Abstract
Differentiated vascular smooth muscle cells (SMCs) retain the capacity to modify their phenotype in response to inflammation or injury. This phenotypic switching is a crucial component of vascular disease, and is partly dependent on epigenetic regulation. An appreciation has been building in the literature for the essential role chromatin remodelling plays both in SMC lineage determination and in influencing changes in SMC behaviour and state. This process includes numerous chromatin regulatory elements and pathways such as histone acetyltransferases, deacetylases, and methyltransferases and other factors that act at SMC-specific marker sites to silence or permit access to the cellular transcriptional machinery and on other key regulatory elements such as myocardin and Kruppel-like factor 4 (KLF4). Various stimuli known to alter the SMC phenotype, such as transforming growth factor beta (TGF-β), platelet-derived growth factor (PDGF), oxidized phospholipids, and retinoic acid, appear to act in part through effects upon SMC chromatin structure. In recent years, specific covalent histone modifications that appear to establish SMC determinacy have been identified, while others alter the differentiation state. In this article, we review the mechanisms of chromatin remodelling as it applies to the SMC phenotype.
Keywords: Phenotypic switching, Histone modification, Vascular development, Vascular disease
1. Introduction
The vascular smooth muscle cell (SMC) plays a vital role in maintenance of vessel homeostasis, blood pressure, and response to injury. These cells evolve during vasculogenesis from migratory, proliferating pericytes secreting extracellular matrix (ECM) proteins and remodelling enzymes investing endothelial tubes into a mature, more quiescent phenotype, characterized by their expression of muscular contractile proteins.1,2 Unlike skeletal and cardiac myocytes (which are thought of as ‘terminally differentiated’ despite their having significant flexibility), mature SMCs retain significant capability to modulate their phenotype in response to a multitude of environmental cues. In particular, the stimuli associated with vascular injury and diseases, such as atherosclerosis, hypertension, aortic aneurysm formation, and post-angioplasty restenosis.1–4 These ‘de-differentiated’ cells down-regulate SMC marker and contractile proteins [e.g. smooth muscle (SM) α-actin, transgelin, SM-myosin heavy chain (MHC)], and may migrate, proliferate, and again secrete ECM and remodelling factors, among other behaviours. The promoter/enhancer regions of these SMC differentiation markers contain common cis elements, among them multiple CC(A/T-rich)6GG (CArG) elements and a transforming growth factor (TGF)-β control element.
This activity is responsible in large part for many of the specific manifestations of the vascular lesions described above, although it may also help to minimize structural failure of the vessel. The accompanying review by Owens et al. in this issue delves more completely into these aspects of SMC phenotypic change, the variety of forms that such change may take, and many of the mechanisms behind these alterations. In this review, we shall limit our discussion to the role that chromatin remodelling plays in phenotypic switching in vascular SMCs.
As implied above, SMC phenotypic switching is a varied and complex process. Evidence has been steadily accruing that epigenetic regulation is a vital element in the determination of SMC differentiation state, particularly in the areas of histone acetylation and methylation.5–7 It is necessary for SMC differentiation that serum response factor (SRF)—an ubiquitous protein capable of activating transcription for many gene subclasses—be allowed to bind to SMC marker promoters and denied to growth and proliferation genes, and that the latter remain silenced, while the cells are quiescent. Just as critical to SMC plasticity is the removal of inhibitory/chromatin compacting complexes from SMC-marker-gene promoters, such as certain histone deacetylases (HDACs) and other factors such as Kruppel-like factor 4 (Klf4).6 These mechanisms are also well established in skeletal and cardiac muscle differentiation and hypertrophy.8,9
2. Chromatin and remodelling
Eukaryotic nucleic DNA is packaged into repeating units of chromatin, composed of nucleosomes. In these particles, 145–147 DNA base pairs are wrapped around an octameric core that contains two molecules each of histones H2A, H2B, H3, and H4. Higher order structural stabilization of the core is achieved by DNA and by the linker histone H1.10–12 The amino-terminal portions of core histones contain flexible protease-sensitive tails, which are evolutionarily conserved sites for numerous post-translational modifications, including acetylation, methylation, phosphorylation, ubiquitylation, and ADP-ribosylation. Any given tail may be modified in multiple ways, in multiple locations.13–15 These modifications are correlated with DNA replication, chromatin assembly, and transcription, by permitting or denying access to the cellular machinery responsible for those activities.11,16 In general, acetylation of histones is thought to be transcriptionally activating, while mono-, di-, and tri-methylation may cause silencing or activation depending on which particular tail lysine residues are modified.
Strahl et al.17 proposed a ‘histone code’ through which combinations of specific residue modifications might regulate specific biological outcomes. Numerous enzymes and enzymatic complexes [e.g. histone acetyltransferases (HATs), HDACs, histone methyltransferases, and DNA methyltransferases (HMTs and DNMTs)] are known to regulate transcription through chromatin modification, and various proteins and protein complexes have been identified that ‘read’ the code and recruit transcription factors or repressors to specific genes. These mechanisms are believed to be partly responsible not only for the wide variety of organ-specific lineages that are found in multicellular organisms, but also for differentiation and trans-differentiation of cells.
3. Chromatin-based regulation is a key determinant of SMC phenotypic state
While numerous articles have addressed the molecular mechanisms behind SMC lineage determination and differentiation, less information generally is known with regard to the ways in which chromatin remodelling affects these processes. It is clear that large-scale changes in chromatin remodelling-related gene transcription may occur during SMC differentiation.18,19 Using microarray profiling of the A404 in vitro model, in which multi-potent P19 A404 cells differentiate into SMC when treated with retinoic acid (RA) over a 96-h time period, our lab found widespread regulation of chromatin modifying and remodelling genes. Within the first 48h of RA treatment, 17.6% of all chromatin remodelling genes identified on the array by ontology showed significant changes in transcription from untreated cells. This number then dramatically increased by the 96-h treatment time point, to >60%. Interestingly, while the numbers of positively and negatively regulated chromatin remodelling genes started at similar levels at 48h, by far the majority of the per cent increase was driven by down-regulated genes. These changes represented numerous classes of epigenetic regulators, including HATs, HDACs, HMTs, DNMTs, and others.
