Abstract
Drought is the major environmental stress that limits cotton (Gossypium hirsutum L.) production worldwide. LOS5/ABA3 (LOS5) encodes a molybdenum co-factor and is essential for activating aldehyde oxidase, which is involved in abscisic acid (ABA) biosynthesis. In this study, a LOS5 cDNA of Arabidopsis thaliana was overexpressed in cotton cultivar Zhongmiansuo35 (Z35) by Agrobacterium tumefaciens-mediated transformation. The transformation and overexpression of AtLOS5 were assessed by PCR and RT-PCR analysis. Detached shoots of transgenic cotton showed slower transpirational water loss than those of Z35. When pot-grown 6-week-old seedlings were withheld from watering for 3 d, transgenic cotton accumulated 25% more endogenous ABA and about 20% more proline than Z35 plants. The transgenic plants also showed increased expression of some drought-responding genes such as P5CS and RD22, and enhanced activity of antioxidant enzymes such as superoxide dismutase, peroxidase, and ascorbate peroxidase. Their membrane integrity was considerably improved under water stress, as indicated by reduced malondialdehyde content and electrolyte leakage relative to control plants. When the pot-grown plants were subjected to deficit irrigation for 8 weeks (watering to 50% of field capacity), transgenic plants showed a 13% increase in fresh weight than the wild type under the same drought condition. These results suggest that the AtLOS5 transgenic cotton plants acquired a better drought tolerance through enhanced ABA production and ABA-induced physiological regulations.
Keywords: abscisic acid, AtLOS5, antioxidant enzymes, drought tolerance, proline, transgenic cotton
Introduction
Cotton (Gossypium hirsutum L.) is an important commercial crop grown worldwide as a source of fibre and edible oil. As a glycophytic plant, cotton shows higher drought and salt tolerance than other major crops such as wheat and rice, so it is classified as a drought- and salt-tolerant crop. Drought stress greatly impacts on cotton growth and limits fibre yield and lint quality, so plant breeding has been used to improve drought tolerance of cotton. Compared with conventional selection in cotton breeding, genetic transformation technology to improve agronomic traits and economic characteristics of crops by incorporating exogenous genes encoding the desired transgenic traits has become an efficient way to accelerate the breeding process (Zhang et al., 2011).
Drought stress, a major environmental stress that negatively impacts on growth and production of crops worldwide, induces a range of physiological and biochemical responses in plants. It triggers expression of stress-related genes, accumulation of metabolites such as abscisic acid (ABA) or osmotically active compounds, and synthesis of specific proteins (Shinozaki and Yamaguchi-Shinozaki, 2007). In fact, there have been many efforts in crops to improve drought tolerance and productivity under water-limiting conditions.
Hundreds of genes induced under drought conditions have been identified, cloned, and used as candidate genes in genetic engineering. These examples include proteins that function in abiotic stress tolerance, such as late embryogenesis abundant proteins, and key enzymes for osmolyte biosynthesis, and regulatory proteins involved in signal transduction regulation or stress-responsive gene expression, such as transcription factors and protein kinases (Shinozaki and Yamaguchi-Shinozaki, 2007).
ABA, classified as a stress hormone, plays significant roles in the regulation of plant growth and development and in plant responses to environmental stresses (Xiong et al., 2001; Xiong and Zhu, 2003). Genes involved in ABA biosynthesis have been cloned and characterized in Arabidopsis thaliana and other plant species (Xiong and Zhu, 2003; Taylor et al., 2005). These genes included zeaxanthin epoxidase (ZEP; Marin et al., 1996), which catalyses the epoxidation of zeaxanthin to produce epoxycarotenoid, 9-cis-epoxycarotenoid dioxygenase (NCED; Schwartz et al., 1997), which catalyses the cleavage reaction of epoxycarotenoids to produce xanthoxin, and abscisic aldehyde oxidase (AAO; Seo et al., 2000), which catalyses the final step of ABA biosynthesis whereby ABA aldehyde is converted to ABA. Moreover, LOS5/ABA3 (LOS5) is involved in the regulation of ABA biosynthesis by encoding molybdenum co-factor sulfurase, which is required by aldehyde oxidase in the last step of ABA biosynthesis in plants (Bittner et al., 2001; Xiong et al., 2001).
The enzymes involved in ABA biosynthesis have been investigated transgenically to improve plant stress tolerance. For example, transgenic Arabidopsis overexpressing AtZEP under drought stress showed increased leaf and lateral root development and longer primary roots compared with control plants, and exhibited much higher expression of the endogenous stress-responsive genes RD29A and Rab18 than wild-type plants under salt stress (Park et al., 2008). When subjected to drought stress, overexpression of NCED improved drought tolerance and caused an increase in endogenous ABA or a reduction in transpiration rate from leaves in transgenic A. thaliana (Iuchi et al., 2001), transgenic tobacco (Nicotiana plumbaginifolia) (Qin and Zeevaart, 2002), transgenic creeping bentgrass (Agrostis palustris) (Aswath et al., 2005), and transgenic tomato (Solanum lycopersicum) (Thompson et al., 2000, 2007; Tung et al., 2008) with improved tolerance. AtLOS5-overexpressing transgenic rice (Oryza sativa) (Xiao et al., 2009) and transgenic tobacco (Nicotiana tabacum) (Yue et al., 2011) under water-deficit conditions had better drought tolerance than non-transgenic controls.
Based on these studies, AtLOS5 was introduced into cotton by Agrobacterium tumefaciens-mediated transformation under the control of a superpromoter, and several independent transgenic lines were produced in this study. To test the function and potential use of AtLOS5 in improving drought tolerance of cotton, transformed phenotypes of transgenic cotton were evaluated by determining stress-related physiological and biochemical parameters under water-deficit conditions in a growth chamber.
Materials and methods
Construction of the binary vector
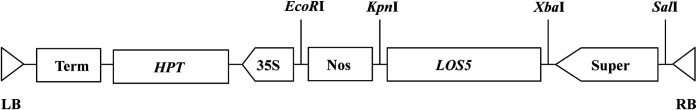
A modified pCAMBIA1300 vector was constructed by adding the superpromoter, which consisted of three copies of the octopine synthase enhancer in front of the manopine synthase promoter, as a SalI–XbaI fragment, into the pCAMBIA1300 binary vector containing a hygromycin phosphotransferase gene for resistance to hygromycin as a selectable marker. The LOS5 cDNA of A. thaliana ecotype Columbia (Xiong et al., 2001) was provided by Dr J.K. Zhu (University of California, Riverside, CA) and cloned as an XbaI–KpnI fragment downstream of the superpromoter in the modified pCAMBIA1300 (Fig. 1). The recombinant plasmid, pCAMBIA1300-LOS5, was introduced into A. tumefaciens strain EHA105 by freeze-thawing and used for plant transformation.
