Abstract
In plants and other organisms, glutathione (GSH) biosynthesis is catalysed sequentially by γ-glutamylcysteine synthetase (γECS) and glutathione synthetase (GSHS). In legumes, homoglutathione (hGSH) can replace GSH and is synthesized by γECS and a specific homoglutathione synthetase (hGSHS). The subcellular localization of the enzymes was examined by electron microscopy in several legumes and gene expression was analysed in Lotus japonicus plants treated for 1–48 h with 50 μM of hormones. Immunogold localization studies revealed that γECS is confined to chloroplasts and plastids, whereas hGSHS is also in the cytosol. Addition of hormones caused differential expression of thiol synthetases in roots. After 24–48 h, abscisic and salicylic acids downregulated GSHS whereas jasmonic acid upregulated it. Cytokinins and polyamines activated GSHS but not γECS or hGSHS. Jasmonic acid elicited a coordinated response of the three genes and auxin induced both hGSHS expression and activity. Results show that the thiol biosynthetic pathway is compartmentalized in legumes. Moreover, the similar response profiles of the GSH and hGSH contents in roots of non-nodulated and nodulated plants to the various hormonal treatments indicate that thiol homeostasis is independent of the nitrogen source of the plants. The differential regulation of the three mRNA levels, hGSHS activity, and thiol contents by hormones indicates a fine control of thiol biosynthesis at multiple levels and strongly suggests that GSH and hGSH play distinct roles in plant development and stress responses.
Keywords: γ-Glutamylcysteine synthetase, (homo)glutathione synthetase, immunogold localization, legumes, phytohormones, plant stress
Introduction
The thiol tripeptide glutathione (GSH; γGlu–Cys–Gly) is a major water-soluble antioxidant and redox buffer in plants, animals, and microorganisms (Meister, 1994; Wild and Mulcahy, 2000; Foyer and Noctor, 2011). In plants, GSH also performs critical functions in cell cycle regulation, plant development, sulphur transport and storage, stress response, and heavy metal detoxification (Maughan and Foyer, 2006). In legumes, the structurally related tripeptide homoglutathione (hGSH; γGlu–Cys–βAla) may partially or completely replace GSH with presumably the same functions (Frendo et al., 2001; Matamoros et al., 2003).
The synthesis of GSH is accomplished in two sequential ATP-dependent reactions catalysed by γ-glutamylcysteine synthetase (γECS) and glutathione synthetase (GSHS), whereas the synthesis of hGSH shares the same first enzyme and then requires a specific homoglutathione synthetase (hGSHS). The biochemical properties of the three thiol synthetases have been determined (Macnicol, 1987; Hell and Bergmann, 1990; Iturbe-Ormaetxe et al., 2002; Jez and Cahoon, 2004). However, there are still uncertainties about their subcellular localizations. Early reports using purified organelles from leaves of spinach (Spinacia oleracea), pea (Pisum sativum), and runner bean (Phaseolus coccineus) concluded that the three enzymes are located in the chloroplasts and cytosol (Klapheck et al., 1988; Hell and Bergmann, 1990), but subsequent studies with nodules and leaves of common bean (Phaseolus vulgaris) and cowpea (Vigna unguiculata) reported that γECS is in the chloroplasts and plastids and that at least some GSHS and hGSHS isoforms are present in the cytosol of nodule host cells (Moran et al., 2000). Recently, cellular and molecular analyses have also indicated that, in Arabidopsis, γECS is localized exclusively in the plastids, whereas GSHS occurs as a mixture of plastidic and cytosolic isoforms that are encoded by two transcript populations of the same gene (Wachter et al., 2005). The more sensitive and precise technique of immunogold electron microscopy has not been used so far to study the subcellular localization of thiol synthetases and, in particular, of hGSHS in legume tissues.
Likewise, information on the regulation of the genes involved in thiol biosynthesis is scarce and in some cases contradictory. Expression of γECS and GSHS remains invariant in Arabidopsis suspension cell cultures exposed to cadmium or xenobiotics that elicit a rapid accumulation of GSH (May et al., 1998). In contrast, treatment of Arabidopsis with metals known to mobilize GSH for phytochelatin synthesis increased coordinately the transcription of γECS and GSHS (Xiang and Oliver, 1998). A strong increase in γECS expression was also observed in leaves and roots of Indian mustard (Brassica juncea) exposed to cadmium (Schäfer et al., 1998; Wachter et al., 2005). Even less is known about the control of hGSHS expression. Only two reports have examined to date the effects of environmental cues or signal molecules on hGSHS expression. Thus, treatment of Medicago truncatula plants with compounds that release nitric oxide (NO), a key signalling molecule in plants (Neill et al., 2003), induced expression of γECS and GSHS, but not of hGSHS, in roots (Innocenti et al., 2007). Similarly, common bean plants treated with H2O2 showed upregulation of γECS and hGSHS in nodules, whereas treatments with cadmium, sodium chloride, or jasmonic acid (JA) had no effect (Loscos et al., 2008).
A better understanding of the regulation of GSH and hGSH biosynthesis in legumes during the stress response requires a precise determination of the subcellular localization of the enzymes and a quantitative expression analysis of the genes involved. In the present work, two objectives were pursued. First, polyclonal antibodies against γECS and hGSHS were produced to immunolocalize both proteins in legumes, taking advantage of the superior resolution of electron microscopy over subcellular fractionation or light microscopy localization techniques. Second, the expression pattern of the three thiol synthetase genes was determined in the model legume Lotus japonicus supplied with several hormones and related compounds that are involved in stress signalling (Fujita et al., 2006; Balbi and Devoto, 2008). This part of the study was focused on roots as they responded more rapidly to hormones than the leaves and it avoided the complication of different rates of hormone transport to the shoot. Nodulated plants were included to determine whether the nodulation status could alter the response of thiol synthesis to the hormonal treatments. These experiments were of interest because ethylene, ABA, JA, and SA inhibit nodulation, possibly as a mechanism to control nodule number (Stacey et al., 2006; Sun et al., 2006; Ding et al., 2008; Tominaga et al., 2009), whereas CK activates nodule formation (González-Rizzo et al., 2006; Tirichine et al., 2007).
Materials and methods
Plant growth and treatments
Nodulated plants of alfalfa (Medicago sativa L. cv. Aragón × Sinorhizobium meliloti 102F78) and common bean (P. vulgaris L. cv. Contender × Rhizobium leguminosarum bv. phaseoli 3622) were grown for 50–55 or 28–30 d, respectively, in pots containing vermiculite under controlled environment conditions (Naya et al., 2007; Loscos et al., 2008). Non-nodulated and nodulated plants of L. japonicus (Regel) Larsen ecotype MG-20 were grown for 21 and 45 d, respectively, in aerated hydroponic cultures under controlled environment conditions. The two sets of plants were harvested at different ages to compensate for the slower growth of nodulated plants; hence, they had similar weights and physiological ages to non-nodulated plants. Nodules of L. japonicus were produced by inoculation of seedling roots with Mesorhizobium loti R7A. The hydroponic medium was 4 l of 1/4 strength B&D nutrient solution (Broughton and Dilworth, 1971), containing 0 or 1.25 mM NH4NO3 for nodulated or non-nodulated plants, respectively. Root and stem nodules of Sesbania rostrata were produced by inoculation with Azorhizobium caulinodans ORS 571, and plants were grown in pots with vermiculite in a glasshouse for 30 d (James et al., 1996). All leguminous plants were at the vegetative stage when leaves, roots, and nodules were harvested. Plant material to be used for expression analysis of γECS, GSHS, and hGSHS was flash frozen in liquid nitrogen and stored at –80 °C, whereas material to be used for immunolocalization studies was immediately high-pressure frozen (see below).