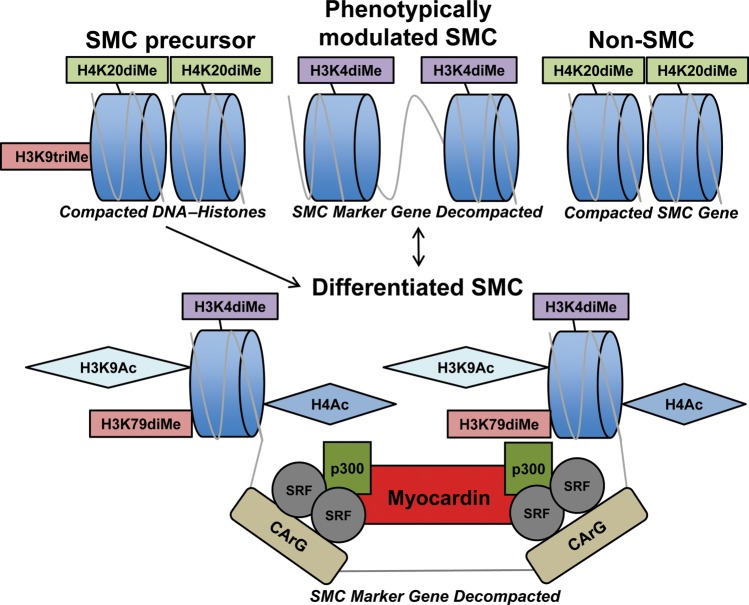
Notably, there are specific histone modifications that appear to support the SMC lineage and alter the ability of the transcriptional regulator SRF to target SMC marker-gene promoters (Figure 1).5–7
Figure 1.
Chromatin remodelling is central to SMC differentiation and phenotypic switching. Demethylation of H3K9 and H4K20 may permit decompaction of SMC marker gene chromatin in SMC stem cell precursors, permitting SMC differentiation to occur.5–7,19,73–79 Non-SMCs, such as embryonic stem cells, endothelial cells, skeletal myoblasts, and 10T1/2 embryonic fibroblasts, retained H4K20diMe in these regions. In contrast, differentiated vascular SMCs displayed several specific histone tail modifications including acetylation (H3K9, H3K14, and H4) as well as methylation (H3K4diMe, H3K79diMe) at SMC marker gene CArG boxes.5–7 These latter modifications likely permit access of myocardin–SRF complexes and the co-activator HAT p300 to SMC-specific CArGs, activating transcription.5–7,26–30,35–40 Phenotypic modulation of SMCs with de-differentiating stimuli leads to removal of the above-described histone modifications from SMC CArG-box chromatin with the exception of H3K4diMe, which may help maintain SMC marker genes as euchromatin and permit re-differentiation under proper conditions.6,7,68,69
4. Roles of various chromatin modulatory classes in SMCs
4.1. Histone acetyltransferases
HATs are well-characterized, covalent histone modification systems, consisting of several protein families. They are classified into two groups (A or B), depending on their mechanism and location of action. The HAT A family members act in the nucleus, transferring acetyl from acetyl-CoA to an ɛ-NH2 group of histone N-tails after nucleosome assembly. HAT A family members are further divided into three yeast homology subgroups—the GNAT family (e.g. PCAF, GCN5L, MORF), the MYST family [e.g. p300/CREB-binding protein (CBP), NCOAT, MOZ], and other [e.g. ATF-2, steroid receptor coactivator 1 (SRC-1), TFIIIC]. In contrast, HAT B family members act in the cytoplasm, transferring acetyl groups to free histones prior to inclusion within chromatin formation.11,20
Previous research suggests that acetylation of SMC-specific promoter loci is crucial for differentiation. Manabe and Owens examined the A404 in vitro SMC differentiation model. They found that SRF, although highly expressed, did not bind CArG-containing regions of SMC genes within intact, untreated A404 chromatin, but rather to the c-Fos CArG promoter region. RA treatment reversed these binding characteristics, and led to histone hyperacetylation in chromatin associated with SMC CArGs.5 Subsequently McDonald et al. identified de novo acetyl-H3K9, -H3K14, and -H4 as distinguishing marks in differentiating SMCs (when compared with non-SMCs, such as embryonic stem cells, endothelial cells, skeletal myoblasts, and 10T1/2 embryonic fibroblasts).6 The acetylation of these locations is likely performed by specific HATs (e.g. p300/CBP) that may also function as co-activators in the process.
CBP and p300 (E1A binding protein p300) are paralogues involved in such varied processes as embryonic development, differentiation, proliferation, and apoptosis.21 Mouse studies have shown them to be ubiquitously expressed during development, and they interact with numerous transcription factors, integrating complex signal transduction pathways. CBP and p300 are necessary factors in skeletal myogenesis and cardiomyogenesis.9,22–25 Several studies have indicated that p300 may also be necessary for SMC differentiation, and likely is essential for phenotypic switching.5,6,26–30 CBP and p300 are present in limiting amounts in mammalian cells, and it is thought that the ability of signalling pathways to regulate transcription may depend on their ability to compete for these factors.31–34 While CBP and p300 are known as HATs, they may themselves act as transcriptional co-factors, and may even acetylate non-histone proteins.21
The transcriptional SRF co-activator myocardin plays a key, although not entirely indispensible, role in SMC differentiation through binding to CArG-box-containing SMC marker genes.28,35–40 SRF has been associated with CBP during c-Fos activation.41 However, p300 is also able to enhance myocardin activity independent of SRF association. SMC-gene activation by myocardin is enhanced when p300 binds to its transcriptional activation domain.28 Notably, while myocardin was found to increase acetylation of H3K9 during SMC differentiation, it did not increase acetyl-H4, implying that a separate factor may assist in this activation step.6 In addition to its myocardin-related role p300 also interacts with the SMC differentiation-promoting transcription factor GATA6, and the combination is known to activate the SM-MHC promoter.29
In our laboratory, we performed studies using a microarray-based approach and the A404 in vitro model of SMC differentiation.5,19 In so doing, we found that over half of the genes identified in the literature (72 genes of 130) as part of the p300 interactome underwent significant regulation during differentiation, suggesting that global alterations in p300-based signalling accompany SMC lineage determination. We also showed that direct chemical inhibition of p300 HAT activity substantially decreases (but does not completely arrest) SMC differentiation. Histone acetylation of H3K9, H4, and H3K14 might be attributable to p300. However, as suggested above, interactions between p300 and myocardin likely also occur independently of acetyltransferase activity.