Fig. 1.
Schematic representation of the T-DNA region of the binary vector pCAMBIA1300-LOS5. LB, left border; RB, right border; HPT, hygromycin phosphotransferase gene; Super, superpromoter; 35S, cauliflower mosaic virus (CaMV) 35S promoter; Nos, nopaline synthase terminator; Term, CaMV 35S terminator.
Cotton transformation
Transformation of the cotton cultivar Zhongmiansuo 35 (Z35) by A. tumefaciens was performed as described previously (Gould and Magallanes-Cedeno, 1998) with minor modifications. Briefly, A. tumefaciens strain EHA105 harbouring pCAMBIA1300-LOS5 was cultured at 28 °C and 250 r.p.m. overnight in a modified Luria–Bertani medium (Sambrook et al., 1989) supplemented with 50 mg l−1 of kanamycin and 50 mg l−1 of rifampicin. The bacteria were collected and resuspended in half-strength liquid MS (Murashige and Skoog, 1962) medium at an optical density of 0.4–0.6 at 600 nm.
The explants (sterile shoot apices of seedlings) were infected with Agrobacterium suspension for 15–20 min and co-cultured on co-cultivation medium (MS salts with 60 μg l−1 of acetosyringone, 100 μg l−1 of kinetin, 30 g l−1 of glucose, and 2.5 g l−1 of Phytagel™) with a piece of sterile filter paper on the surface in the dark for 2 d. They were then transferred on to selection medium (MS salts with 100 μg l−1 of kinetin, 500 mg l−1 of cefotaxime, 15 mg l−1 of hygromycin, 30 g l−1 of glucose, and 2.5 g l−1 of Phytagel) for selection and incubated in a growth chamber at 25 °C (±2 °C) with a light intensity of 50 μmol m−2 s−1 under a long-day photoperiod (16 h light, 8 h dark) for 4–6 weeks. Hygromycin-resistant shoots were transferred on to recovery medium (MS salts with 250 mg l−1 of cefotaxime, 30 g l−1 of glucose, and 2.5 g l−1 of Phytagel) to restore growth. After 2 weeks, the shoots were grafted onto 6-d-old cotton seedlings of Z35 and placed in a mist chamber at high humidity and 25 °C (±2 °C) for 7 d. The plants were gradually hardened in growth chambers, transferred to large pots with soil, and grown to maturity in the greenhouse.
PCR detection and RT-PCR analysis
Genomic DNA was isolated from the young leaves of control and transgenic cotton according to the cetyltrimethylammonium bromide method (Chaudhary et al., 1999). PCR analysis for detection of the LOS5 gene was carried out with specific forward (5′-CTGGGAATGGAACCGTCGTCA-3′) and reverse (5′-GAGCCCGTTTGTAATCCTCGTCTT-3′) primers. PCR amplifications were carried out at 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 60 s, with a final elongation at 72 °C for 10 min. PCR products were separated by 1% (w/v) agarose gel electrophoresis.
For RT-PCR analysis, total RNA was extracted from the young leaves of control and transgenic cotton under normal and water-deficit conditions using a method described previously (Wan and Wilkins, 1994). Synthesis of cDNA was performed using a Takara RNA PCR kit (AMV) (Takara Biotechnology Co., China). The cDNA samples were used to amplify a LOS5 segment with forward (5′-GGGAAAGGGTGGAGGAGT-3′) and reverse (5′-GTAGCCAAACCAAGAGCC-3′) primers. The UBQ7 gene (GenBank accession no. DQ116441) was used as the internal control with specific primers (forward, 5′-GGCCTGATGGGACGAAGA-3′, and reverse, 5′-CAACGTCCAAAGCATCATAGTCA-3′).
Plant growth conditions and drought treatment
Seeds of Z35 and transgenic cotton, the T3 generation of independent lines L5 and L8, were planted in 3.6-litre pots filled with a mixture of vermiculite and peat (1:1) and grown in a growth chamber with a 14 h photoperiod at a 20/30 °C night/day temperature cycle, with a light intensity of 400 μmol m−2 s−1 and at 60% relative humidity. Z35 and each transgenic cotton line had seven pots (replicates). Z35 and transgenic cotton seedlings were cultured with a constant supply of Hoagland nutrient solution diluted 1:8.
Drought treatments were performed with 6-week-old cotton seedlings at the seven to eight leaf stage by withholding irrigation, while the control plants were grown normally. Completely expanded leaves at identical positions of Z35 and transgenic cotton seedlings were harvested 3 d after initiation of the stress treatment for biochemical measurements. Photographs were taken 5 d after initiation of the stress treatment.
In another assay of drought tolerance, seedlings of Z35 and transgenic cotton were grown for 3 weeks in 13-litre pots filled with the vermiculite/peat mixture described above. Seedlings of Z35 and transgenic cotton for the control treatments were watered to 75% of the maximum water-holding capacity of the mixture, while for water-deficit treatment the plants were watered to 50% of the maximum water-holding capacity. These cultural conditions were maintained for another 8 weeks and the samples were then harvested for biomass analysis.
Transpirational water loss
Seeds of Z35 and transgenic cotton, the T3 generation of independent lines L5 and L8, were germinated in sand medium and cultured hydroponically by transferring to pots filled with half-strength Hoagland’s nutrient solution in the growth chamber with 40 % relative humidity. Uniform seedlings cultured for 4 weeks were removed from the nutrient solution, detached at the cotyledonary node, and weighed immediately. They were then placed on filter paper to remove water under the growth chamber conditions and weighed at designated time intervals. Each had six replicates. The rate of transpirational water loss was calculated on the basis of the initial fresh weight of the plants.
Relative water content (RWC), electrolyte leakage, and lipid peroxidation
The RWC of leaves and roots was measured according to the method of Parida et al. (2007). Fully expanded leaves or roots were cut from plants and the fresh weight (FW) was recorded immediately. The fresh parts were then immersed in distilled water for 4 h and the turgid weight (TW) was recorded. Finally, the dry weight (DW) was recorded after drying for 48 h at 80 °C in an oven. RWC was calculated using the formula: RWC (%)=(FW−DW)/(TW−DW)×100.