To investigate the effects of hormones on expression of thiol synthetase genes, L. japonicus plants were treated for up to 48 h with 50 μM of abscisic acid (ABA), gibberellic acid (GA), salicylic acid (SA), JA, indole-3-acetic acid (IAA), 1-aminocyclopropane-1-carboxylic acid (ACC; the immediate ethylene precursor), cytokinins (CK; an equimolar mixture of kinetin and 6-benzyl-aminopurine), or polyamines (PA; an equimolar mixture of spermine, spermidine, and putrescine). Stock solutions of compounds (Sigma-Aldrich) were prepared as follows: 500 mM kinetin and 500 mM 6-benzylaminopurine (each in 200 μl of 1 M NaOH); 500 mM IAA (in 400 μl of 1 M NaOH); 100 mM ABA, ACC, PA, or SA (in 2 ml of ethanol); and 100 mM JA or GA (in 2 ml of dimethylsulphoxide). These volumes were then added to 4 l of the hydroponic solution, which was maintained at pH 6.6 for all treatments. Control plants that had grown simultaneously in hydroponics, and that had been treated with identical concentrations of NaOH, ethanol, or dimethylsulphoxide at the same time points, were used to correct gene expression values of the hormone treatments. Nutrient solution in hydroponics was maintained fully aerated during all the experiments by bubbling air at a flow rate of 160 l h−1 with a Rena Air 200 aquarium pump (Chalfont, Pennsylvania, USA).
To assesss the effects of NO on gene expression, L. japonicus plants were grown for 15 d in 1.5% agar plates (8–10 seedlings per 10 × 10-cm square plate) on modified Fahraeus medium without nitrogen (Boisson-Dernier et al., 2001). Plates were placed vertically under the same controlled environment conditions mentioned above, except that they were placed in the dark during the 24-h treatment. The plates contained a filter paper between the agar and the plants to maintain humidity and to avoid roots entering the agar. The NO-releasing compound S-nitroso-N-acetyl-DL-penicillamine (SNAP; Sigma-Aldrich) was added to the nutrient solution at a concentration of 500 μM, and plants were harvested after 3 and 24 h. The nutrient solution covered only about one-third of the rooting system to prevent anoxia.
Production and purification of recombinant enzymes
The open reading frame of common bean γECS without the signal peptide was amplified by PCR using specific primers (forward, 5'-CCATGGCGAGCCCGCCCACTG-3'; reverse, 5'-GCGGCCGCTAAGACACCCTTAATAAAG-3'). The product was cloned into the pCRII vector (Invitrogen) and the amplified fragment was digested with NcoI and PstI and cloned in a modified expression vector (pMAL-c2*). This plasmid was derived from pMAL-c2 (New England Biolabs, Beverly, USA) by including, within the XmnI multiple cloning site, a 6 × His coding sequence, a thrombin cleavage site, and a NcoI site. The construct in Escherichia coli DH5α cells was sequenced to verify the absence of errors in the open reading frame and was then transferred to BL21(DE3) cells to express the recombinant protein. The fusion protein contained the maltose-binding protein at the N-terminus, followed by the 6 × His tag and the mature γECS protein.
To purify enough recombinant protein for antibody production, cultures (500 ml) were inoculated with 1 ml of a preculture (LB medium with ampicillin) of the recombinant clone that had been grown overnight, and cells were grown at 37 °C until the absorbance at 600 nm reached 0.7–0.8. Expression was then induced with 250 μM isopropyl β-D-1-thiogalactopyranoside for 2 h at 37 °C. Cells were harvested by centrifugation and washed with 20 mM sodium phosphate (pH 7.4) and 0.5 M NaCl, and the pellet was resuspended in 5 ml of wash medium, frozen in liquid nitrogen, and stored at –80 °C. The cell suspension was thawed at 37 °C and sonicated (6 × 30 s). Lysed cells were centrifuged in the cold and the pellet was resuspended in 5 ml of 20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 25 mM imidazole, and 6 M guanidine. After centrifugation, the supernatant was saved and the pellet was resuspended in 5 ml of the same medium. This suspension was sonicated for another 30 s and centrifuged, and the supernatants were pooled.
The recombinant protein was purified in a single step from the pooled supernatants by using a HiTrap Chelating HP (5 ml) column, previously loaded with 100 mM NiSO4 and then washed with two volumes of water, essentially as recommended by the manufacturer (GE Healthcare Bio-Sciences, Uppsala, Sweden). The lyophilized protein (∼6–7 mg) was used to immunize two rabbits and to prepare an affinity column for purification of the monospecific antibody from the antiserum following conventional protocols (BioGenes, Berlin, Germany). Briefly, the protein was coupled to CNBr-activated Sepharose 4 Fast Flow (GE Healthcare Bio-Sciences), the antiserum was loaded on the column, and the monospecific IgG was eluted with 200 mM Gly-HCl buffer containing 250 mM NaCl (pH 2.2). The eluate was immediately adjusted to pH 7.5 with 2 M TRIS-HCl.
A similar procedure was followed to prepare recombinant hGSHS protein using sequence information of pea hGSHS (Iturbe-Ormaetxe et al., 2002) and to purify the corresponding polyclonal antibody.
Immunoblot analyses and immunolocalization of thiol synthetases
Immunoblots to monitor purification of recombinant proteins were performed by using a monoclonal antibody (clone His-1) against the His tag as the primary antibody (dilution 1:3000) and goat anti-mouse IgG conjugated to alkaline phosphatase as the secondary antibody (dilution 1:30000), as described by the supplier (Sigma-Aldrich). Immunoreactive proteins were detected with alkaline phosphatase substrate containing 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium (Sigma-Aldrich).
Immunoblots of plant extracts were performed according to published procedures (Rubio et al., 2009). The secondary antibody was a goat anti-rabbit IgG horseradish peroxidase conjugate (Sigma-Aldrich). The primary antibody was used at a dilution of 1:1000 (γECS) or 1:250 (hGSHS) and the secondary antibody at a dilution of 1:20000 (γECS and hGSHS). Immunoreactive proteins were detected by chemiluminescence using the SuperSignal West Pico or SuperSignal West Femto kits (Thermo Scientific, Rockford, IL, USA).
For immunogold localization, plant material was high-pressure frozen using an EM-PACT (Leica) instrument, and then freeze-substituted and embedded in low-temperature resin (Lowicryl HM23, Polysciences, Warrington, PA, USA) using an EM-AFS (Leica). Details of these procedures were given elsewhere (Rubio et al., 2009).
Expression analyses of thiol synthetases
Total RNA was extracted from roots and leaves with the RNAqueous kit (Ambion, Austin, TX, USA), and mRNA levels were determined by quantitative reverse-transcription PCR analysis using gene-specific primers as described and ubiquitin as the reference gene (Matamoros et al., 2003). The PCR amplification products were confirmed by melting curve analysis and the primer efficiencies, calculated by serial dilutions, were >90%. The number of amplification cycles with respect to ubiquitin (ΔCt) were ∼7–10 for γECS and hGSHS and ∼12–15 for GSHS.
Thiol synthetase activities and thiol contents
Thiol synthetase activities were determined by quantifying the GSH and hGSH produced by GSHS and hGSHS, respectively (Hell and Bergmann, 1988; Matamoros et al., 1999). The enzymes were extracted at 4 °C from 100 mg of roots with 500 μl of a medium consisting of 50 mM TRIS-HCl (pH 8.0), 0.2 mM EDTA, 10 mM MgCl2, and 10% glycerol. The extracts were cleared by centrifugation and depleted of thiols and other endogenous low molecular mass compounds using Vivaspin (10 kDa cut off) ultrafiltration devices (Sartorius, Goettingen, Germany). The reaction mixtures (final volume of 200 μl) contained 100 mM TRIS-HCl (pH 8.5), 50 mM KCl, 20 mM MgCl2, 5 mM ATP, 5 mM phosphoenolpyruvate, 5 units of pyruvate kinase, 5 mM dithioerythritol, 0.5 mM γ-glutamylcysteine, 5 mM Gly (GSHS) or β-Ala (hGSHS), and 100 μl of extract to initiate the reaction. This was terminated after 0 or 60 min at 30 °C by transferring 80-μl aliquots to derivatizing solution, which comprised 300 μl of 200 mM N-(2-hydroxyethyl)piperazine-N'-(3-propanesulphonic acid) and 5 mM diethylenetriaminepentaacetic acid (EPPS/DTPA buffer, pH 8.0), and 120 μl of 7 mM monobromobimane (MBB; Calbiochem). The samples were further incubated for 15 min in the dark and derivatization was stopped by adding 97 μl of 40% acetic acid. Samples were kept at –80 °C until analysis, which was performed by HPLC with fluorescence detection as previously described (Matamoros et al., 1999).
