Abstract
Seed development in Arabidopsis is characterized by stereotypical division patterns, suggesting that coordinated control of cell cycle may be required for correct patterning and growth of the embryo and endosperm. D-type cyclins (CYCD) are key cell cycle regulators with roles in developmental processes, but knowledge regarding their involvement in seed development remains limited. Here, a family-wide gene expression, and loss- and gain-of-function approach was adopted to reveal additional functions for CYCDs in the development of seed tissues. CYCD genes have both discrete and overlapping tissue-specific expression patterns in the seed as revealed by GUS reporter gene expression. Analysis of different mutant combinations revealed that correct CYCD levels are required in seed development. The CYCD3 subgroup is specifically required as its loss caused delayed development, whereas overexpression in the embryo and endosperm of CYCD3;1 or a previously uncharacterized gene, CYCD7;1, variously leads to induced proliferation, abnormal phenotypes, and elevated seed abortion. CYCD3;1 overexpression provoked a delay in embryonic developmental progression and abnormalities including additional divisions of the hypophysis and suspensor, regions where CYCD3 genes are normally expressed, but did not affect endosperm development. Overexpression of CYCD7;1, not normally expressed in seed development, promoted overgrowth of both embryo and endosperm through increased division and cell enlargement. In contrast to post-germination growth, where pattern and organ size is not generally related to division, results suggest that a close control of cell division through regulation of CYCD activity is important during seed development in conferring both developmental rate and correct patterning.
Keywords: Cell cycle, cell division, cyclin D, embryo and endosperm development, embryo patterning, seed
Introduction
Seed development constitutes the first growth phase of angiosperms in which double-fertilization triggers the formation of the embryo and its nourishing tissue, the endosperm. In Arabidopsis, both tissues are formed by a series of highly invariant nuclear and cell division events that are coordinated with cell differentiation and patterning processes to ensure correct growth and morphogenesis (Nawy et al., 2008; Sabelli and Larkins, 2009; Sun et al., 2010).
Despite the increasing knowledge regarding cell type specification in the seed, how cell divisions are regulated and integrated with patterning processes remains largely unresolved. The cell cycle is controlled by cyclin-dependent kinases (CDK) that require a positive regulatory subunit called cyclin for activity (Nieuwland et al., 2009a). The D-type cyclins (CYCD) are conserved between plants and animals and are responsible for triggering the G1/S transition by activating the CYCD-RBR-E2F pathway primarily through their association with the A-type CDK (CDKA) in response to intrinsic and extrinsic signals (Riou-Khamlichi et al., 2000; Dewitte et al., 2007). The primary target of CDK-CYCD complexes is the retinoblastoma-related protein (RBR), which upon phosphorylation leads to the dissociation of E2F transcription factors and the expression of genes required for S-phase entry (Sherr and Roberts, 2004; Gruissem, 2007). Precise regulation of the cell cycle and patterns of cell division are vital for normal embryo development, as evidenced from the severe morphological defects observed in embryos expressing a dominant-negative form of CDKA (Hemerly et al., 2000), an antisense cyclinA3;2 (Yu et al., 2003), or in plants that carry mutations in DNA polymerase ϵ (Jenik et al., 2005).
Arabidopsis has 10 CYCD genes that are classified into six or seven subgroups (Vandepoele et al., 2002; Menges et al., 2007). Previous work have revealed both distinct and functionally redundant roles for CYCDs during post-embryonic development including germination, leaf growth, and stomata and lateral root formation (Masubelele et al., 2005; Dewitte et al., 2007; Kono et al., 2007; Nieuwland et al., 2009b; Sanz et al., 2011). In particular, the CYCD3 gene family has been shown to regulate the contributory cell number through controlling the length of the mitotic window in aerial organs, as well as having a key role in mediating cytokinin responses (Riou-Khamlichi et al., 1999; Dewitte et al., 2007). Recently, CYCD6;1 was shown to lie downstream of the SHORTROOT (SHR) transcription factor in a pathway regulating a formative cell division in the embryonic ground tissue (Sozzani et al., 2010). Partial characterization of CYCD3;2 and CYCD4;1 expression have revealed that both are active in the fertilized ovule and embryo (De Veylder et al., 1999; Swaminathan et al., 2000). Therefore, CYCDs are prime candidates for playing important roles in integrating cell division and patterning processes during embryo and endosperm development.