Electrolyte leakage was determined by relative conductivity as described by Ai et al. (2008). Segments (1 cm2) were obtained from fully unfolded leaves of cotton for the measurements. Electrolyte leakage was calculated by the following formula: electrolyte leakage (%)=L t/L 0×100, where the conductivity measurements L t and L 0 corresponded to the plant leaves before and after boiling in water.
Lipid peroxidation was estimated as the content of malondialdehyde (MDA), as described by Ai et al. (2008). Leaf segments (0.5 g) were homogenized in 5 ml of 5% (w/v) trichloroacetic acid solution and centrifuged at 10 000g for 10 min. The mixture containing 1 ml of sample supernatant, 4 ml f 20% trichloroacetic acid, and 0.5% (w/v) f thiobarbituric acid was heated at 95 °C for 30 min, quickly cooled, and centrifuged at 10 000g for 10 min. MDA content was determined in a spectrophotometer at 532 nm (A 532) and corrected for non-specific turbidity at A 600.
ABA and proline content
ABA was extracted by grinding flesh leaves using a pre-chilled mortar and pestle on ice, homogenizing in 80% (v/v) methanol containing 1 mM butylated hydroxytoluence, and centrifuging at 4000g for 20 min. The supernatant liquid was eluted through a Sep-Pak C18 cartridge (Waters Corp.; Milford, MA) to remove polar compounds and then stored at –20 °C for ELISA. Endogenous ABA content was measured by an indirect ELISA technique, as described by Yang et al. (2001).
Free proline content was measured using a method described by Bates et al. (1973). Leaf segments were homogenized with 3% sulfosalicylic acid and the homogenates were centrifuged at 3000g for 20 min. The mixture containing 2 ml of sample supernatant, 2 ml of acetic acid and 2 ml of 2.5% acid ninhydrin was boiled for 30 min, and the absorbance was determined at A 520.
Antioxidative enzyme activities
The activities of antioxidant enzymes were determined by homogenizing 0.5 g of leaf tissue in 4 ml of extraction buffer [50 mM sodium phosphate buffer (pH 7.0), containing 1% (w/v) polyvinylpyrrolidone] using a pre-chilled mortar and pestle on ice. The homogenate was centrifuged at 10 000g for 30 min at 4 °C. The resulting supernatant was collected as a crude enzyme extract and assayed for the activity of superoxide dismutase (SOD), peroxidase (POD), and ascorbate peroxidase (APX), as described by Parida et al. (2004).
Expression of the genes P5CS, RD22, and DREB2B
Expression of the genes P5CS (GenBank accession no. EU417651), RD22 (GenBank accession no. AY464056), and DREB2B (At3g11020) in cotton under normal and water-deficit conditions was analysed by RT-PCR as described above. The primers for these genes were: P5CS forward (5′-CAAGCGGTCCAATGCTAT-3′) and reverse (5′-TGATGATACAATCTGTGTGTGC-3′); RD22 forward (5′-AGGAGGTGGTGGTGTAAACGTCAA-3′) and reverse (5′-ATGAAACACGGATCTCCTCCCGAA-3′); and DREB2B, forward (5′-GCTGAAATTCGTGAACCCAACCGT-3′) and reverse (5′-AGCTTGGCATCCGAACCATAGAGT-3′).
Statistical analysis
All experimental data are the means of at least three independent replicates, and results were determined using analysis of variance. Variation among treatment means were compared using Duncan’s multiple range test (P<0.05).
Results
Generation of transgenic cotton overexpressing AtLOS5
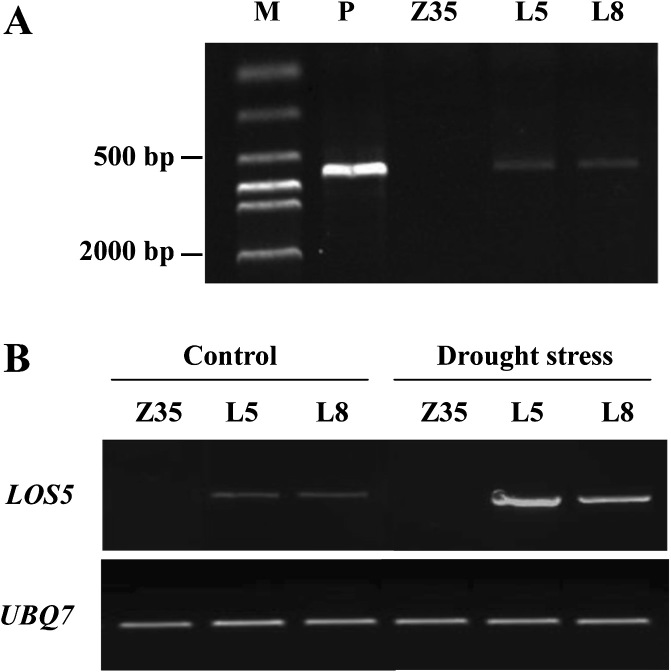
The construct pCAMBIA1300-LOS5 was introduced into cotton Z35 via Agrobacterium mediation, and 24 independent transgenic lines were produced. The primary transformed plants were designated T0 plants, and the seeds from self-fertilization of T0 plants were used to raise T1 progeny. Transgenic cotton harbouring AtLOS5 was screened on MS medium containing 15 mg l−1 of hygromycin. Two dominant lines, L5 and L8, were selected and homozygous T4 transgenic plants were used for drought-tolerance analysis. The presence and integrity of the transgene were confirmed by PCR analysis of genomic DNA with specific primers for AtLOS5 (Fig. 2A). Expression of the AtLOS5 gene was detected by RT-PCR analysis in transgenic cotton under normal and water-deficit conditions, and drought stress increased the expression of AtLOS5 (Fig. 2B).
Fig. 2.
PCR and RT-PCR analyses of LOS5 gene in transgenic cotton. (A) PCR analysis using specific primers for the LOS5 gene to identify two independent transgenic lines. Lane M, DNA marker; lane P, positive control (plasmid pCAMBIA1300-LOS5); Z35, non-transgenic control; lanes L5 and L8, transgenic lines. (B) RT-PCR analysis of LOS5 gene expression in the two transgenic lines under normal and water-deficit conditions. The UBQ7 gene was used as an internal control.