Thiol tripeptides (GSH and hGSH) were extracted from 100 mg of roots with 200 μl of 200 mM methanesulphonic acid containing 0.5 mM DTPA. The extracts were cleared by centrifugation and 50 μl of supernatant was mixed with 23 μl of 4 mM dithioerythritol, 100 μl of EPPS/DTPA buffer (pH 8.0), and 2 μl of 5 M NaOH. The mix was incubated for 1 h at room temperature and 50 μl of 7 mM MBB was added and left for 15 min in the dark. Derivatization was stopped by adding 90 μl of 20% acetic acid. The samples were centrifuged and the thiol derivatives were quantified by HPLC with fluorescence detection (Matamoros et al., 1999). The low concentrations of GSH were accurately determined by HPLC coupled to mass spectrometry (MS). Samples were analysed by liquid chromatography-tandem MS (LC-MS/MS) using a linear LTQ ion trap equipped with a micro-electrospray ionization source (ThermoFisher, San Jose, CA, USA). A 20 μl-aliquot was diluted to 40 μl with 1% formic acid prior to instrumental analysis and loaded on a chromatographic system consisting of a C18 preconcentration cartridge (Agilent Technologies, Barcelona) connected to a 10 cm long × 150 μm i.d. C18 column (Vydac, IL, USA). The separation was done at 1 μl min−1 in a 30 min acetonitrile 0–40% gradient (solvent A: 0.1% formic acid; solvent B: acetonitrile with 0.1% formic acid). The HPLC system comprised an Agilent 1200 capillary pump, binary pump, thermostated microinjector, and microswitch valve. The LTQ instrument was operated in the positive ion mode with a spray voltage of 2 kV. The spectrometric analysis was performed in a targeted mode, acquiring a full MS/MS scan of the precursor ions of GSH (m/z=498.2) and hGSH (m/z=512.2). The quantification was performed using extracted ion chromatograms of the optimum MS/MS transitions in terms of sensitivity (GSH, 498.2→435.2; hGSH, 512.2→449.2).
Results
Subcellular localization of thiol synthetases in legumes
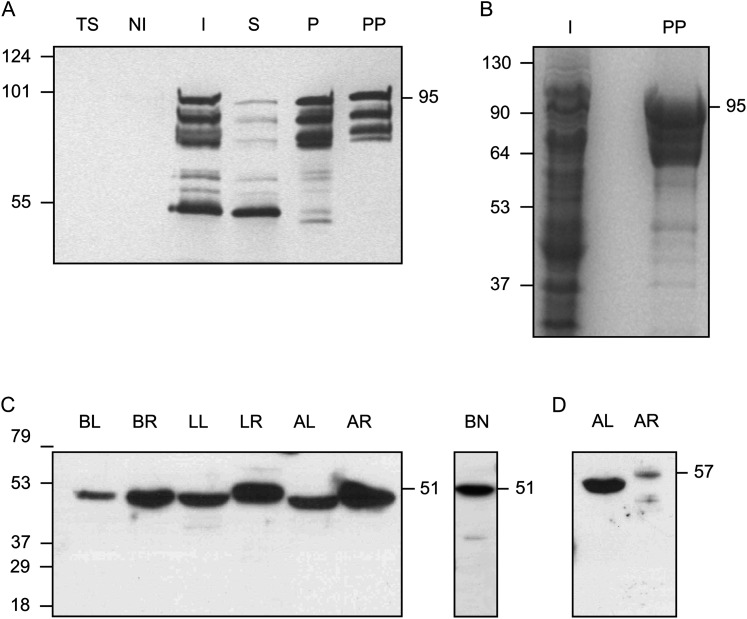
To perform immunolocalization studies of thiol synthetases in leaves, roots, and nodules of some crop and model legumes, it was necessary to purify the proteins and generate antibodies. Preliminary attempts to purify the enzymes directly from legume tissues were unsuccessful as they were found at a low concentration and γECS was particularly labile during extraction. This could explain the lack of any previous immunolocalization of thiol synthetases. Thus, γECS and hGSHS were expressed in E. coli as fusion proteins to enhance their expression and/or solubility. The recombinant proteins had a His tag and were purified with metal-chelating columns. The presence of several protein bands in the preparation of purified γECS (Fig. 1A) can be detected when using the pMAL expression system and is not due to contaminants but to the formation of truncated proteins by partial proteolysis of the fusion protein (Riggs, 2000). This can also be inferred from the fact that these proteins contain the His tags (lane PP, Fig. 1A). In addition, a protein band of ∼50 kDa was observed in the induced and soluble fractions (Fig. 1A, lanes I and S), which was attributed to the maltose-binding protein-tagged protein based on its expected molecular mass and high solubility and stability (Riggs, 2000). This protein product may have originated by proteolysis of the whole fusion protein but its identity was not verified. The fusion protein containing γECS accounted for >95% of the total protein, judging from densitometric analysis of the Coomassie-stained gel (Fig. 1B, lane PP). Recombinant γECS and hGSHS were used to produce antisera, and the polyclonal monospecific antibodies were affinity purified. The γECS antibody recognized a single protein band (51 kDa) in extracts of leaves and roots of all legumes examined; the same immunoreactive protein band could be observed in nodules (Fig. 1C).
Fig. 1.
Expression and purification of γECS and immunoblots of γ-glutamylcysteine synthetase (γECS) and homoglutathione synthetase (hGSHS) in legumes. (A) Purification of the fusion protein between the γECS from bean and the maltose-binding protein from Escherichia coli. Immunoblot using a monoclonal antibody against the His tag: TS, untransformed E. coli BL21 cells (10 μg protein); NI, non-induced transformed cells (10 μg protein); I, induced transformed cells (10 μg protein); S, supernatant of lysed induced cells (10 μg protein); P, pellet of lysed induced cells (2.5 μg protein); PP, purified recombinant enzyme (0.5 μg protein). (B) SDS gel stained with Coomassie. I, induced transformed cells (50 μg protein); PP, purified recombinant enzyme (10 μg protein). The expected molecular mass of the fusion protein is ∼95 kDa. (C, D) Immunoblots of γECS in several organs of representative legumes (C) and hGSHS in alfalfa (D) (20 μg protein). BL, bean leaves; BR, bean roots; BN, bean nodules; LL, Lotus japonicus leaves; LR, Lotus japonicus roots; AL, alfalfa leaves; AR, alfalfa roots. Detection was by chemiluminescence with the SuperSignal West Pico (A–C) or Femto (D) kits. Molecular masses (kDa) of the protein markers are shown on the left and apparent molecular masses (kDa) of the proteins are given on the right.
However, the hGSHS antibody recognized a single protein band at the expected mass (∼57 kDa) in extracts of alfalfa leaves and roots (Fig. 1D) but the corresponding immunoreactive protein was not seen in extracts of L. japonicus or common bean (data not shown). Therefore, immunogold localization studies of γECS were carried out with several legumes but those of hGSHS were limited to alfalfa. The hGSHS protein band in alfalfa leaves was clearly more abundant than in roots and the protein in roots showed a slightly higher apparent molecular mass (Fig. 1D). Because the amino acid sequences of GSHS and hGSHS in both M. truncatula and L. japonicus share 77% identity and a similar value is expected for the two proteins of alfalfa, the possibility cannot be ruled out that the hGSHS antibody also recognizes GSHS. However, hGSHS activity is ∼10–13-fold higher than GSHS activity in alfalfa leaves (4.86 ± 0.27 versus 0.50 ± 0.05 nmol min−1 (g fresh weight)−1) and roots (6.87 ± 1.33 versus 0.53 ± 0.10 nmol min−1 (g fresh weight)−1). Therefore, it is concluded that the antibody recognizes hGSHS and that GSHS is present at negligible amounts in alfalfa leaves and roots.
