Abstract
The life cycle of flowering plants alternates between a predominant sporophytic (diploid) and an ephemeral gametophytic (haploid) generation that only occurs in reproductive organs. In Arabidopsis thaliana, the female gametophyte is deeply embedded within the ovule, complicating the study of the genetic and molecular interactions involved in the sporophytic to gametophytic transition. Massively parallel signature sequencing (MPSS) was used to conduct a quantitative large-scale transcriptional analysis of the fully differentiated Arabidopsis ovule prior to fertilization. The expression of 9775 genes was quantified in wild-type ovules, additionally detecting >2200 new transcripts mapping to antisense or intergenic regions. A quantitative comparison of global expression in wild-type and sporocyteless (spl) individuals resulted in 1301 genes showing 25-fold reduced or null activity in ovules lacking a female gametophyte, including those encoding 92 signalling proteins, 75 transcription factors, and 72 RNA-binding proteins not reported in previous studies based on microarray profiling. A combination of independent genetic and molecular strategies confirmed the differential expression of 28 of them, showing that they are either preferentially active in the female gametophyte, or dependent on the presence of a female gametophyte to be expressed in sporophytic cells of the ovule. Among 18 genes encoding pentatricopeptide-repeat proteins (PPRs) that show transcriptional activity in wild-type but not spl ovules, CIHUATEOTL (At4g38150) is specifically expressed in the female gametophyte and necessary for female gametogenesis. These results expand the nature of the transcriptional universe present in the ovule of Arabidopsis, and offer a large-scale quantitative reference of global expression for future genomic and developmental studies.
Keywords: Female gametophyte, MPSS, ovule, pentatricopeptide-repeat proteins, transcriptional activity
Introduction
The life cycle of plants alternates between a predominant sporophytic (diploid) phase—that initiates with the fusion of male and female gametes, and includes all stages of embryogenesis, seed germination, and vegetative growth—and a gametophytic (haploid) phase that takes place deeply embedded in specialized reproductive organs. In the ovule of most flowering plants including Arabidopsis, the gametophytic phase occurs in synchrony with the development and growth of the integuments, the sporophytic tissues of the ovule that protect the female gametophyte during its formation. Before integument initiation, the young ovule primordium of Arabidopsis differentiates a single subepidermal cell that undergoes meiosis giving rise to four haploid products, the megaspores. Following meiosis, a single surviving megaspore differentiates and expands before undergoing three consecutive rounds of mitotic divisions in a syncitium. At the end of these mitotic rounds, haploid nuclei within the female gametophyte cellularize to form seven cells: an egg cell, two synergids, a binucleated central cell, and three antipodals.
While close to 29 000 predicted genes have been identified in Arabidopsis, it has been suggested that only ∼1000 are likely to be involved in the formation of the female gametophyte (Drews and Yadegari, 2002; García Hernández et al., 2002). Numerous gametophytic mutations have been identified and shown to affect post-meiotic developmental events, including haploid nuclear proliferation, cellularization, and progamic functions occurring prior to the onset of zygote formation (Moore et al., 1997; Drews et al., 1998; Brukhin et al., 2011); in addition, an important subclass of gametophytic mutations result in maternal effects that cause obvious defects in seed development (Grossniklaus et al., 1998; Pagnussat et al., 2005). Even if these studies have allowed the identification of numerous genes involved in female gametogenesis, the intimate association between the diploid and haploid cells that develop within the ovule complicates our general understanding of the crucial mechanisms that control female gametophyte development. While the precise moment for the initiation of the gametophytic phase has not been determined, growth of the integuments is synchronized with the different stages of gametogenesis, suggesting that these two distinct groups of cells are involved in a cross-talk of signals important for coordinating their growth and differentiation. Recent studies have shown that non-cell-autonomous mechanisms controlled by sporophytic cells are crucial for gametic cell specification (Duran-Figueroa and Vielle-Calzada, 2010; Olmedo-Monfil et al., 2010), confirming the importance of sporophytic–gametophytic communication during ovule development (Robinson-Beers et al., 1992; Kelley and Gasser, 2009; Bencivenga et al., 2011).
Large-scale transcriptional analysis of the female gametophyte has been hindered by the small size of the Arabidopsis ovule and a lack of reliable techniques for the isolation of sufficient amounts of unaltered sporophytic or gametophytic cells. To partially overcome these difficulties, the combination of genetic strategies that compare transcripts present in wild-type and mutant ovules lacking a female gametophyte with microarray profiling technologies has allowed the identification of a significant number of genes expressed in gametophytic cells. Ovules of sporocyteless (spl; Yang et al., 1999)—also referred to as nozzle (nzz; Schiefthaler et al., 1999)—coatlicue (coa), or determinant infertile1 (dif1) that all lack a female gametophyte have been used in comparisons with wild-type ovules to identify genes likely to be expressed in the female gametophyte (Yu et al., 2005; Johnston et al., 2007; Jones-Rhoades et al., 2007; Steffen et al., 2007). Whereas the molecular nature of the gene affected in COA remains unknown, NZZ/SPL encodes a MADS-like transcription factor (TF) acting early in the ovule primordium (Yang et al., 1999), and DIF1 encodes a meiotic homologue of the Schizosaccharomyces pombe REC8/RAD21 cohesin genes (Bhatt et al., 1999). Although these strategies have identified several hundred gene candidates as being expressed in the female gametophyte, confirmation of their specific expression in haploid cells has been obtained for only 69 of them, through either promoter::reporter gene fusions or in situ hybridization. In addition, a recent study based on laser-capture microdissection (LCM) of cytological sections from fully differentiated gametophytic cells has provided a gene expression map that suggests similarities between plant and animal gametes (Wuest et al., 2010).
Massively parallel signature sequencing (MPSS)—a next-generation sequencing-based method of gene expression assessment—was used to conduct a quantitative large-scale transcriptional analysis of Arabidopsis ovules. A new micro-aspirator system allowed fast isolation of mRNA samples to compare the universe of transcripts present in wild-type and spl ovules lacking a female gametophyte. In wild-type ovules, transcripts from 9775 annotated genes were detected and quantified, in addition to >2200 new antisense transcripts and several hundred expressed signatures corresponding to unannotated intergenic regions. A total of 1301 genes showed 25-fold reduced or null expression in spl as compared with the wild-type, and 33 of them were experimentally validated using a combination of genetic and molecular approaches; a group of 28 of these genes were confirmed to act preferentially in the female gametophyte or to be dependent on the presence of a female gametophyte in order to be expressed in sporophytic cells of the ovule. Among 18 genes encoding pentatricopeptide-repeat proteins (PPRs) that show transcriptional activity in wild-type but not spl ovules, CIHUATEOTL (At4g38150) is specifically expressed in the female gametophyte and necessary for the initiation of female gametogenesis. By expanding the universe of genes and non-coding RNAs present in the ovule of Arabidopsis, these results offer a large-scale quantitative compendium of global expression as a reference for subsequent studies aiming at the characterization of gene regulatory networks acting during female reproductive development.
Material and methods
Plant material and growth conditions
All seeds of Arabidopsis thaliana were germinated in Murashige and Skoog (MS) medium under short-day conditions (16 h light/8 h dark) at 25 °C. Seedlings were then transplanted to soil and grown in a greenhouse under long-day conditions. Wild-type Columbia-0 (Col-0) and the spl allele nzz 1-3 (Schiefthaler et al., 1999) were used for ovule sample collection.
Micro-aspiration of ovules and RNA extraction
For ovule collection, fully differentiated and unpollinated gynoecia were dissected on an adherent platform with fine hypodermal needles to strip the carpels by hand and expose the ovules attached to the septum. When 4–5 gynoecia were dissected, they were immediately micro-aspirated and frozen for storage. A total of 5000 ovules were collected for each genotype (wild-type and spl). Details of the micro-aspirator device are provided in Supplementary Fig. S1 available at JXB online.
