1172. [DOI] [PMC free article] [PubMed]
- Taylor, S. S., Yang, J., Wu, J., Haste, N. M., Radzio-Andzelm, E. & Anand, G. (2004). Biochim. Biophys. Acta, 1697, 259–269. [DOI] [PubMed]
- Valiev, M., Kawai, R., Adams, J. A. & Weare, J. H. (2003). J. Am. Chem. Soc. 125, 9926–9927. [DOI] [PubMed]
- Walsh, D. A., Angelos, K. L., Van Patten, S. M., Glass, D. B. & Garetto, L. P. (1990). Peptides and Protein Phosphorylation, edited by B. E. Kemp, pp. 43–84. Boca Raton: CRC Press.
- Yang, J., Ten Eyck, L. F., Xuong, N.-H. & Taylor, S. S. (2004). J. Mol. Biol. 336, 473–487. [DOI] [PubMed]
- Zheng, J., Knighton, D. R., Ten Eyck, L. F., Karlsson, R., Xuong, N.-H., Taylor, S. S. & Sowadski, J. M. (1993). Biochemistry, 32, 2154–2161. [DOI] [PubMed]
- Zheng, J., Trafny, E. A., Knighton, D. R., Xuong, N., Taylor, S. S., Ten Eyck, L. F. & Sowadski, J. M. (1993). Acta Cryst. D49, 362–365. [DOI] [PubMed]
- Zhou, J. & Adams, J. A. (1997). Biochemistry, 36, 2977–2984. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: LT PKA–MgATP–IP20, 4dh1
PDB reference: LT PKA–Mg2ATP–IP20, 4dh3
PDB reference: RT PKA–Mg2ADP·PO4–IP20, 4dh5
PDB reference: LT PKA–Mg2AMPPNP–IP20, 4dh7
PDB reference: RT PKA–Mg2AMPPNP–IP20, 4dh8