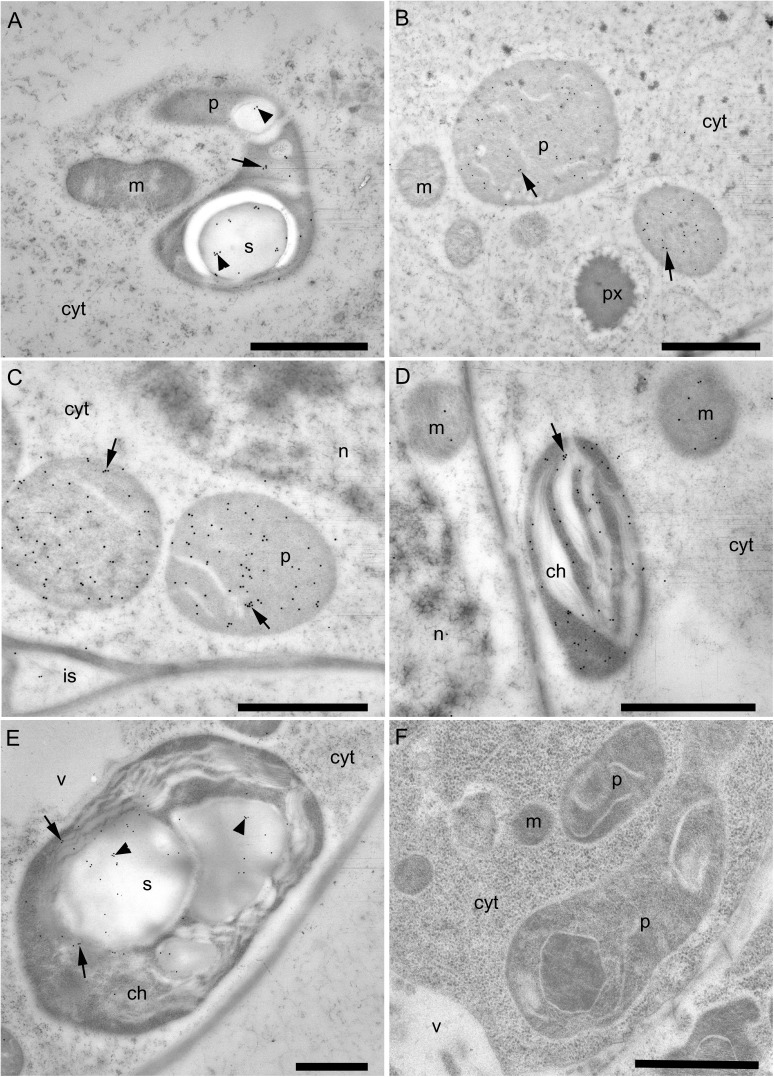
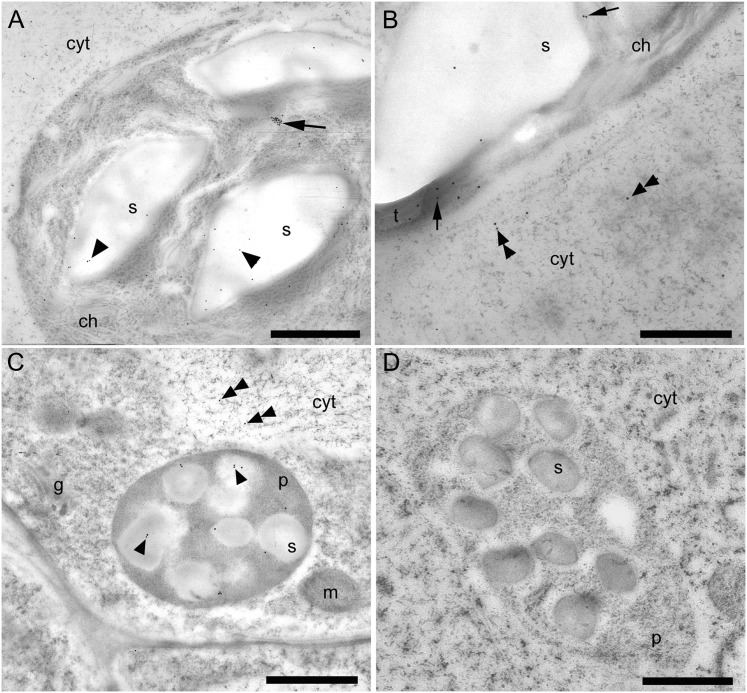
The novel immunolocalization of γECS and hGSHS in legumes entailed sample processing by high-pressure freezing, freeze substitution, and embedding at low temperature. This method optimizes the preservation of protein epitopes in leaves and nodules, thus allowing for a more precise immunolocalization (Rubio et al., 2009). For γECS immunolocalization, two representative crop legumes (common bean and alfalfa) and two model legume species (S. rostrata and L. japonicus) were selected. The tropical legume S. rostrata was also included in this study because it is a model for stem nodulation and the immunolocalization of γECS in the photosynthetic stem nodules was of interest in relation to O2 regulation (James et al., 1996). All three typical plant organs (roots, nodules, and leaves) were examined in detail for most of these species with identical results. Therefore, only a summary of results is presented in Fig. 2. The γECS protein was localized in the amyloplasts of common bean roots (Fig. 2A) and nodules (Fig. 2B). Immunolabelling was also observed in the amyloplasts of S. rostrata root nodules (Fig. 2C) and in the chloroplast thylakoid membranes of stem nodules (Fig. 2D). In alfalfa leaves, γECS was localized to the chloroplasts and much of the labelling was on the starch grains as well as on the thylakoid membranes (Fig. 2E). As a negative control, preimmune serum was used instead of the antibody and in this case no labelling was observed (Fig. 2F). The hGSHS protein was mainly localized on starch grains within chloroplasts of alfalfa leaves (Fig. 3A, B) and plastids of alfalfa roots (Fig. 3C), although there was some sparse labelling within the cytoplasm (Fig. 3A–C). No labelling was detected when the antibody was substituted for preimmune serum (Fig. 3D).
Fig. 2.
Immunogold localization of γ-glutamylcysteine synthetase (γECS) in roots, nodules, and leaves of several legumes. (A) Immunogold-labelled amyloplast (arrow) in a bean root tip, including labelling of starch grains (arrowheads). (B) Immunogold-labelled plastids (arrows) in a bean nodule. (C) Immunogold-labelled plastids (arrows) in a Sesbania rostrata root nodule. (D) Immunogold-labelled chloroplast (arrow) in a S. rostrata stem nodule. (E) Immunogold-labelled chloroplast (arrows) in an alfalfa leaf; note the relatively high-density labelling of the starch grains (arrowheads). (F) Plastids in a bean root tip treated with preimmune serum substituted for the γECS antibody (negative control). cyt, cytoplasm; ch, chloroplast; g, golgi; is, intercellular space; m, mitochondrion; n, nucleus; p, plastid; px, peroxisome; s, starch grain; v, vacuole. Bars, 1 μm.
Fig. 3.
Immunogold localization of homoglutathione synthetase (hGSHS) in leaves (A, B) and roots (C, D) of alfalfa. (A) Immunogold labelling of the interior of a chloroplast, which includes the chloroplast itself (arrow) and the starch grains (arrowheads). (B) Higher magnification of a chloroplast illustrating labelling of some of the thylakoids (arrows) and also sparse labelling in the adjacent cytoplasm (double arrowheads). (C) Amyloplasts in roots showing immunogold labelling of the starch grains (arrowheads) and very sparse labelling in the cytoplasm (double arrowheads). (D) Amyloplast in roots treated with preimmune serum substituted for the hGSHS antibody (negative control). cyt, cytoplasm; ch, chloroplast; g, golgi; m, mitochondrion; p, plastid; s, starch grain; t, thylakoids. Bars, 1 μm (A, C, D) and 500 nm (B).