This study investigates the roles for CYCDs in Arabidopsis seed development and reveals that CYCDs have both distinct and overlapping functions in the formation of seed tissues. A rate-limiting requirement for CYCD3 genes in the normal rate of progression through embryo development was observed. However, ectopic expression of CYCD genes in either embryo or endosperm did not accelerate normal development, but rather induced developmental abnormalities. It is concluded that correct coordination of division processes is required for normal developmental patterning.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia was used as the wild type. Promoter reporter (β-glucuronidase; GUS) gene transgenic lines pCYCD1;1:GUS, pCYCD2;1:GUS, pCYCD3;1:GUS, pCYCD3;2:GUS, pCYCD3;3:GUS, pCYCD4:1:GUS, and pCYCD6;1:GUS were constructed as described (Cockcroft, 1998; Masubelele et al., 2005; Dewitte et al., 2007; Sozzani et al., 2010). The pCYCB1;1:CYCB1;1DB-GUS line was constructed as described (Colón-Carmona et al., 1999). The loss-of-function insertion mutant lines cycd1;1, cycd2;1cycd4;1, cycd3;1cycd3;2cycd3;3, and cycd6;1 were as described (Masubelele et al., 2005; Dewitte et al., 2007; Sozzani et al., 2010). The cycd7;1 loss-of-function insertion mutant is from the INRA-Versailles collection (FLAG 498H08). All mutant lines have been confirmed as representing null alleles. The ACT pRPS5A:GAL4 and EF pUAS:GFP-GUS-intron lines have been described previously (Weijers et al., 2003).
Plasmid construction
All promoter reporter gene plasmids were constructed using the Gateway system (Invitrogen). Promoters for CYCD4;2 (2538 bp), CYCD5;1 (2440 bp), and CYCD7;1 (2549 bp) were amplified using the following primer pairs: pCYCD4;2, 5′-CACCATGTCTCATTCTGTTTC-3′ and 5′-TTGTAGCTTTCTTTCGATCTATAC-3′ pCYCD5;1, 5′-CACCTGGTCCCTCATCTTGACT-3′ and 5′-GCGGCGGAGATAGAAGTGTT-3′; pCYCD7;1, 5′-CACCACTCTTCTTGTTCTTCCTTGTAG-3′ and 5′-TAAGGTATTCTACTCCTCACTCTCGG-3′. Fragments were subcloned into pKGWFS7 (Karimi et al., 2002). For GAL4 under control of FWA promoter (Kinoshita et al., 2004), the GAL4 coding sequence was amplified from pBIN Gal4–mGFP5er (Haseloff, 1999) using primers that added BamHI and BglII sites at the 5′ end of the start codon and the 3′ end of the stop codon, respectively: 5′-GGATCCATGAAGCTCCTGTC-3′ and 5′-AGATCTACCCACCGTACTCG-3′. The fragment was cloned into pCR2.1-TOPO (Invitrogen) giving pCR2.1-GAL4. All overexpression plasmids for use in the mGAL4:VP16/UAS two-component gene expression system (Haseloff, 1999) were constructed using conventional DNA cloning. For GAL4 under control of pFWA, GAL4 was isolated as a ∼700-bp BamHI/BglII fragment from pCR2.1-GAL4 and subcloned into BamHI-digested pBCH2-PFWA:ΔFWA-GFP (Kinoshita et al., 2004) giving pBCH2-PFWA:GAL4. For CYCD3;1 (plus eGFP-GUS-intron (GGi)) under control of pUAS, CYCD3;1 was isolated as a ∼1220-bp BamHI/SacI fragment (CYCD3;1 cDNA plus a Cab22L leader sequence) from pUD3.1 and subcloned into BamHI/SacI-digested pSDM7021 (Weijers et al., 2003) giving pSDM-D3.1. Next, the GGi cassette was isolated as a ∼3100-bp EcoRI fragment from pSDM7021 and subcloned into EcoRI-digested pSDM-D3.1 giving pSDM-D3.1-GGi. For the CYCD7;1 construct, CYCD7;1 cDNA (∼1030 bp) was amplified from floral tissue using primers that added BamHI and SacI sites at the 5′ end of the start codon and the 3′ end of the stop codon, respectively: 5′-GGATCCATGGATAATCTACTCTG-3′ and 5′-GAGCTCCTAAATGTAATTTGACAT-3′. The remaining steps were as described for CYCD3;1.