Substantial literature suggests that the activity of CBP and p300 depends in part on their phosphorylation state and their regulation by RA receptors. Such signalling may trigger the formation of an ‘activated’ p300 subpopulation with increased differentiation-gene (rather than growth-gene) specificity, affecting phenotype choice.33,42–46 As an example, RA-induced differentiation of F9 cells caused p300 (but not CBP) protein levels to decrease during differentiation, due to increased degradation by the ubiquitin-proteasome pathway. This was accompanied by a significant increase in per-molecule HAT activity, and with protein kinase A-mediated p300 phosphorylation.46
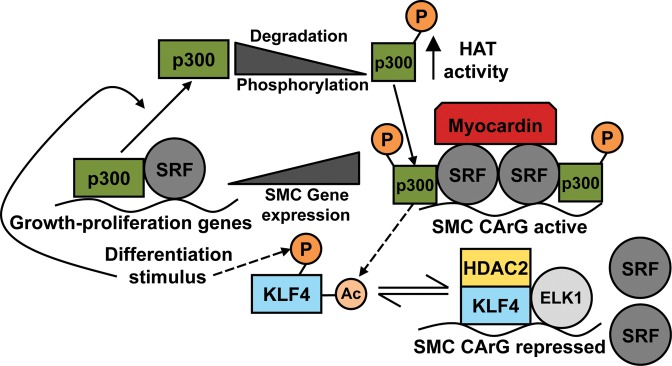
In a similar vein, A404 SMC differentiation with RA caused a progressive decrease in total p300 protein levels despite only minimal changes in p300 transcription.19 The same was observed in a model of human SMC re-differentiation using serum starvation, and siRNA knockdown of p300 expression in both models was found to accelerate SMC differentiation. This suggests that triggering of SMC differentiation may depend in part upon a complex combination of decreases in total p300 levels and activating covalent modifications, leading to migration of the factor from growth-based pathways to myocardin-SRF and thence to SMC differentiation-specific genes (Figure 2).
Figure 2.
Proposed role for the histone acetyltransferase p300 in regulation of SMC differentiation and phenotypic switching.5,6,26–30 Growth/proliferation and differentiation signalling pathways compete for limited amounts of p300.31–34 Inhibition of p300 HAT activity decreases SMC differentiation.19 Activity of p300 depends in part on its phosphorylation state. SMC differentiation stimuli may trigger partial p300 degradation, and the formation of a p300 subpopulation with increased HAT activity per-molecule and SMC marker gene specificity, thereby affecting phenotype choice.19,33,42–46 SMC gene activation by myocardin-serum response factor (SRF) is enhanced by p300 binding.28 KLF4 negatively regulates SMC marker genes in culture and disease.57 Pro-SMC differentiation stimuli (e.g. retinoic acid) lead to KLF4 phosphorylation, and subsequent acetylation by p300 (dotted-line arrows), altering KLF4 binding characteristics and relieving transcriptional repression of SMC genes.6,69,83 Proliferative stimuli lead to de-phosphorylation of KLF4, deacetylation by HDAC2, association with ELK1, ejection of SRF, and repression of SMC markers.
There is some suggestion that other HATs might also be involved in SMC phenotypic determination. While several HATs (including CBP) showed significant down-regulation with SMC differentiation, Myst3 showed progressive up-regulation throughout.18,19 Little is known about this factor at this time apart from its involvement in monocytic leukaemia and haematopoietic stem cells.47
The arginine methyltransferase Prmt2 is known to bind to RA receptors RARα and RXRα.48,49 It also is known to interact with another HAT, SRC-1. There is a possibility that Prmt2 and SRC-1 might relate to RA-mediating triggering of A404 SMC differentiation. Prmt2 transcription is up-regulated early in the process and then remains unchanged for the remainder.19 Given the specific acetylation that accompanies SMC differentiation, further examination of the roles of various HATs in SMC state change is clearly warranted.
4.2. Histone deacetylases
Eukaryotic HDACs are divided into four classes. Class I HDACs (1–3, 8) are typically found in the nucleus, and are similar to yeast RPD3. Class II HDACs resemble yeast HDA1, and are further divided into IIA (−4, −5, −7, −9) located in either the nucleus or cytoplasm, and IIB (−6, −10) generally found in the cytoplasm. Class III HDACs, known as sirtuins (SIRTs 1–7), are NAD-dependent enzymes similar to yeast SIR2 proteins.50 Class IV is an atypical group that encompasses functional characteristics of Class I and II, and includes HDAC11.51 In general, these proteins are thought to inhibit transcription through removal of key acetyl groups, leading to chromatin compaction and preventing binding of transcriptional machinery. In addition to their effects on histones, some HDACs perform protein deactylation in the cytosol.