Transcriptional regulation of thiol synthetases in response to hormones and nitric oxide
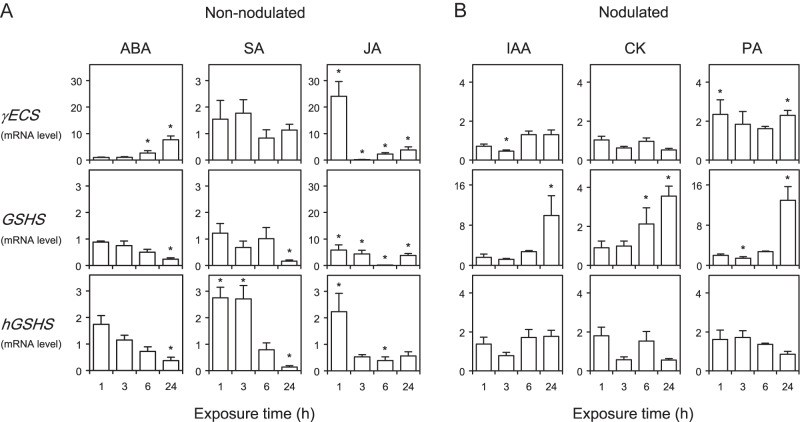
A first type of experiment, aimed at investigating the short-term transcriptional regulation of the thiol biosynthetic pathway, was conducted by exposing L. japonicus plants to hormones and stress-related compounds. This legume species was chosen because its thiol synthetase genes have been characterized and their expression levels determined in various plant tissues (Matamoros et al., 2003). Hormones were provided to plants in the hydroponic medium at a physiologically relevant concentration and the mRNA levels of the three thiol synthetases were quantified in roots, the initial target of hormonal action in the time frame of a few hours. In order to keep these time-course experiments within manageable limits, three hormones (ABA, SA, and JA) were applied to non-nodulated plants (Fig. 4A) and another three hormones (IAA, CK, and PA) to nodulated plants (Fig. 4B). As will be described later, this study was nevertheless complemented with other experiments in which each of the six hormones was provided for 48 h to both non-nodulated and nodulated plants. Initial studies also included GA and ACC, but these compounds were found not to have any meaningful effect on gene expression in roots of non-nodulated plants (data not shown).
Fig. 4.
Time-course patterns of expression of thiol synthetase genes in roots of Lotus japonicus in response to hormones. (A) Non-nodulated plants were supplied with 50 μM of abscisic acid (ABA), jasmonic acid (JA), or salicylic acid (SA) in the rooting medium. (B) Nodulated plants were supplied with 50 μM of indole-3-acetic acid (IAA), cytokinins (CK), or polyamines (PA) in the rooting medium. Steady-state mRNA levels of γ-glutamylcysteine synthetase (γECS), glutathione synthetase (GSHS), and homoglutathione synthetase (hGSHS) were normalized to ubiquitin and expressed relative to those of control plants. These were treated during the same time and with identical concentrations of NaOH (IAA and CK), ethanol (ABA, SA, and PA), or dimethylsulphoxide (JA) to those used to prepare the stock solutions of hormones. The mRNA levels of control plants were given a value of 1. All data are means ± SE of four or five replicates, corresponding to RNA extractions from different roots of two series of plants grown independently (two or three replicates per series). Asterisks denote upregulation (>2-fold) or downregulation (<0.5-fold) of the genes.
The exposure of roots to SA did not affect γECS mRNA levels, slightly upregulated hGSHS after 1–3 h, and strongly downregulated GSHS and hGSHS after 24 h (Fig. 4A). In sharp contrast, JA triggered a coordinated response of the γECS, GSHS, and hGSHS mRNA levels. Thus, JA caused upregulation of the three genes after 1 h of treatment, and this induction was followed by a transient downregulation after 3 or 6 h and by the subsequent recovery of mRNA levels to at least control values after 24 h (Fig. 4A). The hormones ABA and PA are major components of the signalling network for abiotic stress (Bouchereau et al., 1999; Fujita et al., 2006). However, they affected differently the expression of thiol synthetase genes in the roots (Fig. 4). The application of ABA resulted in upregulation of γECS after 6–24 h and in downregulation of GSHS and hGSHS after 24 h. By contrast, exogenous supply of PA to the rooting medium had very minor or no effects on γECS and hGSHS mRNA levels, but strongly activated GSHS after 24 h. In plants, auxins and CK are required, among other functions, for the development of root and shoot meristems (Dello Ioio et al., 2008). In the short-term, the application of IAA and CK increased the GSHS mRNA level by ∼10-fold and 3-fold, respectively, and had virtually no effects on the other two genes (Fig. 4B).
Because several hormones, including ABA and PA, are known to induce NO synthesis (Neill et al., 2003; Tun et al., 2006), the effects of an NO-releasing compound, SNAP, on the mRNA levels of thiol synthetases were examined. The current study found that γECS and GSHS were induced after 3 and 24 h of application of SNAP, whereas the hGSHS mRNA level remained unaffected (Supplementary Fig. S1, available at JXB online).
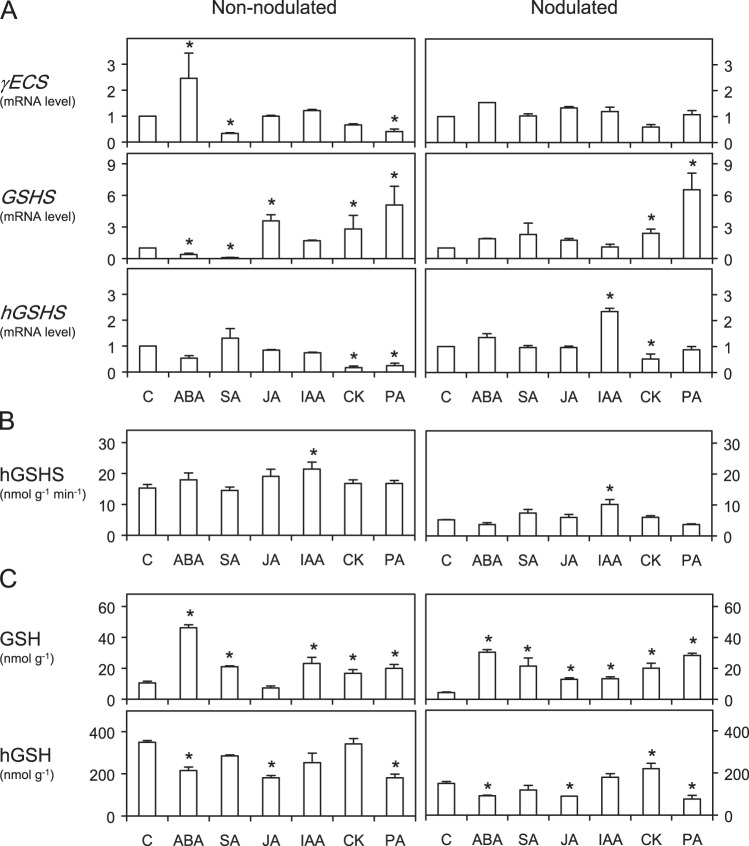
Effect of hormones on thiol synthetase transcripts and activities and on thiol contents of roots
A second type of experiment was performed to study regulatory mechanisms of the thiol biosynthetic pathway. Both non-nodulated and nodulated plants were used for comparison and the exposure time was prolonged to 48 h, so that the effects of hormones on the mRNA levels of thiol synthetase genes (Fig. 5A) could be reflected in the corresponding enzyme activities (Fig. 5B) and thiol contents (Fig. 5C) of the roots. However, the accurate quantification of GSH in roots required the use of HPLC-MS because GSH accounted for only ∼3% of the total thiol tripeptides for both non-nodulated and nodulated plants. Furthermore, the extremely low GSHS levels precluded a reliable assessment of the effects of hormones on this enzyme activity in the roots. The HPLC-MS method also served to confirm hGSH values obtained using HPLC-fluorescence. Both sets of data showed a high correlation (r2>0.90, n=40–60) and therefore, for simplicity, only the hGSH contents obtained by HPLC-fluorescence are presented in Fig. 5.