Plant transformation and selection of lines
Recombinant constructs were introduced into wild-type Arabidopsis by floral dipping (Clough and Bent, 1998). Single-insert homozygous T3 lines were isolated by screening on MS media containing either kanamycin (50 μg/ml) or phosphinothricin (20 μg/ml). All transgenic lines generated in this study underwent normal plant development and so were considered suitable for analysis. To preselect high-expressing pFWA:GAL4 lines, at least six independently transformed lines were compared for GAL4 transcript levels in siliques at 3 days after pollination (dap) by quantitative real-time reverse-transcriptase PCR (QRT-PCR). To preselect high-expressing pUAS:CYCD3;1 and pUAS:CYCD7;1 lines, at least six independently transformed plants were crossed with the pFWA:GAL4 line and compared for GUS activity during endosperm development.
Quantitative real-time reverse transcriptase PCR
Relative transcript abundance was measured in siliques at 3 dap using QRT-PCR as described (Dewitte et al., 2003). The following primer pairs were used: CYCD3;1, 5′-GCAAGTTGATCCCTTTGACC-3′ and 5′-CAGCTTGGACTGTTCAACGA-3′; CYCD7;1, 5′-GATCCATGGATAATCTACTCTG-3′ and 5′-GAGCTCCTAAATGTAATTTGACAT-3′; GAL4, 5′-GGATCCATGAAGCTCCTGTC-3′ and 5′-AGATCTACCCACCGTACTCG-3′; ACTIN, 5′-GAAGAACTATGAATTACCCGATGGGC-3′ and 5′-CCCGGGTTAGAAACATTTTCTGTGAACG-3′.
Microscopy and histology
Histochemical staining for GUS activity was performed as described (Jefferson et al., 1987). Stained and unstained seeds were cleared and mounted prior to microscopic examination as described (Stangeland and Salehian, 2002). Whole-mount preparations of seeds were examined and photographed using a OPTIPHOT-2 microscope (Nikon, Tokyo, Japan) equipped with differential interference contrast optics and a digital camera.
Phenotypic analyses
Developmental progression was performed by recording the percentage of seeds at each embryo stage (Jürgens and Mayer, 1994) at 2, 3, 4, 5, 7, and 9 dap. Seeds were staged by hand-pollination and approximately 150 seeds were scored for each line at each time point. All lines were assessed for frequency of aborted seed in siliques at 10 dap (n > 360). For analyses of CYCD3;1 and CYCD7;1 overexpression, homozygous pUAS:CYCD3;1 and pUAS:CYCD7;1 lines were crossed to pRPS5A:GAL4 (Weijers et al., 2003) and pFWA:GAL4 using the latter two as female parents. All lines were scored for the presence of abnormal seed morphological characteristics.
Analysis of expression patterns in Genevestigator
The Genevestigator V3 microarray expression database (Hruz et al., 2008; www.genevestigator.com) was used (with Anatomy tool) to extract the relative expression levels of the CYCD genes across embryo, suspensor, endosperm (combined analysis), peripheral endosperm, chalazal endosperm, and micropylar endosperm tissues. Results were displayed using a log2 scale and each expression value represents the average expression level over a set of tissues from combined microarray experiments.