Drought tolerance in AtLOS5-overexpressing cotton
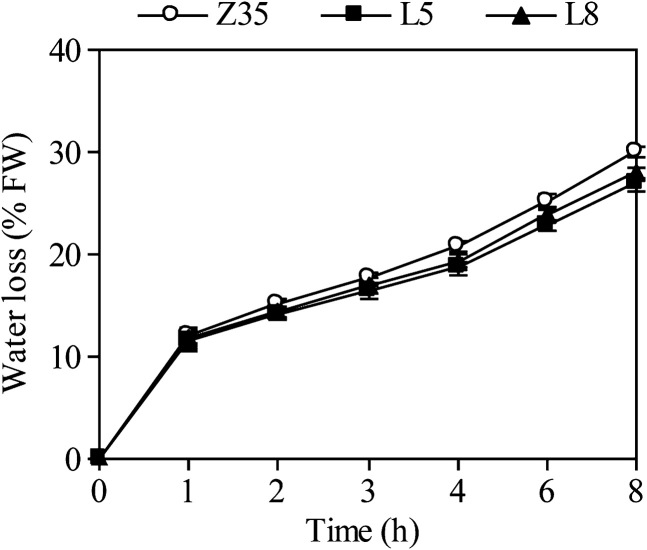
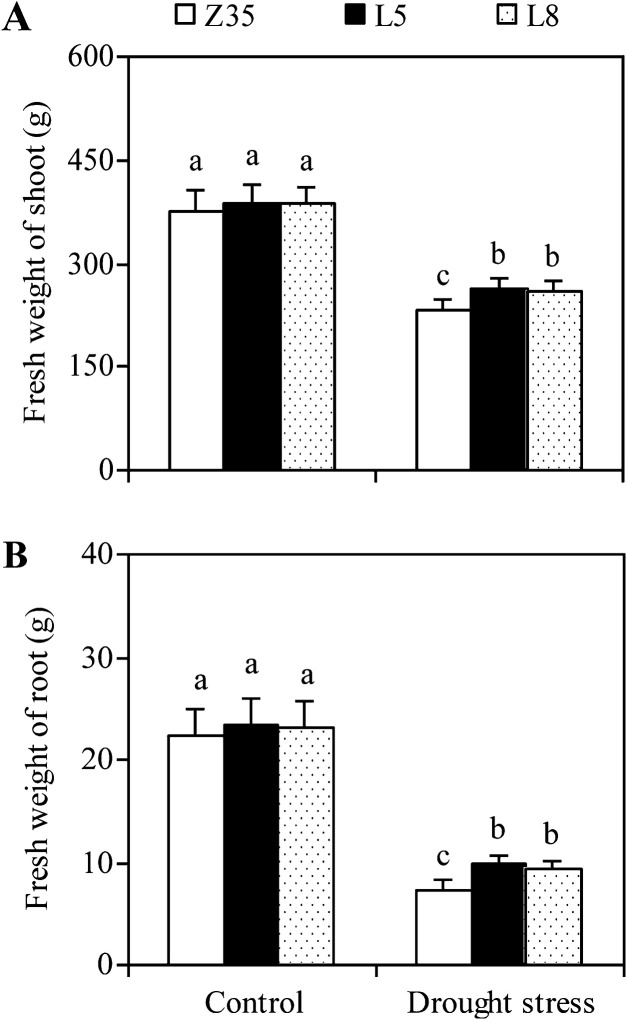
Measurement of transpirational water loss by detached plants showed that transgenic seedlings overexpressing AtLOS5 lost water more slowly than Z35 cotton in the same period under normal conditions (Fig. 3). The visible growth and morphology of non-transgenic Z35 and transgenic cotton were similar under natural (non-stressed) conditions over a 6-week period (Fig. 4). When subjected to drought stress for 5 d, Z35 and transgenic cotton showed differing degrees of wilting – Z35 plants wilted severely, while transgenic cotton wilted only partially. Non-transgenic Z35 and transgenic cotton had similar FWs of shoots and roots under control conditions (Fig. 5). However, the transgenic cotton plants maintained a 13% higher FW of shoots and a 30% higher FW of roots than that Z35 cotton after 8 weeks of water deficit.
Fig. 3.
Comparison of rates of transpirational water loss from detached shoots of non-transgenic Z35 and the transgenic cotton lines L5 and L8. Water loss is expressed as a proportion of the initial FW. Values are means ±standard error (SE) for six plants.
Fig. 4.
Six-week-old seedlings of non-transgenic Z35 and the transgenic cotton lines L5 and L8 subjected to drought stress for 5 d. LOS5-overexpressing cotton showed less wilting compared with Z35 plants.
Fig. 5.
Biomass of non-transgenic Z35 and the transgenic cotton lines L5 and L8 grown in the pots under normal and water-deficit conditions. Three-week-old seedlings were watered to 50 % of the maximum water-holding capacity of the pots for 8 weeks as water-deficit treatment. For the control, seedlings were watered to 75 % of this value. Values are means ±SE, Values with the same letter were not significantly different according to Duncan’s multiple range tests (P<0.05).