Differentiation of both skeletal and cardiac myocytes depends on relief from Class II HDAC inhibition, and substantial evidence also exists for a similar role in SMCs.52 Trichostatin A (TSA), a selective inhibitor of Class I and II HDACs, inhibits SMC proliferation, accelerates differentiation in P19 cells, and stimulates acetylation at the transgelin locus in fibroblasts.26,53,54 In addition, we found that TSA accelerated SMC differentiation in A404 cells treated with RA, leading to comparatively increased levels of acetyl-H4.19
Some of the data on HDACs in SMCs are inconsistent, and likely depend to some extent on the model being evaluated. Qiu et al.55 overexpressed HDACs 1–6, and found that all of them suppressed transactivation of transgelin by Smad3 and myocardin in 10T1/2 cells and PAC1s (a pulmonary artery-derived SMC line). Another study, however, showed that Class II HDACs 4 and 5 suppressed the ability of myocardin to activate SM α-actin and transgelin, while Class I HDACs 1 and 3 had no effect.28 Notably, they also observed that A7r5 SMCs expressed HDACs1–2, and 4–7, but not HDAC3 or 9. We found that A404 cells, in contrast, expressed both Hdac3 and 9. However, Hdac3 was suppressed with differentiation while Hdac9 showed progressive up-regulation over time. Hdac6 essentially disappeared with SMC differentiation. We also noted a significant decrease in Hdac5 transcription during differentiation, consistent with relief of suppression of myocardin.19
A recent study looked at rat aortic SMCs in culture and a murine femoral artery wire injury model.56 They found that serum feeding (a mitogenic, de-differentiation stimulus) induced HDACs 1–3 transcription in cultured SMC, while siRNA or chemical inhibition of HDACs 1–3 using Scriptaid (similar to TSA) prevented serum-induced SMC proliferation. HDAC inhibition with Scriptaid prevented retinoblastoma (Rb) protein phosphorylation, regulated cyclin-dependent kinase inhibitors p21(Cip1) and p27(Kip), and decreased cyclin D1 expression. HDAC inhibition in vivo decreased neointima formation and expression of cyclin D1 in the wire injury model.
KLF4 is known to negatively regulate SMC differentiation in culture and disease models.57 KLF4 overexpression assays lead to loss of marker expression in SMCs, a process associated with the appearance of the H4 deacetylase HDAC2.6 Notably, in A404 cells Hdac2 showed minimal change in expression during SMC differentiation, again likely due to differences between the experimental models.19
HDAC3, unique among Class I HDACs, forms a transcriptional repressor complex with NCoR or SMRT. Deletion of HDAC3 from the murine neural crest (but not HDACs 1, 2 or 8) results in significant vascular abnormalities and perinatal lethality with defective SMC differentiation.58
HDAC7 appears to undergo alternative splicing during SMC differentiation from embryonic stem cells, leading to increased SRF-myocardin binding and activation of SMC gene expression.59 HDAC7 has also been found to be required for Sp1-induced expression of SMC marker genes.60
While chromatin deacetylation is the primary role ascribed to HDACs in SMCs, other regulatory mechanisms have also been attributed to them. Atypically for its class, Class I HDAC8 in SMCs appears to be a cytosolic marker of smooth muscle differentiation in human tissues.61 HDAC8 is co-expressed in vivo with SM α-actin and SM-MHC, and is distributed along the stress fibres. Further, in human SMCs HDAC8, but not HDAC1 or HDAC3, is enriched in cytoskeleton-bound protein fractions.62 Waltregny et al. have suggested, therefore, that HDAC8 may regulate the contractile capacity of SMCs.
The Class III HDAC Sirtuin 1 (Sirt1) is a protein and histone deacetylase known for inhibiting differentiation of skeletal muscle via its direct involvement with Pcaf and p300 suppression through deacetylation.63,64 Its role in SMC phenotype determination is likely very complex. Sirt1 showed early and persistent down-regulation throughout A404 SMC differentiation.19 Another member of the same family, Sirt3, was also significantly down-regulated. These data might suggest that relief from sirtuin suppression would enable p300 activation, with expected pro-differentiation effects in SMCs. However, Sirt1 was found to be down-regulated with injury in a murine carotid ligation and wire injury model, and overexpression of the protein in a transgenic mouse markedly inhibited formation of neointima.65 Sirt1 overexpression in vitro also inhibited SMC proliferation and migration and induced cell cycle arrest. Further, resveratrol, a small-molecule activator of SIRT1, has also been shown to block neointima formation after arterial injury in a rabbit model.66 A recent review pointed out that SIRT1 is a master regulator, with a large list of biologically important targets and interactions, many of which are known players in SMC phenotypic modulation.67 As such, it may be active on several levels in this process.
4.3. Histone methylation-demethylation, and DNA methylation
Methylation and demethylation of histones represents a powerful epigenetic mechanism leading to gene activation or silencing. Beyond their findings of SMC-specific histone acetylation at SMC marker gene loci, McDonald et al. identified certain histone methyl-lysine patterns that were able to distinguish SMCs from non-SMCs, specifically H3K4 dimethylation and H3K79 dimethylation.7 In the same vein, non-SMCs showed H4K20 dimethylation (a transcriptional silencing modification) in SMC CArG-box chromatin, while digestion experiments showed that SMC gene promoter chromatin was nuclease-accessible in SMC, but not in non-SMCs. Dimethylated H3K4 and dimethylated H3K79 seem also to enable recruitment of the SRF/myocardin complex to SMC marker CArG boxes.5–7,26,28
As mentioned above, treatment of SMCs with de-differentiating stimuli (PDGF-BB, injury, oxidized phospholipids) leads to removal of various acetyl-histone modifications from SMC CArG-box chromatin. It also leads to SMC marker gene histone demethylation at H3K79. However, H3K4diMe remains after these stimuli are applied, implying that this modification may be used to track cells from SMC lineage, distinguishing them from stem cells, and perhaps permitting the cells to re-differentiate into SMCs under more favourable conditions.6,7,68,69
Removal/absence of silencing methyl-histone modifications is likely central to SMC differentiation. There are two additional family members of myocardin, the myocardin-related transcription factors A and B (MRTFA and MRTFB) believed to have similar actions to myocardin.70 These proteins are necessary for SMC differentiation in vivo and may have a key role in SMC phenotypic switching in disease.71,72 A study from Lockman et al.73 found that the H3K9 histone demethylase Jmjda1 bound all three myocardin family members. Overexpression of Jmjda1 in 10T1/2 cells decreased mono- and di-methyl H3K9, and stimulated the transgelin and SM α-actin promoters.
Several other HMTs from the SET family showed significant regulation during SMC differentiation of A404 cells.19 One set member, Ehmt2/G9a, is thought to mono- and di-methylate H3K9.74 Consistent with this, Ehmt2 was down-regulated in our study. Setdb1/ESET, which directs H3K9 tri-methylation, was also significantly down-regulated early in SMC differentiation.75 In contrast, Suv39h1 (also known to cause H3K9 tri-methylation) was up-regulated—but very late in the SMC differentiation process.76 This suggests that, once acetylated, SMC-specific H3K9 may no longer present a target for methylation.