Results
Arabidopsis CYCD genes are differentially expressed during seed development
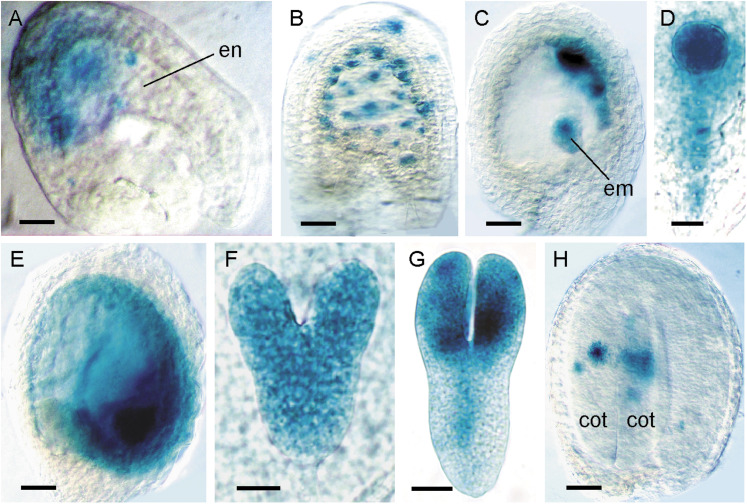
The Arabidopsis CYCD family comprise 10 members that group into six or seven clades (Vandepoele et al., 2002; Menges et al., 2007). To examine the expression of CYCDs during seed development, transgenic plants were analysed that express the β-glucuronidase (GUS) reporter gene under control of each CYCD promoter. For each construct, at least 10 independent lines were compared for GUS activity and, with few exceptions, all showed consistent and reproducible patterns of expression. To correlate CYCD expression and mitotic cell cycle activity throughout seed development, the CYCD reporters were compared with patterns of expression revealed using the mitotic cell division reporter pCYCB1;1:CYCB1;1DB-GUS (Colón-Carmona et al., 1999). Uniform activity of the CYCB1;1 reporter was seen during the first nuclear divisions in the early syncytial endosperm of fertilized ovules (Fig. 1A), which persisted until late syncytial endosperm stages (Fig. 1B), gradually becoming restricted to mitotic domains corresponding to the peripheral and micropylar endosperm (Fig. 1C). Throughout early embryo development, GUS staining was uniform from the one-cell stage up until the globular stage with activity also visible in the suspensor (Fig. 1D). During the onset of endosperm cellularization around the heart stage of embryogenesis, CYCB1;1 activity was observed in the endosperm (Fig. 1E), with uniform staining in mitotically active heart-stage embryos (Fig. 1F), which became localized to the cotyledons, shoot apex, and provasculature in torpedo-stage embryos (Fig. 1G). In mature seeds, GUS activity was restricted to infrequent divisions of the embryo, with no visible activity in the endosperm, corresponding to the cessation of mitotic activity in the seed and the transition to seed maturation and dormancy (Fig. 1H). These results demonstrate that the CYCB1;1 reporter is an ideal marker of mitotic proliferation during seed development.
Fig. 1.
Expression analysis of pCYCB1;1:CYCB1;1DB-GUS during seed development. In all panels, seeds are oriented with the chalazal pole to the left and the micropylar pole to the right. (A) Fertilized ovule with GUS activity localized to dividing nuclei of an early syncytial endosperm (en). (B) Late syncytial endosperm-stage seed containing globular-stage embryo with GUS staining in dividing endosperm nuclei and integuments. (C) Late syncytial-stage seed with expression in the peripheral endosperm domain and embryo (em). (D) Globular-stage embryo with uniform GUS activity in the embryo proper and suspensor. (E) Early cellularized endosperm-stage seed containing heart-stage embryo with strong expression in micropylar endosperm and embryo. (F) Uniform GUS expression in a heart-stage embryo. (G) Torpedo-stage embryo with GUS activity in the cotyledons, shoot apex and provascular tissue. (H) Mature embryo seed stage with GUS staining restricted to dividing cells of the cotyledons (cot). Bars = 20 μm (A, F), 50 μm (B, C), 12 μm (D), 100 μm (E, H), 25 μm (G).