MPSS signature sequencing
MPSS was performed as described in Brenner et al. (2000) by Lynx Therapeutics/Solexa (Hayward, CA, USA). Signatures for a given library were produced in multiple sequencing runs and in two types of sequencing reactions (Brenner et al., 2000; Meyers et al., 2004b ); sequencing reactions and runs were combined to calculate a single normalized abundance for each signature observed in both the wild-type and the spl library. All raw and normalized data are available at http://mpss.udel.edu/at.
Analysis of MPSS data
All MPSS signatures that matched Arabidopsis genomic sequence were analysed following a previously described classification scheme (Meyers et al., 2004a ). In essence, all potential MPSS signatures (∼858 000, all derived from a DpnII restriction site plus the adjacent 13 bases, with a second signature derived from the complementary strand) were extracted from the A. thaliana genome. The position of each potential signature was compared with that of genes in the TAIR annotation version 8.0 (www.arabidopsis.org) and assigned to a class based on the position relative to exons and open reading frames (Meyers et al., 2004b ). Two filters were applied to the MPSS data to remove potentially inaccurate signatures and filter out the subset of signatures that are not expressed at significant levels. A first filter identified signatures found in only one MPSS sequencing run across all available libraries (‘unreliable’ signatures). Because all MPSS libraries consisted of at least four sequencing runs from the same tissue sample, this filter removed signatures that might result from random sequencing errors (the error rate for MPSS is estimated at 0.25% per base; Meyers et al., 2004a ). A second filter identified and eliminated those signatures expressed in any A. thaliana MPSS library at <4 TPM (transcripts per million; ‘non-significant’ signatures). This filter is called significant because 4 TPM is different from 0 TPM with P < 0.005, whereas 1–3 TPM is not significantly different from 0 TPM (P > 0.005). Signatures that are reliable but not significant could represent weakly expressed transcripts (Meyers et al., 2004b ).
Statistical analysis
The data set consisted of gene tags (MPSS results) for 13 454 Arabidopsis loci in the wild-type and spl mutant. The total number of gene tags was 1 507 669 for the wild-type and 1 511 244 for spl. The analysis was performed by the Fisher’s exact test for contingency tables (Fisher, 1922) as recommended by Auer and Doerge (2010). The Fisher’s exact test has as null hypothesis the independence between the criteria of classification that in this case is equivalent to the hypothesis of having the same level of expression in both treatments. The original data matrix of 13 450 rows (loci) by two columns (treatments; wild-type and spl) was collapsed into 13 450 (2×2) contingency tables as recommended by Auer and Doerge (2010) using the function ‘Fisher.full.test’ of the TRANOVA R package (Garcia-Ortega, unpublished results). All analyses were performed using R (R Development Core Team, 2011). The different number of significant genes for different values of alpha was obtained by using Bonferroni’s correction. AlphaB gives a family-wise error rate.
Genetic screen and semi-sterility analysis
SALK lines were ordered from the Salk Institute Genomic Analysis Laboratory (La Jolla, CA, USA) through the Ohio Arabidopsis Stock Centre (Alonso et al., 2003). Defective female gametophytes fail to initiate seed formation, resulting in semi-sterility. Mutations affecting female gametophyte development were identified in a screen for reduced fertility shown by the presence of >20% undeveloped ovules in green siliques (Moore et al., 1997). Insertional lines were grown and planted as full-sib families of 15 individuals, and no less than eight plants per family were visually scored for semi-sterility by quantifying undeveloped ovules in at least five developing siliques. Positive candidates were confirmed by detailed cytological analysis (see below). All lines screened are included in Supplementary Table S6 at JXB online.
Reverse transcription-PCR (RT-PCR)
Total RNA was isolated using Trizol (Invitrogen) from fully differentiated unfertilized ovules ground in liquid nitrogen. Approximately 5 μg of total RNA were treated with 5 U of RNase-free DNase (Boehringer-Mannheim) in 1× DNase buffer (Invitrogen) containing 20 mM MgCl2 for 15 min at room temperature and heat inactivated at 65 °C for 10 min. RNA was reverse transcribed using 20 pmol of an oligo(dT) primer (Sigma) in a 50 μl reaction containing 1× RT PCR buffer (Invitrogen), 3 mM MgCl2, 0.5 mM of dNTPs, 2.6 mM dithiothreitol, and 200 U of Superscript II reverse transcriptase (Invitrogen). RNA was pre-incubated with the oligo(dT) primer and dNTPs at 65 °C for 10 min followed by incubation at 42 °C for 2 h; 1 μl of the cDNA samples was used for PCR amplification with 2 mM MgCl2, 0.2 mM of each dNTP, 1 U of Taq DNA polymerase (Invitrogen), 1× PCR buffer, and 20 pmol of each primer for 30 cycles at an annealing temperature of 60 °C. A list of primer sequences is provided in Supplementary Table S9 at JXB online.
Reporter gene fusions and plant transformation
To construct the promoter::GUS fusions, genomic fragments corresponding to regulatory regions of eight genes were amplified by PCR using the primers described in Supplementary Table S6, and inserted upstream of the β-glucuronidase (GUS) gene in plasmid pBI101 (Jefferson et al., 1987). Genomic fragments included up to 2.5 kb of the regulatory sequence and 500 bp of sequence upstream of the transcriptional initiation codon (5′-untranslated region or exon). For plant transformation, vectors were transferred into Agrobacterium tumefaciens strain GV 2260 (McBride and Summerfelt, 1990). Transformations were performed on wild-type Col-0 by floral dipping procedures (Clough and Bent, 1998). Seeds obtained from the T0 promoter::GUS transformants were in vitro germinated in MS medium containing 50 μg ml−1 kanamycin. Primer sequences and cleavage sites that were used for cloning in pBI101 are provided in Supplementary Table S10 at JXB online.
Whole-mount preparations and histological analysis
Wild-type and mutant gynoecia were dissected longitudinally with hypodermic needles (1 ml insulin syringes; Becton Dickinson) and fixed with FAA buffer (50% ethanol, 5% acetic acid, and 10% formaldehyde), dehydrated in increasing ethanol concentration, cleared in Herr’s solution [phenol:chloral hydrate:85% lactic acid:xylene:clove oil (1:1:1:0.5:1)], and observed with a Leica microscope (Wetzlar, Germany) under Nomarski optics. GUS staining assays were conducted as described in Vielle-Calzada et al. (2000).
Results
MPPS signatures from wild-type and sporocyteless ovules
A vacuum-based micro-aspirator device that allows the dissection of large amounts of intact ovules was developed to isolate developing ovules of Arabidopsis efficiently in a timely manner. Ovules are aspirated through a 10 μl capillary tube connected to a 1.5 ml vacuum chamber that serves to freeze them instantly in liquid nitrogen or dry ice. A motorized pump connected to a 25 litre storage tank allows air extraction from the vacuum chamber, ensuring the steady pressure of 0.3–0.6 kg cm−2 necessary to detach an ovule from the placental tissue by transversally sectioning its funiculus (for more details, see the Materials and methods, and Supplementary Fig. S1 at JXB online). Using this device, 4000–5000 complete ovules could be isolated in <2 h with no contamination from placental or carpel tissue (Supplementary Fig. S2), with an average yield of 12 μg of total RNA per 1000 harvested ovules.
Following this procedure for sample collection, and to assess the complexity of the ovule transcriptome in A. thaliana, MPSS, the first developed next-generation sequencing strategy (Brenner et al., 2000), was used. MPSS was performed on mRNA isolated from fully differentiated ovules before pollination. The number of 17 bp signatures sequenced was 2 699 824 for the wild-type and 2 775 817 for the spl library, for a total of 25 875 (wild-type) and 24 865 (spl) distinct signatures. Sequencing runs were merged and normalized to obtain an estimation of the abundance of each distinct signature in TPM. To take into consideration only signatures with the highest confidence, signatures observed at <4 TPM were disregarded (‘non-significant’ signatures, following a previously established nomenclature; Meyers et al., 2004a , b ); those observed in a single sequencing run were also eliminated (‘unreliable’ signatures; Meyers et al., 2004a , b ). After MPSS sequencing, the complexity of both libraries was similar to the complexity of previously reported MPSS libraries from other Arabidopsis tissues (Meyers et al., 2004b ). Their abundance was spread over four orders of magnitude (Table 1): whereas >65% signatures were found in the range of 4–100 TPM, only ∼ 1% were expressed at levels >1000 TPM (Table 2).