Indeed, one study established that the Polycomb protein Suz12 is required for tri-methylation of H3K9 in differentiated mammalian stem cells (independent of the SET domain-containing Ezh2, with which it forms a complex to tri-methylate H3K27). Knockdown of SUZ12 in human cells caused a reduction in both H3K27triMe and H3K9triMe, and altered the distribution of HP1alpha.77 In keeping with these findings, A404 differentiation led to early and persistent down-regulation of Suz12.
In SMC differentiation of A404 cells, we also noted down-regulation of two chromobox homologues, Cbx1 and Cbx3.19 The protein products of these genes (HP1beta and HP1gamma) recognize tri-methylated H3K9 and mediate silencing through chromatin remodelling to heterochromatin.78,79 This suggests that H3K9 demethylation at SMC-marker-gene promoters may trigger conversion to transcriptionally viable euchromatin during SMC differentiation.
Methylation of DNA at CpG islands leads to heritable chromatin silencing. Vire et al. showed that the silencing pathways of the Polycomb group and DNA methyltransferase systems are mechanically linked.80 We showed that SMC differentiation is associated with highly significant down-regulation of the DNA methyltransferases 3a and 3b.19 This suggests that relief from Dnmt3a- and Dnmt3b-associated DNA methylation may prevent HDAC recruitment to SMC-specific genes during differentiation via methyl-CpG-binding proteins such as Mecp2 and Mbd2.
5. Selected factors known to regulate SMC chromatin structure
5.1. Kruppel-like factor 4
KLF4 acts as a negative regulator of SMC differentiation both in vitro and in disease models.57 It accomplishes this in several ways, but in part through its impact upon SMC-gene chromatin. The effects of PDGF-BB, PDGF-DD, and oxidized phospholipids, all negative regulators of SMC marker genes in numerous models, appear to be dependent on Klf4.69,81,82 This leads to an interaction between Elk1 and Klf4, which together bind to and suppress SMC markers. In this context, the SM α-actin gene is H4 hypoacetylated by HDAC2 and HDAC5, the latter of which has been found to interact directly with Klf4.69 Interestingly, despite PDGF-BB and RA having opposite effects on SMC proliferation and differentiation, they both induce KLF4 expression. However, these substances change the acetylation state of KLF4 in different ways, causing it to preferentially bind to different regions within the transgelin/SM22α promoter.83 RA treatment leads to phosphorylation of KLF4, facilitating its acetylation by p300, while PDGF-BB treatment causes KLF4 de-phosphorylation, permitting deacetylation by HDAC2. These modifications then alter the regional binding preferences of KLF4. This finding further implicates elements known to be involved in histone modification in the regulation of KLF4 suppression of SMC-marker genes.
MicroRNAs represent another epigenetic mechanism exerting control over SMC differentiation state, exerting post-transcriptional repression through targeted mRNA degradation. This topic is more thoroughly reviewed elsewhere in this issue. However, it should be noted that murine SMC differentiation from embryonic stem cells (ESCs) in vitro has been found to be dependent on miR-1, which directly binds to and inhibits KLF4 transcription.84 Additionally, TGF-β and BMP4 have been found to rapidly down-regulate KLF4 through induction of miRs-143 and -145.85 Modulation of KLF4 appears to be a prerequisite for induction of contractile genes by TGF-β and BMP4. In a similar vein, miR-10a appears to be a key positive mediator of ESC-to-SMC differentiation in response to RA, acting at least in part through suppression of HDAC4.86
5.2. BRG1/SWI/SNF
As mentioned previously, other members of the myocardin family act as unique co-activators with SRF to induce SMC-specific gene activation. MRTFA [MKL (megakaryoblastic leukaemia)/myocardin-like 1] can activate transcription of transgelin, telokin, and SM α-actin, but not other SRF-dependent proliferation genes.70–72 The SWI/SNF complex is an ATP-dependent chromatin remodelling macromolecule, and is composed of 7–11 components including either of the ATPase subunits BRM (Brahma) or BRG1 (Brahma-related gene 1). In models of atherosclerosis and in-stent restenosis, BRG1 in particular has been found to be up-regulated in SMCs in vivo.87 Further, SMC marker gene induction in the murine heart by LIM-only CRP2 (a SMC differentiation co-factor) appears to be dependent on its binding to BRG1.88 BRG1 or BRM appears to be critical for SMC-specific gene activation by MRTFA.89 It has been suggested that weak binding of SRF to SMC marker gene CArGs recruits MRTFA, which then recruits the SWI/SNF complex to decompact local chromatin and permit differentiation.
5.3. PRISM
PRISM/PRDM6 (PR domain in smooth muscle) is a chromatin-remodelling zinc finger protein in the PRDM (PRDI-BF1 and RIZ homology domain) family, and is found in SMCs, particularly during murine embryogenesis in the cardiac outflow tract and the descending aorta.90 PRISM interacts with Class I HDACs and the histone methyltransferase EHMT2/G9a (euchromatic histone-lysine N-methyltransferase 2) in SMCs. Overexpression of PRISM in vitro induced SMC proliferation, repressed myocardin and GATA6, while siRNA knockdown of PRISM induced SMC differentiation and slowed proliferation. While PRISM does not have intrinsic HMT activity, it may recruit EHMT2 (a H3K9 di- and H3K27 di-/tri- histone methyltransferase mentioned previously) to SMC regulatory regions and silence SMC marker expression.
5.4. Butyrate
A dietary HDAC inhibitor, butyrate, inhibits SMC proliferation.91 SMCs exposed to butyrate show increased SMC CArG H3K9 acetylation and H3K4 dimethylation, and decreased H3S10 phosphorylation and H3K9 dimethylation. Untreated cells display the opposite covalent modifications. It has been suggested that, through these alterations, butyrate differentially alters G1-specific cell cycle proteins to cause SMC proliferation arrest, with failure to inhibit Rb activity.