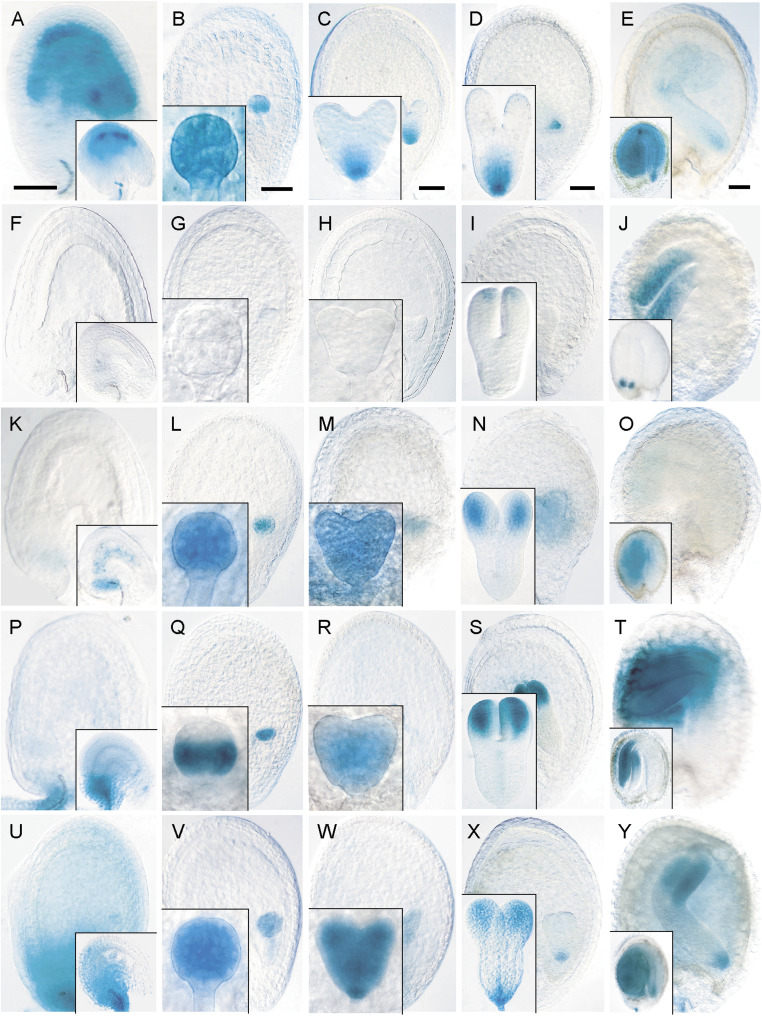
pCYCD1;1:GUS activity was restricted to the innermost integument layer in young seeds (Fig. 2A), whereas in early embryos, expression was uniform up to the globular stage (Fig. 2B), which became restricted to the incipient quiescent centre by the heart stage through to the mature stage (Fig. 2C–E). Global staining was observed in mature embryos, with particularly strong expression in meristems and cotyledons (Fig. 2E). pCYCD2;1:GUS was never detected in the endosperm (Fig. 2F–J), whereas staining was visible in cotyledons after the torpedo stage (Fig. 2I, J). pCYCD3;1:GUS expression was observed in the early endosperm and in the group of transfer cells in the chalazal phloem-unloading domain (Fig. 2K). In the embryo, expression was uniform up until the heart stage, becoming restricted to the cotyledons and shoot apical meristem in mature embryos (Fig. 2L–O). Weak expression was noted in the suspensor up until the heart stage (Fig. 2K–M).
Fig. 2.
Expression analysis of CYCD1;1 to CYCD3;3 during seed development: localization of GUS expression for pCYCD1;1:GUS (A–E), pCYCD2;1:GUS (F–J), pCYCD3;1:GUS (K–O), pCYCD3;2:GUS (P–T), and pCYCD3;3:GUS (U–Y). (A, F, K, P, U) Fertilized ovule (inset) to octant embryo (early syncytial endosperm) seed stage. (B, G, L, Q, V) Late syncytial endosperm stage containing globular-stage embryo (inset). (C, H, M, R, W) Early cellularized endosperm stage containing heart-stage embryo (inset). (D, I, N, S, X) Late cellularized endosperm stage containing torpedo-stage embryo (inset). (E, J, O, T, Y) Bent cotyledon to mature embryo (inset) seed stage. In all panels, seeds are oriented with the chalazal pole to the left and the micropylar pole to the right. Bars = 50 μm.
pCYCD3;2:GUS and pCYCD3;3:GUS had overlapping expression patterns in the seed. Both lacked activity in the endosperm (Fig. 2P–Y) but were uniformly active in the embryo before the globular stage, after which pCYCD3;2:GUS was restricted to the central and basal domains, with strong staining in the ground tissue, whereas pCYCD3;3:GUS remained uniformly expressed (Fig. 2Q, V). Both were expressed throughout heart-stage embryos with strong activity in the provasculature and root and shoot apices (Fig. 2R, W). In torpedo-stage embryos, both were expressed in provasculature and in the upper stem-cell tier of the root meristem with stronger staining throughout the cotyledons, although pCYCD3;3:GUS showed additional activity in the basal root pole (Fig. 2S, X). The patterns of expression persisted into the mature embryo stage (Fig. 2T, Y). Both lines showed weak activity in the suspensor. The expression patterns observed for all CYCD3 genes in the embryo showed considerable overlap with those obtained with the CYCB1;1 reporter (compare Fig. 1D, F–H with Fig. 2L–O, Q–T, V–Y).