Leaf water status and membrane integrity in AtLOS5-overexpressing cotton
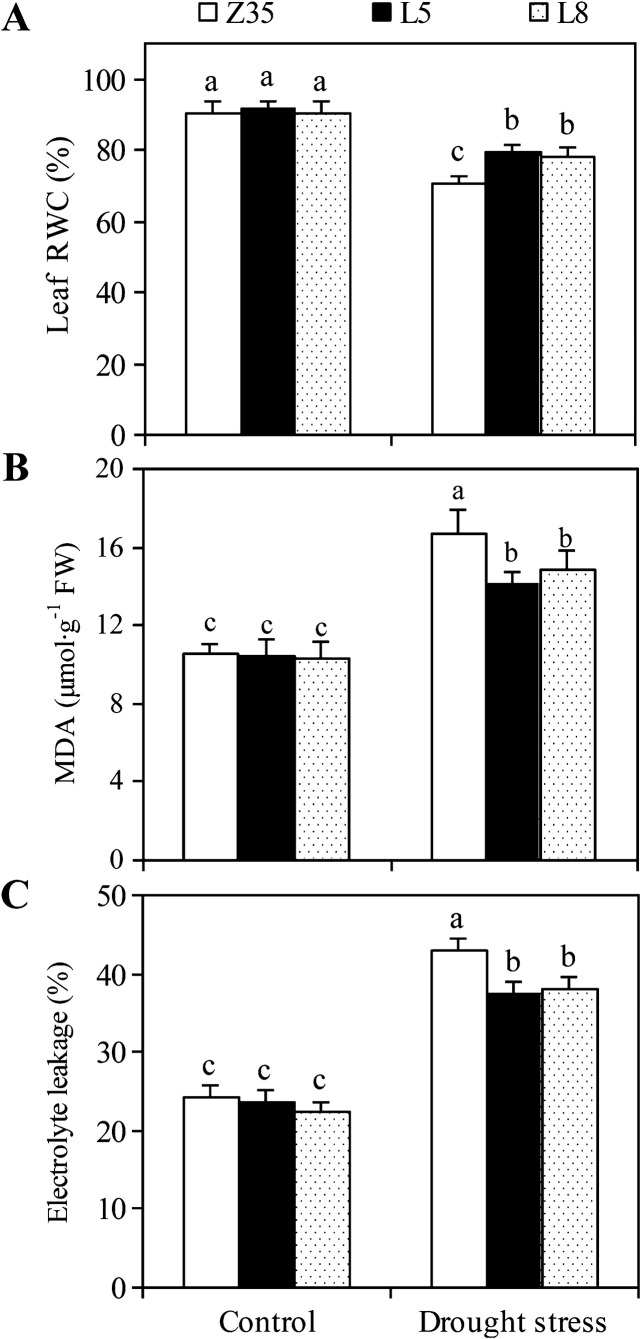
The RWC in leaves of Z35 and transgenic cotton under normal conditions were similar (Fig. 6). However, transgenic cotton maintained a higher RWC of 79% compared with Z35 plants with a RWC of 70%.
Fig. 6.
Leaf RWC (A), MDA content (B), and electrolyte leakage (C) in leaves of non-transgenic Z35 and the transgenic cotton lines L5 and L8 under normal and water-deficit conditions. Values are means ±SE. Values with the same letter were not significantly different according to Duncan’s multiple range tests (P<0.05).
Drought stress increased oxidative damage in the cotton leaves, as indicated by membrane lipid peroxidation, measured as MDA content (Fig. 6B). Drought stress enhanced MDA production in the leaves of both Z35 and transgenic cotton compared with the control plants. MDA production without drought stress was similar in transgenic and Z35 cotton, whereas the MDA content in drought-stressed plants was 14% lower in the leaves of transgenic cotton compared with Z35 plants.
The background level of electrolyte leakage without drought stress was similar for Z35 and transgenic cotton seedlings (Fig. 6C). Drought stress greatly increased electrolyte leakage in Z35 leaves by 76% and in transgenic cotton by 64%.
Antioxidant enzymes in AtLOS5-overexpressing cotton
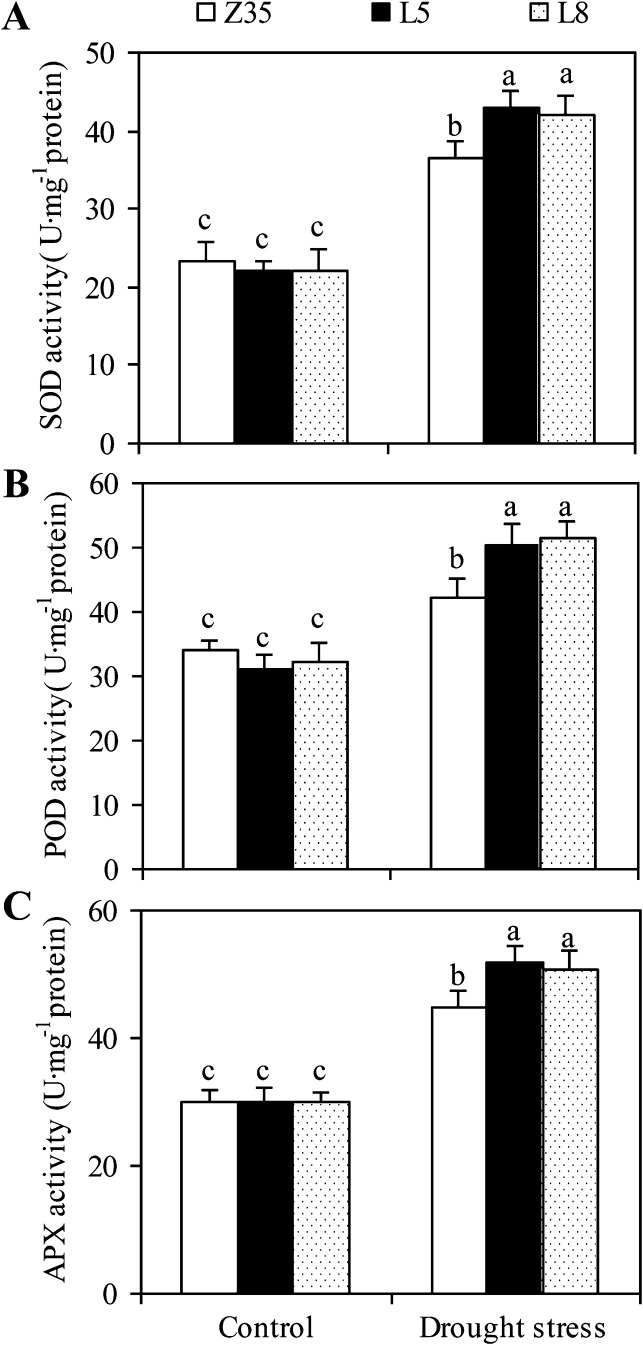
Baseline levels of the antioxidant enzymes SOD, POD, and APX under non-stressed conditions were similar for transgenic and Z35 cotton (Fig. 7). However, drought stress markedly increased the activities of the antioxidant enzymes in the leaves of both Z35 and transgenic cotton. Transgenic cotton under drought stress exhibited higher activities of SOD by 17%, POD by 21%, and APX by 15% compared with Z35 plants.
Fig. 7.
Activities of SOD (A), POD (B), and APX (C) in leaves of non-transgeneic Z35 and the transgenic cotton lines L5 and L8 under normal and water-deficit conditions. Values are means ±SE. Values with the same letter were not significantly different according to Duncan’s multiple range tests (P<0.05).
ABA and proline in AtLOS5-overexpressing cottons
Accumulation of endogenous ABA and proline under normal conditions was similar for both Z35 and transgenic cotton (Fig. 8). Drought stress greatly increased the endogenous ABA and proline contents in the leaves of both Z35 and transgenic cotton, but transgenic cotton accumulated 25% more endogenous ABA and 26% more proline than Z35 cotton.