Fig. 5.
Effects of hormones on the mRNA levels of thiol synthetase genes (A), homoglutathione synthetase (hGSHS) activity (B), and thiol contents (C) in roots of Lotus japonicus. No glutathione synthetase (GSHS) activity could be detected in any of the root extracts. Non-nodulated and nodulated plants were supplied for 48 h with 50 μM of hormones in the rooting medium. Steady-state mRNA levels of γ-glutamylcysteine synthetase (γECS), glutathione synthetase (GSHS), and homoglutathione synthetase (hGSHS) were normalized to ubiquitin mRNA levels and expressed relative to those of control plants (C). These were treated for 48 h with identical concentrations of NaOH (IAA and CK), ethanol (ABA, SA, and PA), or dimethylsulphoxide (JA) to those used to prepare the stock solutions of hormones. The mRNA levels of control plants were given a value of 1. Data of mRNA levels are means ± SE of four or five replicates, corresponding to RNA extractions of different roots from two series of plants grown independently (two or three replicates per series). Asterisks denote upregulation (>2-fold) or downregulation (<0.5-fold) of the genes. Values of thiol contents and enzyme activity of control plants were obtained from roots harvested immediately before the hormone treatments. Data of thiol contents and enzyme activity are means ± SE of four or six replicates, corresponding to extractions of different roots from two series of plants grown independently (two or three replicates per series). Asterisks denote that the means of the hormone treatments are significantly different from the control at P < 0.05 based on Student’s t-test.
The γECS mRNA level did not change after application of most hormonal treatments for 48 h. This gene was only slightly upregulated with ABA and downregulated with SA and PA in non-nodulated plants (Fig. 5A). By contrast, the expression of GSHS was markedly affected, particularly in non-nodulated plants. Notably, this gene was activated by CK and PA in both non-nodulated and nodulated plants, and was upregulated by JA and downregulated by ABA and SA in non-nodulated plants (Fig. 5A). The response of hGSHS was quite different, showing downregulation with CK and PA in non-nodulated plants and activation by IAA and downregulation by CK in nodulated plants (Fig. 5A). In roots of non-nodulated plants, the decreases of hGSHS mRNA levels with CK or PA were not accompanied by lower hGSHS activities (Fig. 5B). The same occurred in roots of nodulated plants treated with CK. In both types of plants exposed to ABA, JA, or PA, there was a decrease in hGSH content despite no detectable variation in hGSHS activity, suggesting consumption or mobilization of the thiol in the roots (Fig. 5B, C).
Discussion
The subcellular localization of the GSH and hGSH biosynthetic pathway is an important aspect of thiol metabolism because these thiol tripeptides have multiple crucial functions and compartmentation of the enzymes would afford additional regulatory mechanisms in plants under physiological or stressful conditions (Bergmann and Rennenberg, 1993). The greater accuracy of immunolocalization has enabled the current study to clarify the previous contradictory reports on thiol localization. Early studies based on enzyme activity assays in isolated organelles led the authors to conclude that γECS, GSHS, and hGSHS are located in the plastids and cytosol (Klapheck et al., 1988; Hell and Bergmann, 1990). Further work using reporter gene fusions and immunocytochemistry in Arabidopsis and Indian mustard, two members of the Brassicaceae family, indicated that γECS is confined to the plastids (Wachter et al., 2005; Pasternak et al., 2008). The immunogold labelling data presented here reveal that γECS is limited to plastids with no cytosolic localization (Fig. 2). Interestingly, gold particles marking the presence of the γECS (Fig. 2A, E) and hGSHS (Fig. 3A, C) proteins were relatively abundant on the starch grains within the leaf chloroplasts and root amyloplasts. This localization strongly suggests a connection between (h)GSH biosynthesis and regulation of starch metabolism, possibly involving changes in redox-sensitive steps, and it is consistent with a proteomic study in which two enzymes involved in thiol synthesis, Cys synthase [O-acetylserine(thiol)lyase] and γECS, were detected in amyloplasts of wheat (Triticum aestivum) endosperm (Balmer et al., 2006). A link between thiols and starch metabolism is also supported by the immunolocalization of glutathione peroxidase on starch grains in leaf chloroplasts and root and nodule plastids of L. japonicus and S. rostrata (Ramos et al., 2009).
In this work, the hGSHS protein was found in the chloroplasts and root proplastids with lower amounts in the cytosol (Fig. 3), whereas in previous studies most or all hGSHS activity was detected in the cytosol (Klapheck et al., 1988; Moran et al., 2000). Because hGSHS is encoded by a single gene in legumes (Frendo et al., 2001; Matamoros et al., 2003), the cytosolic and plastidic isoforms derive from the same gene. In fact, both GSHS and hGSHS of L. japonicus contain sequences encoding potentially plastid transit peptides (Matamoros et al., 2003). Therefore, the current results show that the final step of GSH and hGSH biosynthesis in legumes occurs in the plastids and cytosol, and that in both cases γ-glutamylcysteine is provided as substrate for GSHS and hGSHS by the chloroplasts in leaves and by the proplastids and amyloplasts in roots and nodules.
Two types of experiments were performed to examine in detail the regulatory mechanisms of thiol synthesis in response to hormones. As far as is known, such mechanisms have been investigated until now only for JA (Xiang and Oliver, 1998) and SA (Pucciariello et al., 2009), probably because these compounds as well as GSH metabolism are directly associated with plant defence (Wingate et al., 1988; Beckers and Spoel, 2006; Fujita et al., 2006; Balbi and Devoto, 2008). However, previous reports have employed experimental approaches and plant systems different from those used here.
The strong upregulation of γECS and GSHS after exposure of non-nodulated plants of L. japonicus to JA for only 1 h (Fig. 4A) is fully consistent with the coordinated and rapid response of these genes to JA in Arabidopsis grown in soil or liquid cultures (Xiang and Oliver, 1998). This initial gene activation by JA in both model plants might be related to a function of GSH in their responses to biotic stress, as this thiol rapidly induces transcription of typical defence genes, such as phenylalanine ammonia-lyase and chalcone synthase (Wingate et al., 1988). On the other hand, Pucciariello et al. (2009), using roots of M. truncatula deficient in (h)GSH, concluded that the thiol concentration modulates the SA-signalling pathway. In the present paper, it is shown that SA regulates, in turn, thiol biosynthesis. The antagonistic effects of SA and JA on the GSHS mRNA levels of non-nodulated roots of L. japonicus are obvious after 24–48 h, with an almost complete disappearance of the transcript after 48 h of SA treatment (Figs. 4A and 5A). Notably, these effects on mRNA levels were not accompanied by corresponding changes in the GSH content, which was even enhanced in the case of SA (Fig. 5C). Therefore, a post-transcriptional activation of GSHS and/or mobilization of GSH from leaves to roots may occur in plants after exogenous supply of SA. The post-transcriptional regulation of GSHS activity would provide a second controlling step of thiol biosynthesis, as the γECS activity of Arabidopsis is known to be regulated by redox changes of key Cys residues (Hicks et al., 2007; Gromes et al., 2008). By contrast, SA or JA did not affect hGSHS mRNA or activity levels (Fig. 5A, B), indicating a completely independent regulation of GSHS and hGSHS and, probably, of the enzyme activities.