Table 1.
MPSS signatures in wild-type and sporocyteless ovules of Arabidopsis
| Class | Descriptiona | Wild-type ovules (%) | spl ovules (%) |
| 1 | Exon—sense strand | 9687 (57.3%) | 9304 (59%) |
| 2 | 500 bp 3'-UTR | 4195 (24.8%) | 3464 (22%) |
| 3 | Exon—antisense strand | 2241 (13.2%) | 2267 (14.4%) |
| 4 | Unnanotated region | 283 (1.67%) | 259 (1.64%) |
| 5 | Intron–sense strand | 143 (0.8%) | 117 (0.7%) |
| 6 | Intron—antisense strand | 18 (0.1%) | 24 (0.15%) |
| 7 | Splice site—sense strand | 326 (1.9%) | 315 (2%) |
Based on TAIR 8.0.
Table 2.
Quantitative distribution of gene expression in vegetative and reproductive organs of Arabidopsis
| 4–8 TPM | 9–24 TPM | 25–50 TPM | 51–100 TPM | 101–1000 TPM | >1000 TPM | Total | |
| Inflorescencea | 2878 (24.72%) | 3497 (30.4%) | 2046 (20.66%) | 1213 (10.42%) | 1498 (12.87%) | 151 (1.3%) | 11 283 |
| Leafa | 2006 (18.59%) | 3363 (31.17%) | 2420 (22.43%) | 1489 (13.88%) | 1383 (12.81%) | 120 (1.1%) | 10 781 |
| Roota | 1810 (15.85%) | 3400 (29.78%) | 2515 (22.03%) | 1784 (15.62%) | 1770 (15.50%) | 139 (1.22%) | 11 418 |
| Wild-type ovule | 1305 (13.35%) | 2380 (24.35%) | 2028 (20.75%) | 1800 (18.41%) | 2118 (21.68%) | 143 (1.46%) | 9774 |
| sporocyteless ovule | 1207 (13.75%) | 2241 (25.52%) | 1748 (19.91%) | 1419 (16.16%) | 2017 (22.97%) | 148 (1.69%) | 8780 |
The results are given as the number of genes (%).
From Meyers et al. (2004a).
Previously uncharacterized transcripts identified by MPSS
All MPSS signatures were compared with the public Arabidopsis genomic sequence information to relate the expression data to specific genomic positions, including annotated genes (TAIR version 8.0). In wild-type ovules, a total of 19 034 signatures were matched to unique locations in the genome, 1442 signatures mapped to duplicated locations, and 4959 remained with no match to the genome. Unmatched signatures have been previously shown to derive from spliced 3' ends that have not yet been identified, transcripts found in regions of the genome not yet sequenced, sequencing errors, or non-Arabidopsis RNA contaminants (Meyers et al., 2004a). The number of MPSS signatures that mapped uniquely to the genome was 16 893 and 15 750 for the wild-type and spl library, respectively. Because these MPSS signatures are derived from specific locations at the 3' end of the corresponding mRNA molecule, each distinct signature corresponds to a distinct transcript (see the Materials and methods for details). All signatures mapping to a unique sequence of the Arabidopsis genome were grouped into one of seven classes according to their genomic location. As expected, in both libraries the most abundant class corresponded to transcripts produced by sense strand expression from protein-coding mRNAs (classes 1, 2, 5, and 7). Among these, the most abundant class corresponds to signatures derived from exon sequences (class 1; Table 1), followed by signatures that mapped within 500 bp downstream of the STOP codon of a previously annotated coding sequence (class 2). Signatures assigned to class 5 or class 7 are less numerous, as they map within intronic sequences (suggesting an alternative polyadenylation event that truncates a previously defined open reading frame) or expand over a previously identified splicing site encompassing a pre-defined intron. Additionally, numerous unique sequences identified novel antisense transcripts and unannotated intergenic transcribed regions not previously described as being expressed in the ovule. The most abundant group is composed of 2241 (wild-type) and 2267 (spl) antisense signatures that mapped to previously annotated exons (class 3). An additional small group of antisense signatures maps to previously annotated introns (class 6). Signatures in class 3 or class 6 may correspond to novel natural antisense transcripts (NATs) reminiscent of those found in other organs of Arabidopsis (Terryn and Rouze, 2000; Wang et al., 2006; Ron et al., 2010). Finally, 283 (wild-type) and 269 (spl) signatures correspond to distinct intergenic positions in the genome that contain unannotated transcripts expressed in the ovule (class 4).
Even if the MPSS procedure included a DNase treatment of RNA samples before processing, it was confirmed that signatures corresponding to intergenic regions represented real transcripts and not contaminating DNA by determining that 91.5% (260 signatures) of the wild-type and 89.9% (232 signatures) of spl are detected at significant levels in MPSS libraries from other tissues (Supplementary Tables S1, S2 at JXB online), indicating that these intergenic regions are indeed consistently transcribed in Arabidopsis. In addition, to verify the absence of RNA derived from floral organs that could have contaminated the ovule sampling during aspiration, null expression of genes reported as being uniquely active in reproductive organs other than the ovule, including petals (At2g19070), stamens (At1g20130, At1g33430, At2g19070, and At3g27025), pollen (At5g65110), and the transmitting tract or septum (At1g72290, At3g50330, and At5g67060), was confirmed (Higginson et al., 2003; Gremski et al., 2007; Peiffer et al., 2009; Phan et al., 2011; Supplementary Table S3 at JXB online).
Down-regulated genes in sporocyteless ovules
Class 1, 2, 5, and 7 signatures that mapped to annotated genes for each of the two libraries were summed to identify and quantitatively estimate the expression of genes represented by normalized transcriptional variants. For each annotated gene, the sum of these signatures corresponds to the total number of transcripts that were detected in either wild-type or spl ovules, representing a quantitative estimation of its expression. A total of 9774 and 8770 annotated genes were detected at expression levels of at least 4 TPM in wild-type and spl ovules, respectively (Table 2; complete data available at http://mpss.udel.edu/at). In both libraries, the expression levels of these genes spread over the same four orders of magnitude that characterize the total number of individual signatures sequenced in both libraries. As for previously characterized MPSS libraries (Meyers et al., 2004b ), the most represented class includes genes at transcriptional abundances comprised between 9 and 24 TPM (24% and 25% of genes detected in the wild-type and spl, respectively); however, and in contrast to estimates from vegetative tissues, the ovule shows an over-represented class of genes detected at a transcriptional abundance comprised between 100 and 1000 TPM (21.7% and 23% for the wild-type and spl, respectively, compared with ∼13% in root, leaf, or complete inflorescence samples; Table 2). Since this over-represented class is not specific to the wild-type but is also characteristic of spl ovules lacking a female gametophyte, these results suggest that a subgroup of highly transcribed genes is expressed in sporophytic tissues of the ovule irrespective of the presence of the female gametophyte.