5.5. Vascular pathological states; diabetes and thoracic aortic aneurysm
Environmental alterations due to vascular pathobiology may lead to chromatin remodelling in SMC non-marker genes, which may also cause persistent SMC phenotypic state change or predisposition. For example, vascular SMC derived from diabetic mice (db/db) continue to exhibit pro-inflammatory and pro-atherogenic characteristics even when placed in culture. Hyperglycaemia in these SMCs appears to increase miR-125, decreasing levels of target gene Suv39h1 (methyltransferase), and thereby decreasing H3K9-trimethylation-related silencing of SMC inflammatory genes. This, in turn, leads to SMC hypersensitivity to inflammatory stimulation with tumour necrosis factor α.92,93 Similarly, SMCs derived from diabetic mice display low levels of lysine (K)-specific demethylase 1A, a protein that negatively regulates H3K4 dimethylation. These cells show H3K4me2 modification with increased transcription of the inflammatory genes monocyte chemoattractant protein-1, and interleukin-6.94
As another example of this phenomenon, the development of thoracic aortic aneurysms (from various causes) appears related to SMAD2 (SMAD family member 2) dysregulation. Recent work has shown that aortic SMCs in these patients develop constitutive overexpression of SMAD2 through increases in H3K9/14 acetylation and H3K4 methylation of the SMAD2 promotor, leading to changes in SMC phenotype.95
6. Conclusions and future directions
Chromatin remodelling appears to be central to determination of SMC phenotypic state, in permitting or denying access of the transcriptional machinery to SMC marker genes, in recruiting transcription factors and co-activators to these genes, and in durably defining the SMC lineage in the face of phenotypic switching. Also crucial is the recruitment or access denial of specific HDACs and HMTs to histones associated with SMC marker CArG regions. Certain histone tail modifications in these areas favour the differentiated SMC phenotype, while their absence occurs with de-differentiation leading to lower expression of SMC markers. These include acetylation of H3K9, H3K14, and H4, and dimethylation of H3K4 and H3K79 (but not methyl-H4K20 and methyl-H3K9, which are markers of several non-SMCs and SMC precursors—see text above). H3K4diMe in particular appears to be a durable marker of the SMC lineage despite changes in differentiation state.
Factors such as KLF4, PRISM, and BRG1/SWI/SNF, act as key suppressors and mediators during this process, interacting with the myocardin family and being altered by phenotype-modifying stimuli while remodelling chromatin. Later studies must define whether and what additional histone modifications exist that alter or determine SMC phenotype, which triggers and co-factors are involved, and whether the observed modifications are the cause or result of SMC differentiation state change.
The HAT p300 has a large and diverse interactome, is present in limiting amounts among competing signalling processes (e.g. differentiation vs. proliferation and growth), modulates both chromatin and transcription factor activity, and itself acts as a myocardin co-activator. In SMCs undergoing differentiation, a decrease in p300 protein levels accompanied by activating covalent modifications may cause the factor to migrate from growth-based pathways to SMC differentiation-specific pathways. Future work must define more precisely the role of p300 in SMC phenotypic determination, along with those of other potentially contributory HATs, such as MYST3, PRMT2, and SRC-1.
Conflicting data surround the exact function of specific HDACs in determining SMC differentiation state, varying in different models. While it appears that relief from HDAC suppression is required for maintaining SMC differentiation, the role of specific HDACs remains to be established. The most consistent data are found for HDAC2 and HDAC5, which both down-regulate SMC marker gene expression. In contrast, HDACs 3, 7, and 8 appear to be key players in the process of SMC differentiation and in regulation of contractile function. The dynamic role of HDACs in SMC plasticity constitutes a rich area of future research.
Many open questions exist. There remains a need for a clear distinction between pericytes, SMCs, and myofibroblasts, as these cells may overlap significantly in structure, function, and contribution to disease processes. Additionally, whether SMCs can truly transdifferentiate to other cell types is unclear. Ultimately, we seek to make use of SMC phenotypic flexibility in the treatment of vascular injury and disease. Further study of SMC chromatin remodelling may provide some answers, and guide us towards new therapeutic options.
Conflict of interest: none declared.
Funding
This work was supported by research grants from the National Institutes of Health (1P50HL083800-01 to P.S.T., 5K08 HL080567 to J.M.S.), the Deutsche Forschungsgemeinschaft (MA4688/1-1 to L.M.), and the American Heart Association (09POST2260118 to L.M.).
References
- 1.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 3.Ross R. Atherosclerosis: an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 4.Ailawadi G, Moehle CW, Pei H, Walton SP, Yang Z, Kron IL, et al. Smooth muscle phenotypic modulation is an early event in aortic aneurysms. J Thorac Cardiovasc Surg. 2009;138:1392–1399. doi: 10.1016/j.jtcvs.2009.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manabe I, Owens GK. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ Res. 2001;88:1127–1134. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- 6.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 8.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 9.McKinsey TA, Zhang CL, Olson EN. Signaling chromatin to make muscle. Curr Opin Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 10.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 11.Marmorstein R. Protein modules that manipulate histone tails for chromatin regulation. Nat Rev Mol Cell Biol. 2001;2:422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- 12.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 13.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradbury EM. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 15.Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 17.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 18.Spin JM, Nallamshetty S, Tabibiazar R, Ashley EA, King JY, Chen M, et al. Transcriptional profiling of in vitro smooth muscle cell differentiation identifies specific patterns of gene and pathway activation. Physiol Genomics. 2004;19:292–302. doi: 10.1152/physiolgenomics.00148.2004. [DOI] [PubMed] [Google Scholar]