This differential regulation was further underscored by another novel observation. The application of IAA for 24 h to nodulated plants caused a strong activation of GSHS but had no effect on the other two genes (Fig. 4B). After 48 h of treatment, IAA was the only hormone eliciting changes in hGSHS activity in the roots of both non-nodulated and nodulated plants (Fig. 5B). This auxin also caused induction of hGSHS in nodulated plants but no change in the hGSH content (Fig. 5C). These results strongly suggest that GSH and hGSH are not functionally equivalent and that, at least in nodulated plants, the regulation of hGSHS activity by auxin occurs at the transcriptional level.
Interestingly, two ‘classic’ hormones that play key roles in cell division, namely CK (Dello Ioio et al., 2008) and PA (Bouchereau et al., 1999; Theiss et al., 2002), upregulated GSHS after 24–48 h but either did not change or decreased hGSHS mRNA levels (Figs. 4B and 5A), lending credence to a specific function of GSH in this process. Furthermore, the activation of GSHS was accompanied by an increase of GSH in the roots, whereas hGSH remained almost unchanged with CK and decreased with PA (Fig. 5C). The different responses of GSHS and hGSHS cannot be interpreted in terms of a functional compensation between the corresponding thiols because the concentration of GSH in roots and leaves is far too low compared to that of hGSH. The expression pattern of thiol synthetases of roots of nodulated plants supplied for 24 h with PA (Fig. 4B) was similar to that elicited with NO donors after 3–24 h (Supplementary Fig. S1), namely, transcriptional activation of γECS and GSHS but not of hGSHS. This suggests that NO is mediating the effects of PA, consistent with recent reports showing that PA may directly or indirectly regulate NO synthesis (Tun et al., 2006; Yamasaki and Cohen, 2006). These results are in keeping with those reported for M. truncatula roots treated with the NO donors sodium nitroprusside and nitrosoglutathione (Innocenti et al., 2007), and indicate that the differential regulation of GSHS and hGSHS by NO is probably widespread in legumes. The contrasting response of the two genes to NO, which is a crucial signalling molecule in plants and other organisms, is probably biologically relevant and further supports the hypothesis that GSHS and hGSHS play different roles in legumes. This is somewhat surprising taking into account that GSHS and hGSHS display high sequence homology and have originated by duplication in both M. truncatula (Frendo et al., 2001) and L. japonicus (Matamoros et al., 2003).
Contrary to the positive relationship between GSHS transcript levels and GSH contents observed with CK and PA, the two parameters were inversely related in non-nodulated plants treated with ABA (Fig. 5A, C). The increase of GSH in plants following exposure to ABA is difficult to explain in the absence of measurements of γECS and GSHS activities, which could not be assayed because of the lability and low abundance of the enzymes. Another complicating factor was the possible post-translational regulation of γECS by the redox environment (Hicks et al., 2007; Gromes et al., 2008), which may vary with the hormonal treatment. However, the fact that GSH was also increased in nodulated plants with ABA, without any change in γECS and GSHS mRNA levels, indicates a post-transcriptional control of the enzyme activities and/or mobilization or lower consumption of GSH in these plants, as mentioned for SA.
In conclusion, the results demonstrate the existence of subcellular compartmentation of the thiol biosynthetic pathway in legume leaves, roots, and nodules. They also reveal a selective regulation of the three thiol synthetase genes by hormones and NO. Notably, GSHS and hGSHS are differentially regulated by most hormones examined. The contrasting response of the two genes and the two thiols to hormones suggests distinct functions for GSH and hGSH in cell division, organ development, and stress signalling. Moreover, with a few exceptions, the response profiles of the GSH and hGSH contents to the various hormonal treatments are similar in the roots of non-nodulated and nodulated plants, indicating that thiol homeostasis is independent of the nitrogen source.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Effect of NO on the expression of γ-glutamylcysteine synthetase (γECS), glutathione synthetase (GSHS), and homoglutathione synthetase (hGSHS) genes in roots of Lotus japonicus.
Acknowledgments
The authors are grateful to Frank Minchin and two anonymous reviewers for constructive criticism and helpful comments on the manuscript. Marina Gay and Joaquín Navascués were recipients of postdoctoral contracts of the Junta para la Ampliación de Estudios (JAE-Doc) programme, funded by Consejo Superior de Investigaciones Científicas-Fondo Social Europeo.
This work was funded by Ministerio de Ciencia e Innovación-Fondo Europeo de Desarrollo Regional (AGL2008-01298 and AGL2011-24524) and Gobierno de Aragón (group A53).
References
- Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. The New Phytologist. 2008;177:301–318. doi: 10.1111/j.1469-8137.2007.02292.x. [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, DuPont FM, Buchanan BB, Hurkman WJ. Proteome of amyloplasts isolated from developing wheat endosperm presents evidence of broad metabolic capability. Journal of Experimental Botany. 2006;57:1591–1602. doi: 10.1093/jxb/erj156. [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Spoel SH. Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biology. 2006;8:1–10. doi: 10.1055/s-2005-872705. [DOI] [PubMed] [Google Scholar]
- Bergmann L, Rennenberg H. 1993. Glutathione metabolism in plants. In: LJ De Kok, I Stulen, H Rennenberg, C Brunold, WE Rauser, eds, Sulfur nutrition and assimilation in higher plants. The Netherlands: SPB Academic Publishing, 109–123. [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. Hairy roots of Medicago truncatula as tools for studying nitrogen-fixing and endomycorrhizal symbioses. Molecular Plant–Microbe Interactions. 2001;14:693–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- Bouchereau A, Aziz A, Larher F, Martin-Tanguy J. Polyamines and environmental challenges: recent development. Plant Science. 1999;140:103–125. [Google Scholar]
- Broughton WJ, Dilworth MJ. Control of leghaemoglobin synthesis in snake beans. The Biochemical Journal. 1971;125:1075–1080. doi: 10.1042/bj1251075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Scaglia Linhares F, Sabatini S. Emerging role of cytokinin as a regulator of cellular differentiation. Current Opinion in Plant Biology. 2008;11:23–27. doi: 10.1016/j.pbi.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Ding Y, Kalo P, Yendrek C, Sun J, Liang Y, Marsh JF, Harris JM, Oldroyd GED. Abscisic acid coordinates Nod factor and cytokinin signaling during the regulation of nodulation in. Medicago truncatula. The Plant Cell. 2008;20:2681–2695. doi: 10.1105/tpc.108.061739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo P, Hernández-Jiménez MJ, Mathieu C, Duret L, Gallesi D, Van de Sype G, Hérouart D, Puppo A. A Medicago truncatula homoglutathione synthetase is derived from glutathione synthetase by gene duplication. Plant Physiology. 2001;126:1706–1715. doi: 10.1104/pp.126.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signalling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- González-Rizzo S, Crespi M, Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with. Sinorhizobium meliloti. The Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromes R, Hothorn M, Lenherr ED, Rybin V, Scheffzek K, Rausch T. The redox switch of γ-glutamylcysteine ligase via a reversible monomer-dimer transition is a mechanism unique to plants. The Plant Journal. 2008;54:1063–1075. doi: 10.1111/j.1365-313X.2008.03477.x. [DOI] [PubMed] [Google Scholar]
- Hell R, Bergmann L. Glutathione synthetase in tobacco suspension cultures: catalytic properties and localization. Physiologia Plantarum. 1988;72:70–76. [Google Scholar]