The transcriptional abundance of all signatures detected by MPSS was compared between wild-type and spl ovules to identify differentially expressed genes. In homozygous spl individuals, female gametophyte development is arrested at the archesporial cell, before meiosis (Yang et al., 1999). Although in most ovules the female gametophyte is not formed and nucellar cell proliferation is limited, both inner and outer integuments fully differentiate as in wild-type ovules. Detailed cytological observations indicated that, in the Columbia (Col-0) ecotype background, 95% of spl ovules completely lack a female gametophyte, whereas 4% show a defective vacuolated gametophytically derived cell with a variable number of nuclei, and 1% exhibit a normal cellularized female gametophyte. On the basis of this phenotypic criterion, the transcriptional activity of gametophytic genes detected in wild-type ovules should be at least 25 times lower in ovules of spl individuals. By applying this criterion to MPSS data, 612 genes were down-regulated in spl ovules (Supplementary Table S4 at JXB online). For genes showing wild-type expression levels <25 TPM on the basis of significant and reliable signatures, an arbitrary 3-fold criterion allowed the identification of 689 additional down-regulated genes in spl ovules. A Fischer’s exact test indicated that all 1301 candidate genes show a significant difference in their trancriptional abundance between wild-type and spl ovules. The application of Bonferroni’s correction (corresponding to an α=1e-15 in each individual test) indicated that the probability of making one or more false discoveries (family-wise error rate) is 7.43e-20 for this same group of 1301 genes (Supplementary Table S5). At least 84% of these 1301 differentially expressed genes show wild-type expression at abundances <51 TPM, a percentage significantly higher than in the overall MPSS wild-type ovule library (51%; Table 3; Supplementary Table S4). Whereas only 4.2% (55 genes) are detected at transcriptional abundances >100 TPM (compared with 25% in the overall wild-type ovule library), none of them is expressed above 1000 TPM (143 genes in the overall wild-type ovule library). Taken together, these results indicate that candidate genes down-regulated in spl ovules are mostly detected at low levels of transcriptional abundance in the wild-type MPSS ovule library.
Table 3.
Quantitative distribution of MPSS-detected genes in the ovule of Arabidopsis
| 9–24 TPM | 25–50 TPM | 51–100 TPM | 101–1000 TPM | >1000 TPM | Total | |
| No. of genes down-regulated in spl ovulesa | 690 (53%) | 403 (31%) | 153 (11.8%) | 55 (4.2%) | None | 1301 |
| No. of genes expressed in wild-type ovules | 2380 (28.1%) | 2028 (23.94%) | 1800 (21.25%) | 2118 (25.0%) | 143 (1.69%) | 8469 |
Includes only the abundance in wild-type ovules of genes with 25-fold reduced or null expression in sporocyteless ovules (see text for details).
Experimental validation of differentially expressed genes
A combination of loss-of-function genetic screens, RT-PCR, and reporter gene fusions was used to confirm independently that a sample of the 1301 MPSS differentially detected genes were indeed down-regulated in spl ovules as compared with wild-type ovules. In an initial step, 448 publicly available T-DNA lines, each harbouring an insertion in a selected gene to be differentially detected on the basis of the MPSS transcriptional analysis, were screened in a search for semi-sterile phenotypes that could be the consequence of mutations affecting female gametogenesis (Supplementary Table S6 at JXB online). Fully penetrant heterozygous mutations acting at the haploid level (gametophytic mutants) and affecting the viability of the female gametophyte but not the male are expected to show a decrease in seed set of ∼50%. A total of five immature siliques were dissected to identify consistent cases of partial sterility in 6720 adult individuals (15 full-sibling plants per T-DNA line). Positive candidates showing >20% of non-fertilized ovules were selected for a subsequent cytological analysis. Whereas randomly selected insertional lines (T-DNA or transposon-based lines) yield close to 2% of female gametophytic mutants (54 mutants in 2511 lines screened; Brukhin et al., 2011), 4.46% of candidate lines (20 mutants out of 448 lines screened) showed levels of semi-sterility ranging between 26% and 61% (Table 4), confirming that this collection of differentially expressed genes is significantly enriched for genes that are involved in female gametophyte formation or function (χ2=1696.9 > χ2 0.05[1]=3.84). A detailed cytological analysis using conventional clearing techniques revealed that all 20 new mutants showed variable but significant frequencies of ovules exhibiting defects in female gametophyte development, including arrest at different stages of haploid nuclear proliferation, and absence of polar nuclear fusion prior to pollination (Table 4). For six of these mutants, a second independent T-DNA insertion exhibited an equivalent defect, confirming that the disruption of the corresponding gene results in a mutant phenotype. Functional elements affected by these mutants include TFs such as bZip16 that encodes a salt-stress response protein, several genes encoding membrane transporters, and a multidrug and toxic compound extrusion (MATE) efflux protein. In addition to the genetic approach, differential expression was confirmed for 12 MPSS-detected genes by conducting RT-PCR; for seven of them, at least one of the insertional lines described above showed a mutant phenotype (Supplementary Fig. S3). Whereas half of these 12 genes showed expression in wild-type but not in spl ovules, the other half showed preferential expression in the wild-type as compared with spl, indicating that some transcripts corresponding to coding sequences could be present in spl ovules at abundances below the level of detection of the MPSS experiments.
Table 4.
Female gametophytic lethal mutants corresponding to genes down-regulated in sporocyteless ovules
| Gene ID | Predicted function | Insertional line | Transcriptional activity (TPM) | Aborted ovules (%) | Mutant phenotypea | |
| Wild-type ovule | spl ovule | |||||
| At1g43886 | Transposable element | SALK_020396, SALK_084658 | 35 | 1 | 61.22 | 1N FG arrest. Collapsed female gametophyte. |
| At4g38150 | Pentatricopeptide-repeat protein | SALK_098509, SALK_001022 | 28 | 0 | 45.12 | 1N/2N FG arrest. |
| At4g31600 | UDP-glucoronic acid/UDP-N-acetyl galactosamine transporter related | SALK_120828 | 21 | 0 | 55.66 | 1N FG arrest. |
| At1g79410 | AtOCT5 (Arabidopsis thaliana ORGANIC CATION/CARNITINE TRANSPORTER5) | SALK_045609 | 15 | 0 | 58.49 | 1N FG arrest. Unfused polar nuclei. |
| At3g55060 | Unknown protein | SAIL_693_H10 | 34 | 0 | 46.75 | 1N FG arrest. |
| At4g28020 | Unknown protein | SALK_096057 | 95 | 0 | 43.13 | 1N/2N FG arrest. |
| At5g16720 | Unknown protein | SALK_024827 | 29 | 0 | 64.73 | 1N FG arrest. |
| At4g38380 | Antiporter/drug transporter | SAIL_652_D01 | 10 | 0 | 53.66 | 1N FG arrest. |
| At3g04040 | Unknown protein | SALK_093343 | 40 | 0 | 43.25 | 1N FG arrest. |
| At5g49215 | Glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein | SALK_056269, SAIL_109_E10 | 21 | 0 | 44.88 | 1N FG arrest. |
| At2g40950 | BZIP17; DNA binding/transcription activator/transcription factor | SALK_104326, SALK_004048 | 53 | 1 | 40.67 | 1N FG arrest. |
| At1g32310 | Unknown protein | SALK_018488 | 32 | 0 | 50.2 | 1N FG arrest. Absence of female gametophyte. |
| At5g55210 | Unknown protein | SALK_060883c | 41 | 0 | 49 | 1N FG arrest |
| At3g16730 | Unknown protein | SALK_059304 | 21 | 0 | 26.24 | 1N FG arrest. Collapsed female gametophyte. |
| At3g16130 | Unknown protein | SALK_032681 | 29 | 0 | 47.3 | 1N FG arrest. |
| At5g57290 | 60S acidic ribosomal protein P3 (RPP3B) | SALK_120009, SALK_054076 | 43 | 0 | 49.24 | 1N/2N FG arrest. |
| At2g38140 | PSRP4 (PLASTID-SPECIFIC RIBOSOMAL PROTEIN 4) | SALK_129668 | 48 | 0 | 43.9 | 2N/4N FG arrest. |
| At2g32720 | CB5-B (CYTOCHROME B5 ISOFORM B); haem binding | SALK_041099 | 131 | 0 | 56.22 | 1N FG arrest. Collapsed female gametophyte. |
| At2g29660 | Zinc finger (C2H2 type) family protein | SALK_119814 | 35 | 0 | 47.66 | 1N FG arrest. |
| At4g38520 | Protein phosphatase 2C family protein / PP2C family protein | SALK_049798, SALK_049777 | 58 | 0 | 43.11 | 1N/2N FG arrest. |
FG, female gametogenesis; 1N, one-nuclear stage of female gametogenesis; 2N, two-nuclear stage of female gametogenesis; 4N, four-nuclear stage of female gametogenesis.