Fig. 8.
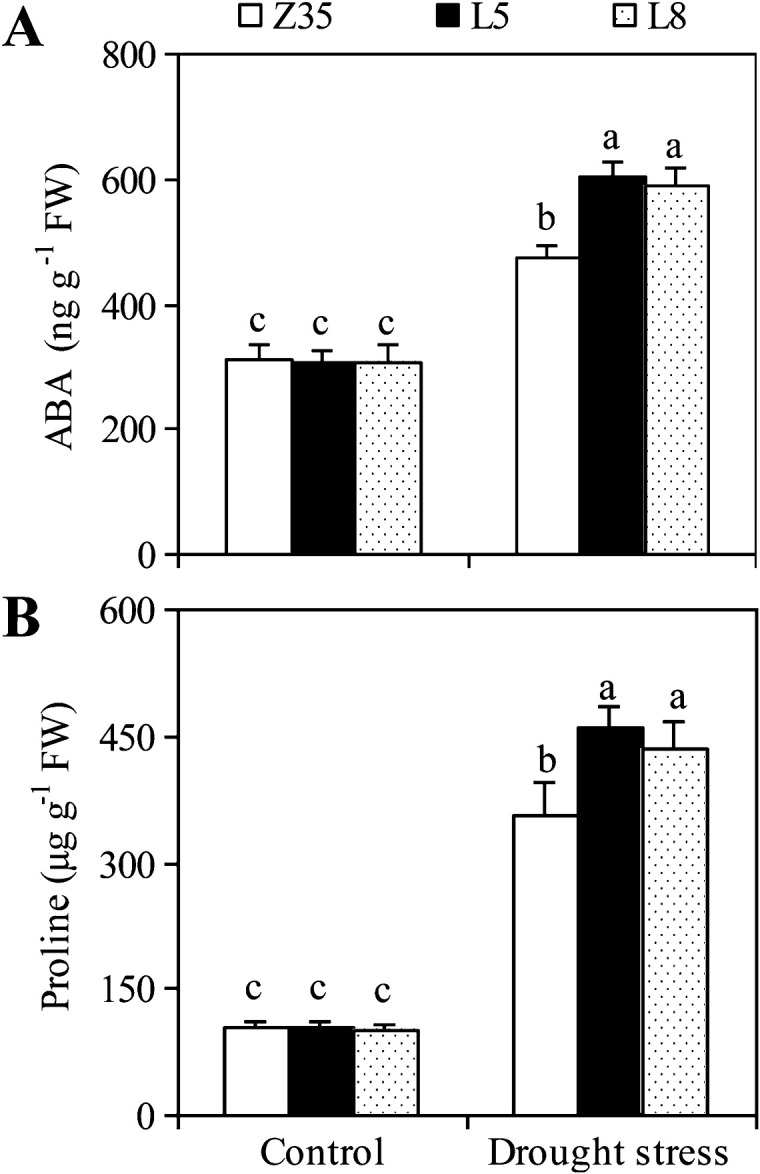
Levels of endogenous ABA (A) and proline (B) in the leaves of non-transgenic Z35 and the transgenic cotton lines L5 and L8 under normal and water-deficit conditions. Values are means ±SE. Values with the same letter were not significantly different according to Duncan’s multiple range tests (P<0.05).
Drought-related gene expression in AtLOS5-overexpressing cotton
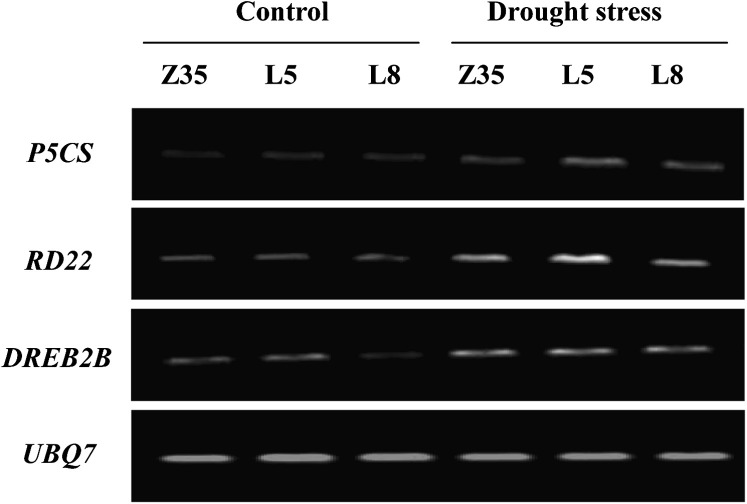
The drought-related genes P5CS, RD22, and DREB2B had low expression under normal conditions, but their expression was greatly promoted by water-deficit stress (Fig. 9). Although expression of RD22 and DREB2B under water-deficit conditions was improved to varying degrees in Z35 and transgenic cotton, the expression of P5CS in AtLOS5-overexpressing cotton, especially in line L5, was higher than in Z35 plants.
Fig. 9.
RT-PCR expression analysis of genes P5CS, RD22, and DREB2B in both non-transgenic Z35 and the transgenic cotton lines L5 and L8 under normal and water-deficit conditions. The gene UBQ7 was used as an internal control.
Discussion
Previous studies have demonstrated that transgenic rice (Xiao et al., 2009) and transgenic tobacco (Yue et al., 2011) showed higher expression levels of AtLOS5 under normal conditions compared with non-transgenic plants. Consistent with these studies, the AtLOS5 gene driven by the superpromoter had higher expression levels in transgenic cotton than in non-transgenic plants under both normal and water-deficit conditions (Fig. 2B). As reported previously in Arabidopsis (Xiong et al., 2001), the transcript levels of AtLOS5 in transgenic cotton were increased significantly in response to drought stress.