- 19.Spin JM, Quertermous T, Tsao PS. Chromatin remodeling pathways in smooth muscle cell differentiation, and evidence for an integral role for p300. PLoS One. 2010;5:e14301. doi: 10.1371/journal.pone.0014301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 22.Polesskaya A, Naguibneva I, Fritsch L, Duquet A, Ait-Si-Ali S, Robin P, et al. CBP/p300 and muscle differentiation: no HAT, no muscle. Embo J. 2001;20:6816–6825. doi: 10.1093/emboj/20.23.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 24.Dai YS, Cserjesi P, Markham BE, Molkentin JD. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J Biol Chem. 2002;277:24390–24398. doi: 10.1074/jbc.M202490200. [DOI] [PubMed] [Google Scholar]
- 25.Poizat C, Sartorelli V, Chung G, Kloner RA, Kedes L. Proteasome-mediated degradation of the coactivator p300 impairs cardiac transcription. Mol Cell Biol. 2000;20:8643–8654. doi: 10.1128/mcb.20.23.8643-8654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu P, Li L. Histone acetylation and recruitment of serum responsive factor and CREB-binding protein onto SM22 promoter during SM22 gene expression. Circ Res. 2002;90:858–865. doi: 10.1161/01.res.0000016504.08608.b9. [DOI] [PubMed] [Google Scholar]
- 27.Aravind L, Koonin EV. SAP: a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 28.Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, et al. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada H, Hasegawa K, Morimoto T, Kakita T, Yanazume T, Sasayama S. A p300 protein as a coactivator of GATA-6 in the transcription of the smooth muscle-myosin heavy chain gene. J Biol Chem. 2000;275:25330–25335. doi: 10.1074/jbc.M000828200. [DOI] [PubMed] [Google Scholar]
- 30.Wang SX, Elder PK, Zheng Y, Strauch AR, Kelm RJ., Jr Cell cycle-mediated regulation of smooth muscle alpha-actin gene transcription in fibroblasts and vascular smooth muscle cells involves multiple adenovirus E1A-interacting cofactors. J Biol Chem. 2005;280:6204–6214. doi: 10.1074/jbc.M409506200. [DOI] [PubMed] [Google Scholar]
- 31.Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, et al. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 32.Partanen A, Motoyama J, Hui CC. Developmentally regulated expression of the transcriptional cofactors/histone acetyltransferases CBP and p300 during mouse embryogenesis. Int J Dev Biol. 1999;43:487–494. [PubMed] [Google Scholar]
- 33.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 34.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 37.Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, et al. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida T, Sinha S, Dandre F, Wamhoff BR, Hoofnagle MH, Kremer BE, et al. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ Res. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida T, Kawai-Kowase K, Owens GK. Forced expression of myocardin is not sufficient for induction of smooth muscle differentiation in multipotential embryonic cells. Arterioscler Thromb Vasc Biol. 2004;24:1596–1601. doi: 10.1161/01.ATV.0000137190.63214.c5. [DOI] [PubMed] [Google Scholar]
- 41.Ramirez S, Ait-Si-Ali S, Robin P, Trouche D, Harel-Bellan A, Ait Si Ali S. The CREB-binding protein (CBP) cooperates with the serum response factor for transactivation of the c-fos serum response element. J Biol Chem. 1997;272:31016–31021. doi: 10.1074/jbc.272.49.31016. [DOI] [PubMed] [Google Scholar]
- 42.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 43.Dietze EC, Troch MM, Bowie ML, Yee L, Bean GR, Seewaldt VL. CBP/p300 induction is required for retinoic acid sensitivity in human mammary cells. Biochem Biophys Res Commun. 2003;302:841–848. doi: 10.1016/s0006-291x(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 44.Ait-Si-Ali S, Ramirez S, Barre FX, Dkhissi F, Magnaghi-Jaulin L, Girault JA, et al. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 45.Kawasaki H, Eckner R, Yao TP, Taira K, Chiu R, Livingston DM, et al. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 46.Brouillard F, Cremisi CE. Concomitant increase of histone acetyltransferase activity and degradation of p300 during retinoic acid-induced differentiation of F9 cells. J Biol Chem. 2003;278:39509–39516. doi: 10.1074/jbc.M307123200. [DOI] [PubMed] [Google Scholar]
- 47.Katsumoto T, Aikawa Y, Iwama A, Ueda S, Ichikawa H, Ochiya T, et al. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20:1321–1330. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott HS, Antonarakis SE, Lalioti MD, Rossier C, Silver PA, Henry MF. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2) Genomics. 1998;48:330–340. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- 49.Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem. 2002;277:28624–28630. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- 50.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 51.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 52.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 53.Minucci S, Horn V, Bhattacharyya N, Russanova V, Ogryzko VV, Gabriele L, et al. A histone deacetylase inhibitor potentiates retinoid receptor action in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1997;94:11295–11300. doi: 10.1073/pnas.94.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamoto H, Fujioka Y, Takahashi A, Takahashi T, Taniguchi T, Ishikawa Y, et al. Trichostatin A, an inhibitor of histone deacetylase, inhibits smooth muscle cell proliferation via induction of p21(WAF1) J Atheroscler Thromb. 2006;13:183–191. doi: 10.5551/jat.13.183. [DOI] [PubMed] [Google Scholar]
- 55.Qiu P, Ritchie RP, Gong XQ, Hamamori Y, Li L. Dynamic changes in chromatin acetylation and the expression of histone acetyltransferases and histone deacetylases regulate the SM22alpha transcription in response to Smad3-mediated TGFbeta1 signaling. Biochem Biophys Res Commun. 2006;348:351–358. doi: 10.1016/j.bbrc.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 56.Findeisen HM, Gizard F, Zhao Y, Qing H, Heywood EB, Jones KL, et al. Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition. Arterioscler Thromb Vasc Biol. 2011;31:851–860. doi: 10.1161/ATVBAHA.110.221952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida T, Kaestner KH, Owens GK. Conditional deletion of Kruppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ Res. 2008;102:1548–1557. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh N, Trivedi CM, Lu M, Mullican SE, Lazar MA, Epstein JA. Histone deacetylase 3 regulates smooth muscle differentiation in neural crest cells and development of the cardiac outflow tract. Circ Res. 2011;109:1240–1249. doi: 10.1161/CIRCRESAHA.111.255067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margariti A, Xiao Q, Zampetaki A, Zhang Z, Li H, Martin D, et al. Splicing of HDAC7 modulates the SRF-myocardin complex during stem-cell differentiation towards smooth muscle cells. J Cell Sci. 2009;122:460–470. doi: 10.1242/jcs.034850. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L, Jin M, Margariti A, Wang G, Luo Z, Zampetaki A, et al. Sp1-dependent activation of HDAC7 is required for platelet-derived growth factor-BB-induced smooth muscle cell differentiation from stem cells. J Biol Chem. 2010;285:38463–38472. doi: 10.1074/jbc.M110.153999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waltregny D, De Leval L, Glenisson W, Ly Tran S, North BJ, Bellahcene A, et al. Expression of histone deacetylase 8, a class I histone deacetylase, is restricted to cells showing smooth muscle differentiation in normal human tissues. Am J Pathol. 2004;165:553–564. doi: 10.1016/S0002-9440(10)63320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waltregny D, Glenisson W, Tran SL, North BJ, Verdin E, Colige A, et al. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility. Faseb J. 2005;19:966–968. doi: 10.1096/fj.04-2303fje. [DOI] [PubMed] [Google Scholar]
- 63.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 64.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 65.Li L, Zhang HN, Chen HZ, Gao P, Zhu LH, Li HL, et al. SIRT1 acts as a modulator of neointima formation following vascular injury in mice. Circ Res. 2011;108:1180–1189. doi: 10.1161/CIRCRESAHA.110.237875. [DOI] [PubMed] [Google Scholar]
- 66.Zou J, Huang Y, Cao K, Yang G, Yin H, Len J, et al. Effect of resveratrol on intimal hyperplasia after endothelial denudation in an experimental rabbit model. Life Sci. 2000;68:153–163. doi: 10.1016/s0024-3205(00)00925-5. [DOI] [PubMed] [Google Scholar]
- 67.Stunkel W, Campbell RM. Sirtuin 1 (SIRT1): the misunderstood HDAC. J Biomol Screen. 2011;16:1153–1169. doi: 10.1177/1087057111422103. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor-BB represses smooth muscle cell marker genes via changes in binding of MKL factors and histone deacetylases to their promoters. Am J Physiol Cell Physiol. 2007;292:C886–C895. doi: 10.1152/ajpcell.00449.2006. [DOI] [PubMed] [Google Scholar]
- 69.Yoshida T, Gan Q, Owens GK. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. Am J Physiol Cell Physiol. 2008;295:C1175–C1182. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci USA. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, et al. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci USA. 2005;102:8916–8921. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol. 2007;292:H1170–H1180. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- 73.Lockman K, Taylor JM, Mack CP. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2007;101:e115–e123. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
- 74.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de la Cruz CC, Kirmizis A, Simon MD, Isono K, Koseki H, Panning B. The polycomb group protein SUZ12 regulates histone H3 lysine 9 methylation and HP1 alpha distribution. Chromosome Res. 2007;15:299–314. doi: 10.1007/s10577-007-1126-1. [DOI] [PubMed] [Google Scholar]
- 78.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 79.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 80.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 81.Pidkovka NA, Cherepanova OA, Yoshida T, Alexander MR, Deaton RA, Thomas JA, et al. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circ Res. 2007;101:792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 82.Thomas JA, Deaton RA, Hastings NE, Shang Y, Moehle CW, Eriksson U, et al. PDGF-DD, a novel mediator of smooth muscle cell phenotypic modulation, is upregulated in endothelial cells exposed to atherosclerosis-prone flow patterns. Am J Physiol Heart Circ Physiol. 2009;296:H442–H452. doi: 10.1152/ajpheart.00165.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu K, Zheng B, Han M, Wen JK. ATRA activates and PDGF-BB represses the SM22alpha promoter through KLF4 binding to, or dissociating from, its cis-DNA elements. Cardiovasc Res. 2011;90:464–474. doi: 10.1093/cvr/cvr017. [DOI] [PubMed] [Google Scholar]
- 84.Xie C, Huang H, Sun X, Guo Y, Hamblin M, Ritchie RP, et al. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 2011;20:205–210. doi: 10.1089/scd.2010.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davis-Dusenbery BN, Chan MC, Reno KE, Weisman AS, Layne MD, Lagna G, et al. down-regulation of Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. J Biol Chem. 2011;286:28097–28110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang H, Xie C, Sun X, Ritchie RP, Zhang J, Chen YE. miR-10a contributes to retinoid acid-induced smooth muscle cell differentiation. J Biol Chem. 2010;285:9383–9389. doi: 10.1074/jbc.M109.095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang QJ, Goddard M, Shanahan C, Shapiro L, Bennett M. Differential gene expression in vascular smooth muscle cells in primary atherosclerosis and in stent stenosis in humans. Arterioscler Thromb Vasc Biol. 2002;22:2030–2036. doi: 10.1161/01.atv.0000042206.98651.15. [DOI] [PubMed] [Google Scholar]
- 88.Chang DF, Belaguli NS, Chang J, Schwartz RJ. LIM-only protein, CRP2, switched on smooth muscle gene activity in adult cardiac myocytes. Proc Natl Acad Sci USA. 2007;104:157–162. doi: 10.1073/pnas.0605635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang M, Fang H, Zhou J, Herring BP. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem. 2007;282:25708–25716. doi: 10.1074/jbc.M701925200. [DOI] [PubMed] [Google Scholar]
- 90.Davis CA, Haberland M, Arnold MA, Sutherland LB, McDonald OG, Richardson JA, et al. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol. 2006;26:2626–2636. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mathew OP, Ranganna K, Yatsu FM. Butyrate, an HDAC inhibitor, stimulates interplay between different posttranslational modifications of histone H3 and differently alters G1-specific cell cycle proteins in vascular smooth muscle cells. Biomed Pharmacother. 2010;64:733–740. doi: 10.1016/j.biopha.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes. 2010;59:2904–2915. doi: 10.2337/db10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reddy MA, Villeneuve LM, Wang M, Lanting L, Natarajan R. Role of the lysine-specific demethylase 1 in the proinflammatory phenotype of vascular smooth muscle cells of diabetic mice. Circ Res. 2008;103:615–623. doi: 10.1161/CIRCRESAHA.108.175190. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Gomez D, Coyet A, Ollivier V, Jeunemaitre X, Jondeau G, Michel JB, et al. Epigenetic control of vascular smooth muscle cells in Marfan and non-Marfan thoracic aortic aneurysms. Cardiovasc Res. 2011;89:446–456. doi: 10.1093/cvr/cvq291. [DOI] [PMC free article] [PubMed] [Google Scholar]