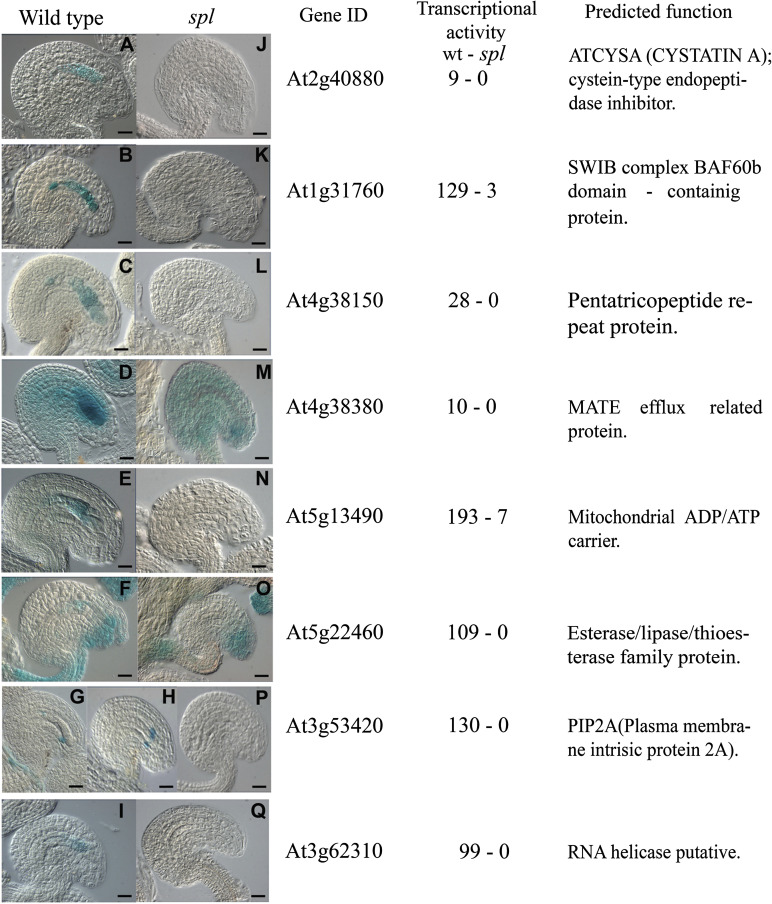
Finally, the in situ pattern of expression of eight MPSS-detected differentially expressed genes was examined during ovule development. For each of them, a promoter::reporter fusion cassette was constructed by cloning a genomic fragment comprised between 0.5 kb and 1.9 kb that included a putative regulatory region and the initial portion of the coding sequence of the corresponding gene in front of uidA (GUS; see the Materials and methods for details). To ensure that the observed patterns were reproducible, GUS expression was examined during wild-type ovule development in at least three independent transgenic lines for each promoter fusion, analysing five or more T2 generation individuals for each line. To determine possible differences from the wild-type, their pattern of expression was examined in homozygous spl ovules lacking a female gametophyte. Transformed lines harbouring promoter fusions corresponding to five of these genes (At1g31760, At2g40880, At3g62310, At4g38150, and At5g13490) showed GUS expression restricted to cells of the female gametophyte (Fig. 1). In fusions with At1g31760, At2g40880, and At4g38150 (encoding a SWIB complex BAF60b domain protein, a cysteine-type endopeptidase inhibitor, and a PPR, respectively) all cells of the fully differentiated female gametophyte exhibited GUS expression. In fusions with At3g62310, GUS expression was confined to the egg apparatus (synergids and egg cell), and with At513490, to the central cell and the antipodals. As expected, for all four lines these patterns were absent in spl ovules lacking a female gametophyte. Transformed lines corresponding to At4g38380 (encoding a MATE efflux protein) showed strong GUS expression in the female gametophyte and weak GUS expression in the surrounding sporopytic cells. In spl ovules, only the weaker sporophytic pattern of expression is maintained, indicating that diploid cells weakly express the corresponding gene independently of the presence of the female gametophyte.
Fig. 1.
Differential patterns of GUS expression in wild-type and sporocyteless ovules. Wild-type and spl mutant ovules were processed for histochemical analysis as described in the Materials and Methods. (A–J) Wild-type ovules; (J–Q) mutant spl ovules. The genomic accession number of each gene is indicated following the Arabidopsis Genome Initiative, and their corresponding transcriptional abundance is given in transcripts per million (TPM). Scale bars: (A and Q), 3 mm=15 μm; (B, D, F, M) 3 mm=15.5 μm; (C) 3 mm=12.5 μm; (E, N) 3 mm=11.25 μm; (G) 3 mm=19.5μm; (H) 3 mm=18 μm; (I, P) 3 mm=17.3 μm; (J, L, O) 3 mm=16.07 μm; (K) 3 mm=11.8 μm.
Two lines showed an unusual differential pattern in wild-type and spl ovules. In the case of fusions with At5g22460 encoding a lipase-esterase/thioesterase family protein, transformant lines showed GUS expression in the egg apparatus and antipodals, but also in the funiculus and the integuments at the micropylar pole (Fig. 1). In contrast, spl ovules lacking a female gametophyte show reduced expression in the funiculus and the integuments. In the case of fusions with At3g53420 encoding the plasma membrane aquaporin PIP2A, GUS expression in wild-type ovules is restricted to one or two cells of the endothelium, in close cellular association with the female gametophyte. This pattern is completely absent in spl ovules. Both lines suggest that the sporophytic expression of the corresponding gene is likely to be dependent on the presence of a female gametophyte or possibly regulated by an SPL-dependent pathway.
Pentatricopeptide-repeat genes are involved in female gametogenesis
All differentially expressed genes were classified according to protein domains available in Pfam, using a cut-off value of E < 0.01 (Finn et al., 2010). A total of 1061 genes (81.5% of the MPSS differentially expressed gene collection) could be grouped in the eight different classes shown in Supplementary Table S7 at JXB online. Whereas genes involved in housekeeping metabolism are predominant (452 genes), 97 code for signalling proteins such as kinase or leucine-rich repeat (LRR) domain proteins, calmodulins, or hormone-related and stress-induced proteins. Another important group is composed of 72 genes encoding TFs, including members of the Myb superfamily (23 genes), APETALA2/ETHYLENE-RESPONSIVE ELEMENT BINDING proteins (AP2/ERBP; six genes), and basic region/leucine zipper motif proteins (bZIP; four genes); only one of these TFs (At1g01860) was previously reported as specifically expressed in the female gametophyte (Wang et al., 2010). Additional classes include 76 genes encoding RNA-binding proteins such as RNA helicases, RNA methyltransferases, or Pumilo-type proteins; 16 genes encoding proteins involved in chromatin remodelling such as histones or histone-interacting proteins required for chromatin packing; and a group of 14 genes that encode exonuclease, methyltransferase, or endonuclease domain proteins. Other classes include genes involved in cell cycle regulation, DNA binding interactions, and organellar processing (see below). Finally, 240 genes do not code for a previously defined Pfam domain and could not be assigned to a specific functional class.