ABA accumulation is a common trait of transgenic plants that overexpress genes involved in ABA biosynthesis, such as AtZEP in transgenic Arabidopsis (Park et al., 2008), NCED in transgenic lines of tomato (Thompson et al., 2000), Arabidopsis (Iuchi et al., 2001), tobacco (Qin and Zeevaart, 2002), and bentgrass (Aswath et al., 2005), or AtLOS5 in transgenic tobacco (Yue et al., 2011). The accumulation of ABA can be explained by a model for stress induction of ABA biosynthesis in Arabidopsis (Xiong et al., 2002). Osmotic stress first induces expression of NCED, an early limiting step in controlling drought stress-induced ABA biosynthesis. Initially, accumulation of ABA potentiates expression of other ABA biosynthesis genes such as AAO, LOS5, and ZEP, which leads to more ABA biosynthesis. This coordinated increase in the transcription of all ABA biosynthesis genes would result in a more rapid and sustained increase in ABA biosynthesis. Therefore, overexpression of AtLOS5 increased ABA accumulation greatly in transgenic cotton under water-deficit conditions (Fig. 8A).
As a major physiological signal, ABA affects many stress-adaptation responses, such as regulation of shoot and root growth and limiting transpiration rate, thereby reducing wilting of plants (Lata and Prasad, 2011). Moreover, ABA modifies gene expression, and a large group of stress-responsive genes are regulated by stress-induced ABA. Overexpression of these genes involved in ABA biosynthesis leads to increased ABA production and reduced leaf transpiration under drought conditions, which consequently increases drought tolerance in transgenic plants.
Transgenic plants under drought conditions showed reduced leaf transpiration in tomato overexpressing LeNCED1 (Thompson et al., 2000), in Arabidopsis overexpressing AtNCED3 (Iuchi et al., 2001) and in tobacco overexpressing PvNCED1 (Qin and Zeevaart, 2002). Detached transgenic tobacco overexpressing AtLOS5 had lower transpirational water loss than control plants under normal conditions (Yue et al., 2011). Moreover, transgenic Arabidopsis overexpressing AtZEP in response to drought had higher FW compared with control plants (Park et al., 2008). Overexpression of AtLOS5 in rice under water-deficit conditions increased relative yield production and spikelet fertility of transgenic plants (Xiao et al., 2009). Consistent with these reports, transgenic cotton overexpressing AtLOS5 showed reduced transpirational water loss (Fig. 3) and enhanced drought tolerance (Fig. 4). Furthermore, transgenic cotton overexpressing AtLOS5 under water-deficit conditions maintained a higher RWC (Fig. 6A) and retained more shoot or root FW than non-transgenic controls (Fig. 5).
Reactive oxygen species triggered by drought stress can attack cellular macromolecules, thereby causing membrane damage or MDA accumulation, and affect protein synthesis and stability in plants. Meanwhile, water-deficit stress-induced ABA triggers induction of the antioxidant defence system and upregulates the activities of antioxidant enzymes such as SOD, CAT, and APX to protect plants from oxidative stress (Jiang and Zhang, 2002). Both transgenic cotton overexpressing AtLOS5 and non-transgenic Z35 plants under water-stress conditions had greatly increased electrolyte leakage and MDA production (Fig. 6B, C) and increased activities of the antioxidant enzymes SOD, POD, and APX (Fig. 7). However, efficient scavenging of reactive oxygen species in transgenic cotton protected membranes and macromolecules better and maintained less electrolyte leakage and MDA content than Z35 plants during drought stress, which contributed to enhanced drought tolerance of transgenic cotton overexpressing AtLOS5.
To further define which stress-responsive genes are regulated by stress-induced ABA, the drought-related genes DREB2B, RD22, and P5CS were analysed in transgenic cotton and Z35 plants under normal or water-deficit conditions. DREB2B, which is involved in osmotic-responsive gene expression in the ABA-independent stress-tolerance pathway, is induced by drought or high-salt stress (Liu et al., 1998). In our studies, water-deficit stress increased the expression of DREB2B in transgenic cotton overexpressing AtLOS5 and in non-transgenic Z35 plants, but there were no obvious differences in DREB2B levels (Fig. 9). Conversely, one ABA-dependent signal transduction pathway in drought and high salinity stress responses, MYC and MYB transcription factors, synthesized following accumulation of endogenous ABA, bound cis elements in the promoter and co-operatively activated the dehydration-responsive gene RD22 (Abe et al., 1997; Shinozaki and Yamaguchi-Shinozaki, 2007). We found that the RD22 gene could be induced by drought or ABA (Fig. 9). Under water-deficit conditions, RD22 was expressed more in AtLOS5-overexpressing cotton, especially in the line L5, than in non-transgenic Z35 plants.
P5CS, which catalyses the first two steps in proline biosynthesis, plays a key role in biosynthesis under osmotic stress (Savoure et al., 1995; Yoshiba et al., 1995). Expression of the P5CS gene was induced by both water-deficit in non-transgenic Z35 cotton and improved endogenous ABA in transgenic cotton overexpressing AtLOS5 (Fig. 9). The increased expression of P5CS further increased proline production in transgenic cotton. Drought stress notably increased proline content in both transgenic cotton overexpressing AtLOS5 and in non-transgenic Z35 plants, but AtLOS5-overexpressing cotton accumulated more proline than Z35 plants (Fig. 8B). Proline accumulates in many plant species in response to environmental stress, such as drought and high salinity, and its accumulation frequently correlates with tolerance to drought or salt stress in plants (Ben et al., 2008; Parida et al., 2008).
In conclusion, overexpression of the AtLOS5 gene in transgenic cotton seedlings improved drought tolerance, as indicated by increased FW in the growth chamber. The improved drought tolerance is probably attributed to enhanced expression of ABA-responsive genes as a result of increased ABA levels under the drought stress. Further study is required to determine field performance of the transgenic cotton.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (No. 30825028). We thank Dr Zhizhong Gong, Professor of College of Biological Sciences at China Agricultural University, and Dr Jiankang Zhu, Professor of Department of Botany and Plant Sciences, University of California, Riverside, for technical advice. We also thank Dr Calvin G. Messersmith, Professor Emeritus of Plant Sciences, North Dakota State University, for technical improvement of the manuscript.