- Hell R, Bergmann L. γ-Glutamylcysteine synthetase in higher plants: catalytic properties and subcellular localization. Planta. 1990;180:603–612. doi: 10.1007/BF02411460. [DOI] [PubMed] [Google Scholar]
- Hicks LM, Cahoon RE, Bonner ER, Rivard RS, Sheffield J, Jez JM. Thiol-based regulation of redox-active glutamate-cysteine ligase from. Arabidopsis thaliana. The Plant Cell. 2007;19:2653–2661. doi: 10.1105/tpc.107.052597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti G, Pucciariello C, Le Gleuher M, Hopkins J, de Stefano M, Delledonne M, Puppo A, Baudouin E, Frendo P. Glutathione synthesis is regulated by nitric oxide in Medicago truncatula roots. Planta. 2007;225:1597–1602. doi: 10.1007/s00425-006-0461-3. [DOI] [PubMed] [Google Scholar]
- Iturbe-Ormaexte I, Heras B, Matamoros MA, Ramos J, Moran JF, Becana M. Cloning and functional characterization of a homoglutathione synthetase from pea nodules. Physiologia Plantarum. 2002;115:69–73. doi: 10.1034/j.1399-3054.2002.1150107.x. [DOI] [PubMed] [Google Scholar]
- James EK, Iannetta PPM, Nixon PJ, Whiston AJ, Peat L, Crawford RMM, Sprent JI, Brewin NJ. Photosystem II and oxygen regulation in Sesbania rostrata stem nodules. Plant, Cell and Environment. 1996;19:895–910. [Google Scholar]
- Jez JM, Cahoon RE. Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana . Journal of Biological Chemistry. 2004;279:42726–42731. doi: 10.1074/jbc.M407961200. [DOI] [PubMed] [Google Scholar]
- Klapheck S, Zopes H, Levels HG, Bergmann L. Properties and localization of the homoglutathione synthetase from Phaseolus coccineus leaves. Physiologia Plantarum. 1988;74:733–739. [Google Scholar]
- Loscos J, Matamoros MA, Becana M. Ascorbate and homoglutathione metabolism in common bean nodules under stress conditions and during natural senescence. Plant Physiology. 2008;146:1282–1292. doi: 10.1104/pp.107.114066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnicol PM. Homoglutathione and glutathione synthetases of legume seedlings: partial purification and substrate specificity. Plant Science. 1987;53:229–235. [Google Scholar]
- Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M. Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiology. 1999;121:879–888. doi: 10.1104/pp.121.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros MA, Clemente MR, Sato S, Asamizu E, Tabata S, Ramos J, Moran JF, Stiller J, Gresshoff PM, Becana M. Molecular analysis of the pathway for the synthesis of thiol tripeptides in the model legume. Lotus japonicus. Molecular Plant–Microbe Interactions. 2003;16:1039–1046. doi: 10.1094/MPMI.2003.16.11.1039. [DOI] [PubMed] [Google Scholar]
- Maughan S, Foyer CH. Engineering and genetic approaches to modulating glutathione network in plants. Physiologia Plantarum. 2006;126:382–397. [Google Scholar]
- May MJ, Vernoux T, Sánchez-Fernández R, Van Montagu M, Inzé D. Evidence for posttranscriptional activation of γ-glutamylcysteine synthetase during plant stress responses. Proceedings of the National Academy of Sciences, USA. 1998;95:12049–12054. doi: 10.1073/pnas.95.20.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A. Glutathione-ascorbic acid antioxidant system in animals. Journal of Biological Chemistry. 1994;269:9397–9400. [PubMed] [Google Scholar]
- Moran JF, Iturbe-Ormaetxe I, Matamoros MA, Rubio MC, Clemente MR, Brewin NJ, Becana M. Glutathione and homoglutathione synthetases of legume nodules. Cloning, expression, and subcellular localization. Plant Physiology. 2000;124:1381–1392. doi: 10.1104/pp.124.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya L, Ladrera R, Ramos J, González EM, Arrese-Igor C, Minchin FR, Becana M. The response of carbon metabolism and antioxidant defenses of alfalfa nodules to drought stress and to the subsequent recovery of plants. Plant Physiology. 2007;144:1104–1114. doi: 10.1104/pp.107.099648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Hancock JT. Nitric oxide signalling in plants. The New Phytologist. 2003;159:11–35. doi: 10.1046/j.1469-8137.2003.00804.x. [DOI] [PubMed] [Google Scholar]
- Pasternak M, Lim B, Wirtz M, Hell R, Cobbett CS, Meyer AJ. Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. The Plant Journal. 2008;53:999–1012. doi: 10.1111/j.1365-313X.2007.03389.x. [DOI] [PubMed] [Google Scholar]
- Pucciariello C, Innocenti G, Van de Velde W, et al. (Homo)glutathione depletion modulates host gene expression during the symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti . Plant Physiology. 2009;151:1186–1196. doi: 10.1104/pp.109.142034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J, Matamoros MA, Naya L, James EK, Rouhier N, Sato S, Tabata S, Becana M. The glutathione peroxidase gene family of Lotus japonicus: characterization of genomic clones, expression analyses and immunolocalization in legumes. The New Phytologist. 2009;181:103–114. doi: 10.1111/j.1469-8137.2008.02629.x. [DOI] [PubMed] [Google Scholar]
- Riggs P. Expression and purification of recombinant proteins by fusion to maltose-binding protein. Molecular Biotechnology. 2000;15:51–63. doi: 10.1385/MB:15:1:51. [DOI] [PubMed] [Google Scholar]
- Rubio MC, Becana M, Kanematsu S, Ushimaru T, James EK. Immunolocalization of antioxidant enzymes in high-pressure frozen root and stem nodules of. Sesbania rostrata. The New Phytologist. 2009;183:395–407. doi: 10.1111/j.1469-8137.2009.02866.x. [DOI] [PubMed] [Google Scholar]
- Schäfer HJ, Haag-Kerwer A, Rausch T. cDNA cloning and expression analysis of genes encoding GSH synthesis in roots of the heavy-metal accumulator Brassica juncea L.: evidence for Cd-induction of a putative mitochondrial γ-glutamylcysteine synthetase isoform. Plant Molecular Biology. 1998;37:87–97. doi: 10.1023/a:1005929022061. [DOI] [PubMed] [Google Scholar]
- Stacey G, McAlvin CB, Kim SY, Olivares J, Soto MJ. Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and. Medicago truncatula. Plant Physiology. 2006;141:1473–1481. doi: 10.1104/pp.106.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Cardoza V, Mitchell DM, Bright L, Oldroyd G, Harris JM. Crosstalk between jasmonic acid, ethylene and Nod factor signalling allows integration of diverse inputs for regulation of nodulation. The Plant Journal. 2006;46:961–970. doi: 10.1111/j.1365-313X.2006.02751.x. [DOI] [PubMed] [Google Scholar]
- Theiss C, Bohley P, Voigt J. Regulation by polyamines of ornithine decarboxylase activity and cell division in the unicellular green alga. Chlamydomonas reinhardtii. Plant Physiology. 2002;128:1470–1479. doi: 10.1104/pp.010896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- Tominaga A, Nagata M, Futsuki K, et al. Enhanced nodulation and nitrogen fixation in the abscisic acid low-sensitive mutant enhanced nitrogen fixation1 of. Lotus japonicus. Plant Physiology. 2009;151:1965–1976. doi: 10.1104/pp.109.142638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun NN, Santa-Catarina C, Begum T, Silveira V, Handro W, Segal Floh EI, Scherer GFE. Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant and Cell Physiology. 2006;47:346–354. doi: 10.1093/pcp/pci252. [DOI] [PubMed] [Google Scholar]
- Wachter A, Wolf S, Steininger H, Bogs J, Rausch T. Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. The Plant Journal. 2005;41:15–30. doi: 10.1111/j.1365-313X.2004.02269.x. [DOI] [PubMed] [Google Scholar]
- Wild AC, Mulcahy RT. Regulation of γ-glutamylcysteine synthetase subunit gene expression: insights into transcriptional control of antioxidant defenses. Free Radical Research. 2000;32:281–301. doi: 10.1080/10715760000300291. [DOI] [PubMed] [Google Scholar]
- Wingate VPM, Lawton MA, Lamb CJ. Glutathione causes a massive and selective induction of plant defense genes. Plant Physiology. 1988;87:206–210. doi: 10.1104/pp.87.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ. Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in. Arabidopsis. The Plant Cell. 1998;10:1539–1550. doi: 10.1105/tpc.10.9.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Cohen MF. NO signal at the crossroads: polyamine-induced nitric oxide synthesis in plants? Trends in Plant Science. 2006;11:522–524. doi: 10.1016/j.tplants.2006.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.