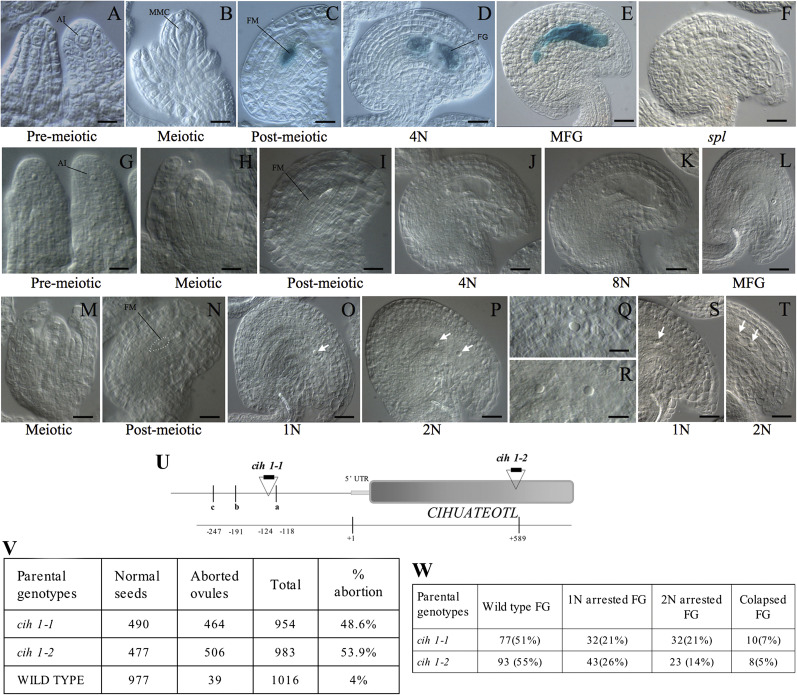
A group of 18 genes encoding PPRs showed consistent MPSS transcriptional activity in wild-type ovules but remained undetected in spl ovules (Supplementary Table S8 at JXB online). PPRs are sequence-specific RNA-binding and processing proteins acting in organelles and having a degenerate 35 amino acid structural motif found in >450 Arabidopsis family members. Although several PPRs are important for embryo development (Cushing et al., 2005; Lu et al., 2011), no PPR genes have been shown to act specifically in the female gametophyte. To determine if at least one of these 18 genes could indeed be specifically expressed in the female gametophyte and not in the rest of the ovule, a promoter::GUS fusion was constructed with a genomic fragment of 564 bp corrresponding to the regulatory region of At4g38150, a PPR gene predicted to act in mitochondria (Lurin et al., 2004). A cytological examination of transformed lines showed that GUS expression initiated in the functional megaspore following meiosis, and was restricted to the developing female gametophyte at subsequent stages of female gametogenesis (Fig. 2). Mutant ovules of homozygous spl individuals did not show GUS expression (Fig. 1), confirming that the activity of At4g38150 is restricted to the female gametophyte. To determine a possible function for At4g38150, ovule development was examined in two independent insertional lines harbouring a T-DNA element inserted in either its regulatory or coding region (Fig. 2). Both heterozygous mutant lines exhibited equivalent defects affecting female gametogenesis. The corresponding gene was named CIHUATEOTL (CIH), after the goddess of fertility in totonac culture (Aguilar-Moreno, 2007). In contrast to wild-type plants in which the haploid functional megaspore undergoes three rounds of mitotic divisions before differentiating a female gametophyte, heterozygous cih-1 and cih-2 individuals exhibited aberrant phenotypes that include a functional megaspore that arrests at the one-nuclear stage and fails to divide mitotically, or a female gametophyte arrested at the two-nuclear stage and not undergoing full differentiation. In both cases, the phenotype results in fully differentiated ovules that show a single conspicuous uninucleated or binucleated cell within the nucellus (Fig. 2). These results indicate that CIH encodes a PPR protein that acts in the female gametophte and is necessary for haploid nuclear proliferation during gametogenesis.
Fig. 2.
CIHUATEOTL is specifically expressed in the female gametophyte and its function is essential for female gametogenesis. Wild-type or cih-1 ovules were either histologically processed for GUS detection, or fixed and cleared for morphological analysis. (A-F) GUS expression in developing ovules of pCIH::GUS transformants. (G–L) Wild-type ovules showing normal female gametophyte development. (M–T) Mutant cih-1 ovules showing defects in female gametogenesis. (O) A mature cih-1 ovule shows a female gametophyte arrested at the one-nuclear stage (1N; arrow). (P) A mature cih-1 ovule shows a female gametophyte arrested at the two-nuclear stage (2N; arrows). (Q) Detail of a cih-1 ovule arrested at the one-nuclear stage. (R) Detail of a cih-2 ovule arrested at the two-nuclear stage. (S) A mature cih-2 ovule arrested at the one-nuclear stage of female gametogenesis (arrow). (T) A mature cih-2 ovule arrested at the 2-nuclear stage of female gametogenesis. (U) Genomic structure of the CIH locus; the position of allelic T-DNA insertions is indicated relative to the transcription initiation site. An I box promoter motif (a), a gibberelin-response motif (b), and a TATA box (c) are found in the regulatory region of CIH. (V) Developing siliques of cih-1 and cih-2 show close to 50% aborted ovules. (W) Frequency of phenotypic classes in cih-1 and cih-2 ovules. Scale bar: (A) 4 mm=7.2 μm; (B, Q, R) 4 mm=10 μm; (C, I, N) 4 mm=13.7 μm; (D) 4 mm=16 μm; (E, O) 4 mm=21 μm; (F) 4 mm=16.4 μm; (G) 4 mm=9.3 μm; (H) 4 mm=10.8 μm; (J) 4 mm=17.4 μm; (K, T) 4 mm=17.7 μm; (L) 4 mm=26.6 μm; (M) 4 mm=14.2 μm; (P, S) 4 mm=19.5 μm.
Discussion
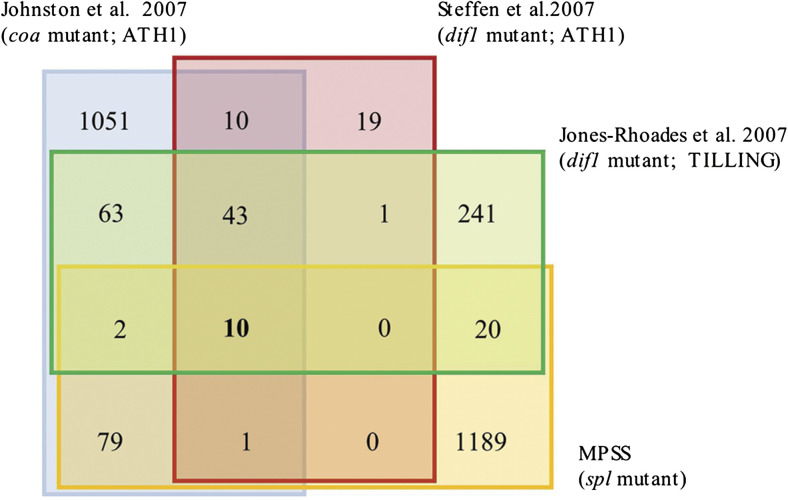
The evolutionary tendency in flowering plants has favoured the prevalence of the diploid sporophyte over a haploid gametophyte that is restricted to a few cells that form in specialized reproductive organs. The ephemeral nature of the female gametophyte deeply embedded within the ovule complicates the study of the genetic and molecular interactions that occur between cells involved in the somatic to reproductive transition. Several hybridization-based microarray profiling experiments have identified distinct collections of candidate genes as being expressed in the female gametophyte, with little overlap among data sets (Fig. 3). Yu et al. (2005) conducted Affymetrix ATH1 microarray experiments to compare the transcripts in ovules undergoing female gametogenesis and fully differentiated ovules; 202 genes were postulated to be expressed in the female gametophyte, but only five were confirmed by reporter gene fusions. A similar Affymetrix ATH1-based approach on a male-sterile background was used to identify genes with reduced expression in ovules of dif1, a recessive sporophytic mutant in which 75% of fully differentiated ovules lack a female gametophyte and 25% contain a cell reminiscent of the megaspore mother cell (Steffen et al., 2007); 43 genes were shown to be expressed in the female gametophyte using translational promoter::GFP (green fluorescent protein) fusions. In addition to conducting Affymetrix ATH1-based microarray profiling in coa, Johnston et al. (2007) complemented the spl-based statistical analysis of Yu et al. (2005) and significantly reduced the baseline cut-off fold change to identify a collection of 1295 genes which were candidates to be expressed in the female gametophyte; the mRNA of 12 genes was specifically localized in the female gametophyte. Using a whole-genome tiling array, Jones-Rhoades et al. (2007) compared the transcriptional activity in ovules between the wild-type and dif1, identifying 382 genes down-regulated in the mutant, and confirming that 11 were specifically expressed in the female gametophyte. Despite these independent efforts, only 10 genes are currently predicted to be specifically expressed in the female gametophyte by all microarray experiments that have used genetic substraction to assess global expression in the Arabidopsis ovule (Fig. 3). Although the sensitivity of these experiments is similar if not equivalent, the strategy has relied on the use of three different mutant backgrounds lacking a functional female gametophyte but having distinct mutant phenotypes prevailing in fully differentiated ovules, and therefore it is likely that some of the differences observed are due to deregulation of non-equivalent transcript collections, including those corresponding to genes that are not necessarily expressed in the female gametophyte, but repressed by the absence of DIF1, COA, or SPL activity.