Glossary
Abbreviations
- AAO
abscisic aldehyde oxidase
- ABA
abscisic acid
- APX
ascorbate peroxidase
- DW
dry weight
- FW
fresh weight
- MDA
malondialdehyde
- NCED
9-cis-epoxycarotenoid dioxygenase
- POD
peroxidase
- RWC
relative water content
- SE
standard error
- SOD
superoxide dismutase
- TW
turgid weight
- ZEP
zeaxanthin epoxidase
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai L, Li ZH, ZX X, Tian XL, Eneji AE, Duan LH. Coronatine alleviates polyethylene glycol-induced water stress in two rice (Oryza sativa L.) cultivars. Journal of Agronomy and Crop Science. 2008;194:360–368. [Google Scholar]
- Aswath CR, Kim SH, Mo SY, Kim DH. Transgenic plants of creeping bentgrass harboring the stress inducible gene, 9-cis-epoxycarotenoid dioxygenase, are highly tolerant to drought and NaCl stress. Plant Growth Regulation. 2005;47:129–139. [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline content for water-stress studies. Plant and Soil. 1973;39:205–207. [Google Scholar]
- Ben HA, Ghanem ME, Bouzid S, Lutts S. An inland and a coastal population of the Mediterranean xerohalophyte species Atriplex halimus L. differ in their ability to accumulate proline and glycinebetaine in response to salinity and water stress. Journal of Experimental Botany. 2008;59:1315–1326. doi: 10.1093/jxb/ern040. [DOI] [PubMed] [Google Scholar]
- Bittner F, Oreb M, Mendel RR. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana . Journal of Biological Chemistry. 2001;276:40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- Chaudhary B, Yasmeen A, Husnain T, Riazuddin S. Mini-scale genomic DNA extraction from cotton. Plant Molecular Biology Reporter. 1999;17:1–7. [Google Scholar]
- Gould JH, Magallanes-Cedeno M. Adaptation of cotton shoot apex culture to Agrobacterium-mediated transformation. Plant Molecular Biology Reporter. 1998;16:1–10. [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis . The Plant Journal. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Jiang MY, Zhang JH. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. Journal of Experimental Botany. 2002;53:2401–2410. doi: 10.1093/jxb/erf090. [DOI] [PubMed] [Google Scholar]
- Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. Journal of Experimental Botany. 2011;62:4731–4748. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain, separate two cellular signal transduction pathways in drought and low temperature-responsive gene expression, respectively, in Arabidopsis . The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana . EMBO Journal. 1996;15:2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiology. 1962;15:474–497. [Google Scholar]
- Park HY, Seok HY, Park BK, Kim SH, Goh CH, Lee BH, Lee CH, Moon YH. Overexpression of Arabidopsis ZEP enhances tolerance to osmotic stress. Biochemical and Biophysical Research Communications. 2008;375:80–85. doi: 10.1016/j.bbrc.2008.07.128. [DOI] [PubMed] [Google Scholar]
- Parida AK, Das AB, Mohanty P. Defense potentials to NaCl in a mangrove, Bruguiera parviflora: differential changes of isoforms of some antioxidative enzymes. Journal of Plant Physiology. 2004;161:531–542. doi: 10.1078/0176-1617-01084. [DOI] [PubMed] [Google Scholar]
- Parida AK, Dagaonkar VS, Phalak MS, Aurangabadkar LP. Differential responses of the enzymes involved in proline biosynthesis and degradation in drought tolerant and sensitive cotton genotypes during drought stress and recovery. Acta Physiologiae Plantarum. 2008;30:619–627. [Google Scholar]
- Parida AK, Dasgaonkar VS, Phalak MS, Umalkar GV, Aurangabadkar LP. Alterations in photosynthetic pigments, protein, and osmotic components in cotton genotypes subjected to short-term drought stress followed by recovery. Plant Biotechnology Reports. 2007;1:37–48. [Google Scholar]
- Qin XQ, Zeevaart JAD. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiology. 2002;128:544–551. doi: 10.1104/pp.010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Savoure A, Jaoua S, Hua XJ, Ardiles W, Van Montagu M, Verbruggen N. Isolation, characterization and chromosomal location of a gene encoding the Δ1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana . FEBS Letters. 1995;372:13–19. doi: 10.1016/0014-5793(95)00935-3. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- Seo M, Koiwai H, Akaba S, Komano T, Oritani T, Kamiya Y, Koshiba T. Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana . The Plant Journal. 2000;23:481–488. doi: 10.1046/j.1365-313x.2000.00812.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Taylor IB, Sonneveld T, Bugg TDH, Thompson AJ. Regulation and manipulation of the biosynthesis of abscisic acid, including the supply of xanthophyll precursors. Journal of Plant Growth Regulation. 2005;24:253–273. [Google Scholar]
- Thompson AJ, Andrews J, Mulholland BJ, et al. Overproduction of abscisic acid in tomato increases transpiration efficiency and root hydraulic conductivity and influences leaf expansion. Plant Physiology. 2007;143:1905–1917. doi: 10.1104/pp.106.093559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Mulholland BJ, Dadswell AR, Blake PS, Symonds RC, Burbidge A, Taylor IB. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. The Plant Journal. 2000;23:363–374. doi: 10.1046/j.1365-313x.2000.00789.x. [DOI] [PubMed] [Google Scholar]
- Tung SA, Smeeton R, White CA, Black CR, Taylor IB, Hilton HW, Thompson AJ. Over-expression of LeNCED1 in tomato (Solanum lycopersicum L.) with the rbcS-3C promoter allows recovery of lines that accumulate very high levels of abscisic acid and exhibit severe phenotypes. Plant, Cell and Environment. 2008;31:968–981. doi: 10.1111/j.1365-3040.2008.01812.x. [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Analytical Biochemistry. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Xiao BZ, Chen X, Xiang CB, Tang N, Zhang QF, Xiong LZ. Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Molecular Plant. 2009;2:73–83. doi: 10.1093/mp/ssn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong LM, Lee H, Ishitani M, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong LM, Lee H, Ishitani M, Zhu JK. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis . Journal of Biological Chemistry. 2002;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- Xiong LM, Zhu JK. Regulation of abscisic acid biosynthesis. Plant Physiology. 2003;133:29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC, Zhang JH, Wang ZQ, Zhu QS, Wang W. Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiology. 2001;127:315–323. doi: 10.1104/pp.127.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Shinozaki K. Correlation between the induction of a gene for Δ1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. The Plant Journal. 1995;7:751–760. doi: 10.1046/j.1365-313x.1995.07050751.x. [DOI] [PubMed] [Google Scholar]
- Yue YS, Zhang MC, Zhang JC, Duan LS, Li ZH. Arabidopsis LOS5/ABA3 overexpression in transgenic tobacco (Nicotiana tabacum cv. Xanthi-nc) results in enhanced drought tolerance. Plant Science. 2011;181:405–411. doi: 10.1016/j.plantsci.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhen JB, Li ZH, Kang DM, Yang YM, Kong J, Hua JP. Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.) Plant Molecular Biology Reporter. 2011;29:626–637. [Google Scholar]