Fig. 3.
Differences in the universe of genes which are candidates to be specifically expressed in the female gametophyte of Arabidopsis. Venn diagram illustrating the distribution of genes which are candidates to be expressed in the female gametophyte on the basis of four independent experiments conducted with three distinct mutant backgrounds (spl, dif1, and coa) and three different platforms (Affymetrix ATH1 microarray, whole genome TILING microarray, and MPSS). The corresponding type of platform and mutant background are indicated for each experiment. (This figure is available in colour at JXB online.)
In this study, a large-scale transcriptional analysis using MPSS allowed the quantification of 1301 genes down-regulated in spl ovules. In addition, the sampling allowed the identification of thousands of antisense signatures originating from previously annotated genes, as well as several hundred signatures corresponding to intergenic regions that are in most cases active at several developmental stages, and in a few cases exclusively transcribed in the ovule (close to 10% of class 4 signatures in each library). The importance of NATs has been overlooked due to their heterogeneity, low expression level, and unknown function. Whereas the abundance of signatures from class 3 and class 6 indicates that this type of non-coding RNA could play an important regulatory role in the ovule, either by being produced within its sporophytic cells or by being actively transported from other organs into reproductive tissues, signatures from class 4 are likely to represent transcripts from non-coding RNA precursors (including microRNAs) that are in most cases also expressed in other vegetative tissues (Supplementary Tables S1, S2 at JXB online); the identification of 60 signatures that are exclusively expressed in ovules (wild-type or spl) suggests that there is a small fraction of non-coding RNAs specific to regulatory mechanisms controlling female reproductive development.
When compared with previous studies aiming at identifying candidate genes to be expressed in the female gametophyte, the data set presented here is most similar to that of Johnston et al (2007), with 92 genes shared among the two data sets (Fig. 3). In contrast, the MPSS-based collection shares only 21 genes with data sets identified by microarray comparisons of wild-type and dif1 ovules (Jones-Rhoades et al., 2007; Steffen et al., 2007), confirming that data set overlap is highly dependent on the mutant background used to conduct a genetic subtraction. Among all MPSS differentially detected genes, 189 (14.5%) are not present in the platform that was used for previous Affymetrix ATH1 microarray profiling experiments comparing the expression of wild-type and spl ovules, and 179 genes (13.7%) exhibited a microarray signal below the level of detection of these same experiments (Yu et al., 2005), demonstrating that 28.2% of the MPSS data set could not have been identified as differentially expressed through Affymetrix ATH1 microarray profiling. Interestingly, 80% of these MPSS differentially detected genes show wild-type levels of expression below 51 TPM, suggesting that low rates of transcriptional abundance prevail within the female gametophyte as compared with the rest of the ovule.
By combining genetic and molecular approaches, 28 of these differentially detected genes were shown to be specifically active in the female gametophyte, and 20 new mutations caused by the alteration of female gametophytic genes were identified. While their expression was detected at late stages of ovule development, most cause an early arrest of nuclear proliferation, indicating that the corresponding genes also act early during female gametogenesis. In addition, five out of eight genes tested show a pattern of expression specific to the female gametophyte; of the three remaining genes, one shows preferential expression in the female gametophyte but is also expressed in sporophytic cells of the ovule, and the remaining two show a reduction in sporophytic expression in ovules lacking a female gametophyte, indicating that although the majority of down-regulated genes in spl ovules are indeed candidates to be specifically active in the female gametophyte, there is a subclass of transcripts that are repressed in sporophytic cells in the absence of a female gametophyte. On the basis of these results, the MPSS collection of differentially expressed genes is likely to include (i) genes that are specifically expressed in the female gametophyte; (ii) genes that are preferentially expressed in the female gametophyte as compared with sporophytic cells of the ovule; (iii) genes that require the presence of a female gametophyte to be expressed in sporophytic cells of the ovule; or (iv) genes that are positively regulated by an SPL-dependent genetic pathway (Fig, 4).
Fig. 4.
Classes of down-regulated genes in the sporocyteless ovule as compared with the wild-type. Each class is represented by a specific example. (A) Genes specifically expressed in the female gametophyte. (B) Genes that are preferentially expressed in the female gametophyte as compared with sporophytic cells of the ovule. (C) Genes that require the presence of the female gametophyte to be expressed in sporophytic cells of the ovule, or genes for which sporophytic expression is dependent on the function of SPL.
Plants contain more PPR genes than other eukaryotes, but the evolutionary and functional significance of this expansion remains elusive (Small and Peeters, 2000; Saha et al., 2007; Schmitz-Linneweber and Small, 2008). Most PPRs are localized in organelles where they regulate gene expression in either the mitochondria or chloroplasts (Fujii and Small, 2011), but there is evidence that PPRs also have roles beyond organelle gene expression through DNA binding activity (Ikeda and Gray, 1999; Mancebo et al., 2001). Although >440 PPR-encoding genes have been annotated in Arabidopsis (Lurin et al., 2004), only a few have been functionally characterized. Eighteen genes encoding PPRs which are expressed in wild-type but not in spl ovules have been identified here. Their low levels of transcriptional abundance, comprised between 9 and 31 TPMs, suggests that these genes are specifically expressed in gametophytic cells of the ovule. These 18 genes are all different from the seven genes encoding PPRs—including EMB175—for which sporophytic mutations have revealed essential roles in embryogenesis (Cushing et al., 2005), and also from AtPPR2 that is necessary for both normal gametogenesis and embryogenesis (Lu et al., 2011). The gametophytic activity of some of these new PPR genes is confirmed by the function of CIH (At4g38150), the first PPR gene found to be specifically expressed in the female gametophyte of Arabidopsis. Although AtPPR2 and CIH are both necessary for mitotic progression during female gametogenesis, mutations in CIH do not cause defects in embryogenesis, indicating that a possible redundant function for these two genes could be restricted to the gametophytic phase. While these results suggest that a specific group of PPRs is prone to act during the gametophytic phase of the life cycle, additional experiments will be required to determine their overall function during female gametophyte development.
New generation sequencing technologies applied to plant model organisms have begun to provide a complementary framework for the elucidation of the functional and evolutionary basis of the alternation of generations. As several projects begin generating a phylogenetic landscape to approach the study of plant evolution on the basis of genomic information (Delwiche et al., 2004), large-scale efforts to characterize global expression in the ovule could be of considerable importance to assemble the massive amount of data necessary to implement computational predictive analysis and modelling. These results suggest that a combination of technological platforms is necessary to explore the transcriptional universe of the ovule and contribute to the overall representation of the female gametophyte transcriptome. With multidisciplinary strategies progressively elucidating a cohesive and articulated understanding of female reproductive development in flowering plants, new opportunities develop to incorporate an overall body of knowledge into the framework of biological mechanisms that shape the evolution of vascular plants.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Micro-aspirator device for ovules of Arabidopsis.
Figure S2. Sample of micro-aspirated wild-type ovules.
Figure S3. RT-PCR of differentially expressed genes in wild-type and sporocyteless ovules.
Table S1. Transcriptional activity of wild-type ovule signatures from unnanotated regions.
Table S2. Transcriptional activity of sporocyteless ovules signatures from annotated regions.
Table S3. Transcriptional activity of non-ovule genes in diverse MPSS collections.
Table S4. List of MPSS differentially expressed genes between wild-type and sporocyteless ovules.
Table S5. Fisher’s exact test for 1301 spl down-regulated genes.
Table S6. Insertional lines screened for female gametophytic defects.
Table S7. Functional classification of MPSS differentially expressed genes between wild- type and spl ovules.
Table S8. PPR genes down-regulated in the sporocyteless ovule.
Table S9. Primers used in RT-PCR experiments.
Table S10. Primers and restriction sites used for promoter::fusion plasmid construction.
Acknowledgments
We thank Christian Haudenschild and Caghan Demirci for help with MPSS data generation, and the SALK Institute and the Ohio Stock Center for providing insertional T-DNA lines. This work was supported by grants from Consejo Nacional de Ciencia y Tecnología (CONACyT), the Howard Hughes Medical Institute (HHMI), and the UC-MEXUS initiative. Work in the Meyers lab is supported by the NSF Plant Genome Research Program. NS-L, M.A-V, ND-F, VP-E, DR-L, and IR-A are recipients of a graduate scholarship from CONACyT. J-PVC is an International Scholar of HHMI.
References
- Aguilar-Moreno M. Handbook of life in the Aztec world. Oxford: Oxford University Press; 2007. [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Auer PL, Doerge RW. Statistical design and analysis of RNA sequencing data. Genetics. 2010;185:405–416. doi: 10.1534/genetics.110.114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S, Colombo L, Masiero S. Cross talk between the sporophyte and megagametophyte during ovule development. Sexual Plant Reproduction. 2011;24:113–121. doi: 10.1007/s00497-011-0162-3. [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. The Plant Journal. 1999;19:463–472. doi: 10.1046/j.1365-313x.1999.00548.x. [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, et al. Gene expression analysis by massivelly parallel signature sequencing (MPSS) on microbead arrays. Nature Biotechnology. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Brukhin VB, Jaciubek M, Bolaños Carpio A, Kuzmina V, Grossniklaus U. Female gametophytic mutants of Arabidopsis thaliana identified in a gene trap insertional mutagenesis screen. International Journal of Developmental Biology. 2011;55:73–84. doi: 10.1387/ijdb.092989vb. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cushing DA, Forsthoefel NR, Gestaut DR, Vernon DM. Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta. 2005;221:424–436. doi: 10.1007/s00425-004-1452-x. [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Andersen RA, Bhattacharya D, Mishler BD, McCourt RM. Algal evolution and the early radiation of green plants. In: Cracraft J, Donoghue MJ, editors. Assembling the tree of life. Oxford: Oxford University Press; 2004. pp. 121–137. [Google Scholar]
- Drews GN, Yadegari R. Development and function of the angiosperm female gametophyte. Annual Review of Genetics. 2002;36:99–124. doi: 10.1146/annurev.genet.36.040102.131941. [DOI] [PubMed] [Google Scholar]
- Drews GN, Lee D, Christensen CA. Genetic analysis of female gametophyte development and function. The Plant Cell. 1998;10:5–17. doi: 10.1105/tpc.10.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán-Figueroa N, Vielle-Calzada JP. ARGONAUTE9-dependent silencing of transposable elements in pericentromeric regions of Arabidopsis. Plant Signaling and Behavior. 2010;5:1476–1479. doi: 10.4161/psb.5.11.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytologist. 2011;191:37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Research. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. Journal of the Royal Statistical Society. 1922;85:87–94. [Google Scholar]
- Garcia-Hernandez M, Berardini TZ, Chen G, et al. TAIR: a resource for integrated Arabidopsis data. Functional and Integrative Genomics. 2002;2:239–53. doi: 10.1007/s10142-002-0077-z. [DOI] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development. 2007;134:3593–3601. doi: 10.1242/dev.011510. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Ikeda TM, Gray MW. Characterization of a DNA-binding protein implicated in transcription in wheat mitochondria. Molecular and Cellular Biology. 1999;19:8113–8122. doi: 10.1128/mcb.19.12.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginson T, Li SF, Parish RW. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. The Plant Journal. 2003;35:177–192. doi: 10.1046/j.1365-313x.2003.01791.x. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Meier P, Gheyselinck J, Wuest SE, Federer M, Schlagenhauf E, Becker JD, Grossniklaus U. Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biology. 2007;8:R204. doi: 10.1186/gb-2007-8-10-r204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Borevitz JO, Preuss D. Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genetics. 2007;7:1848–1861. doi: 10.1371/journal.pgen.0030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Gasser CS. Ovule development: genetic trends and evolutionary considerations. Sexual Plant Reproduction. 2009;22:229–234. doi: 10.1007/s00497-009-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Li C, Wang H, Chen H, Berg H, Xia Y. AtPPR2, an Arabidopsis pentatricopeptide repeat protein, binds to plastid 23S rRNA and plays an important role in the first mitotic division during gametogenesis and in cell proliferation during embryogenesis. The Plant Journal. 2011;67:13–25. doi: 10.1111/j.1365-313X.2011.04569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, et al. Genome-wide analysis of arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. The Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo R, Zhou X, Shillinglaw W, Henzel W, Macdonald PM. BSF binds specifically to the bicoid mRNA 3' untranslated region and contributes to stabilization of bicoid mRNA. Molecular and Cellular Biology. 2001;21:3462–3471. doi: 10.1128/MCB.21.10.3462-3471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KE, Summerfelt KR. Improved binary vectors for Agrobacterium mediated plant transformation. Plant Molecular Biology. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Research. 2004a;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nature Biotechnology. 2004b;22:1006–1011. doi: 10.1038/nbt992. [DOI] [PubMed] [Google Scholar]
- Moore JM, Vielle-Calzada JP, Gagliano W, Grossniklaus U. Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harbor Symposia on Quantitave Biology. 1997;62:35–47. [PubMed] [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish SW. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. The Plant Cell. 2011;23:2209–2224. doi: 10.1105/tpc.110.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Kaushik S, Sakai H, Arteaga-Vazquez M, Sanchez-Leon N, Ghazal H, Vielle-Calzada JP, Mayers BC. A spatial dissection of the Arabidopsis floral transcriptome by MPSS. BMC Plant Biology. 2008;8:43. doi: 10.1186/1471-2229-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: 2011. [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female-sterile mutants. The Plant Cell. 1992;4:1237–1249. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S. Proper regulation of a sperm-specific cis-nat-siRNa is essential for double fertilization in Arabidopsis. Genes and Development. 2010;24:1010–1021. doi: 10.1101/gad.1882810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D, Prasad AM, Srinivasan R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiology and Biochemistry. 2007;45:521–534. doi: 10.1016/j.plaphy.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1999;96:11664–11669. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set of organelle gene expression. Trends in Plant Science. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Small ID, Peeters N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends in Biochemical Sciences. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN. Identification of genes expressed in the Arabidopsis female gametophyte. The Plant Journal. 2007;51:281–292. doi: 10.1111/j.1365-313X.2007.03137.x. [DOI] [PubMed] [Google Scholar]
- Terryn N, Rouzé P. The sense of naturally transcribed antisense RNAs in plants. Trends in Plant Sciences. 2002;5:394–396. doi: 10.1016/s1360-1385(00)01696-4. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Baskar R, Grossniklaus U. Delayed activation of the paternal genome during seed development. Nature. 2000;404:91–94. doi: 10.1038/35003595. [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang C, Hearn DJ, Kang IH, Punwani JA, Skaggs MI, Drews GN, Schumaker KS, Yadegari R. Identification of transcription-factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biology. 2010;10:110. doi: 10.1186/1471-2229-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chua MH, Wang XJ. Prediction of trans-antisense transcripts in Arabidopsis thaliana . Genome Biology. 2006;7:R92. doi: 10.1186/gb-2006-7-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenfuhrer J, von Mering C, Grossniklaus U. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Current Biology. 2010;20:506–512. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes and Development. 1999;13:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HJ, Hogan P, Sundaresan V. Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiology. 2005;139:1853–1869. doi: 10.1104/pp.105.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.