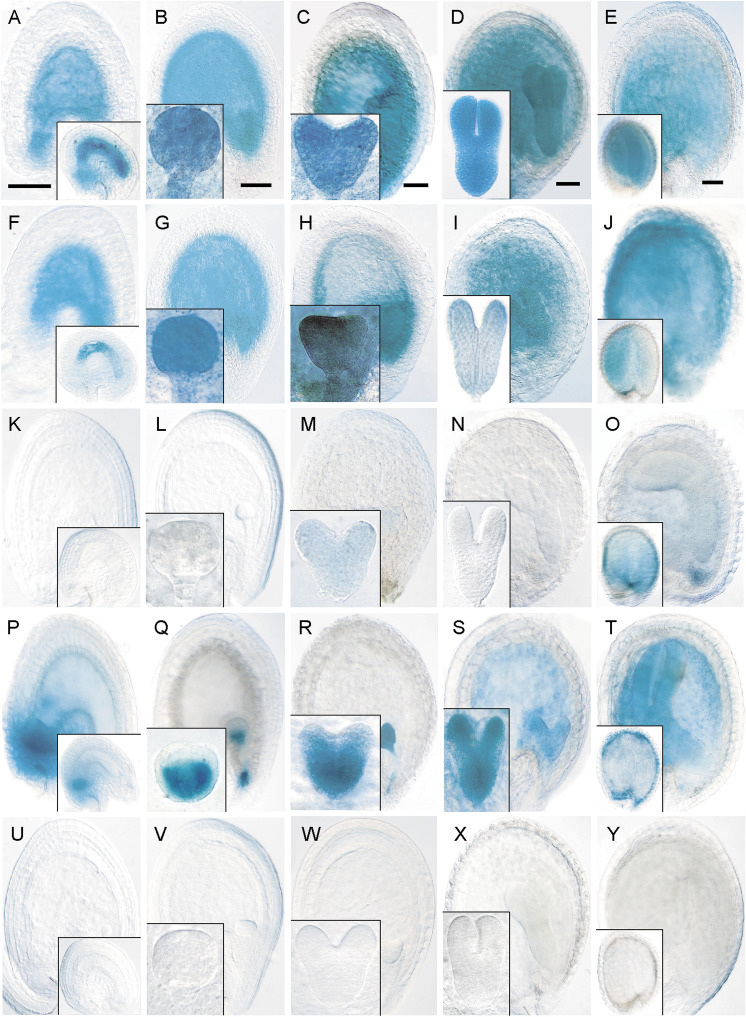
pCYCD4;1:GUS and pCYCD4;2:GUS had overlapping expression patterns with persistent activity throughout proliferative phases in the endosperm and suspensor, and in the phloem-unloading domain and chalazal proliferating tissue (Fig. 3A–J). Both were uniformly expressed in the embryo until the mature stage, after which staining gradually disappeared, starting in the root pole and ending in cotyledons (Fig. 3A–J). The expression patterns observed for both CYCD4 genes in the seed showed striking similarities with those of the CYCB1;1 reporter (compare Fig. 1D, F–H with Fig. 3B–E, G–J). pCYCD5;1:GUS showed only transient activity in heart-stage embryos and endosperm in mature seeds (Fig. 3K–O). pCYCD6;1:GUS was expressed in the endosperm after cellularization and during remaining seed stages with staining also in the chalazal proliferating tissue (Fig. 3P–T). pCYCD6;1:GUS expression was uniform in the early embryo and suspensor, which became more restricted in the ground tissue layer by the globular stage (Fig. 3Q). In heart- and torpedo-stage embryos, staining was strong in the provasculature and cotyledons (Fig. 3R, S) which declined in mature embryos (Fig. 3T). In contrast, pCYCD7;1:GUS expression was never detected in seeds (Fig. 3U–Y). The expression patterns observed for all CYCDs during endosperm, embryo, and peripheral seed tissue development using GUS reporters are summarized in Figs. 4 and 5.
Fig. 3.
Expression analysis of CYCD4;1 to CYCD7;1 during seed development: localization of GUS expression for pCYCD4;1:GUS (A–E), pCYCD4;2:GUS (F–J), pCYCD5;1:GUS (K–O), pCYCD6;1:GUS (P–T), and pCYCD7;1:GUS (U–Y). (A, F, K, P, U) Fertilized ovule (inset) to octant embryo (early syncytial endosperm) seed stage. (B, G, L, Q, V) Late syncytial endosperm stage containing globular-stage embryo (inset). (C, H, M, R, W) Early cellularized endosperm stage containing heart-stage embryo (inset). (D, I, N, S, X) Late cellularized endosperm stage containing torpedo-stage embryo (inset). (E, J, O, T, Y) Bent cotyledon to mature embryo (inset) seed stage. In all panels, seeds are oriented with the chalazal pole to the left and the micropylar pole to the right. Bars = 50 μm.
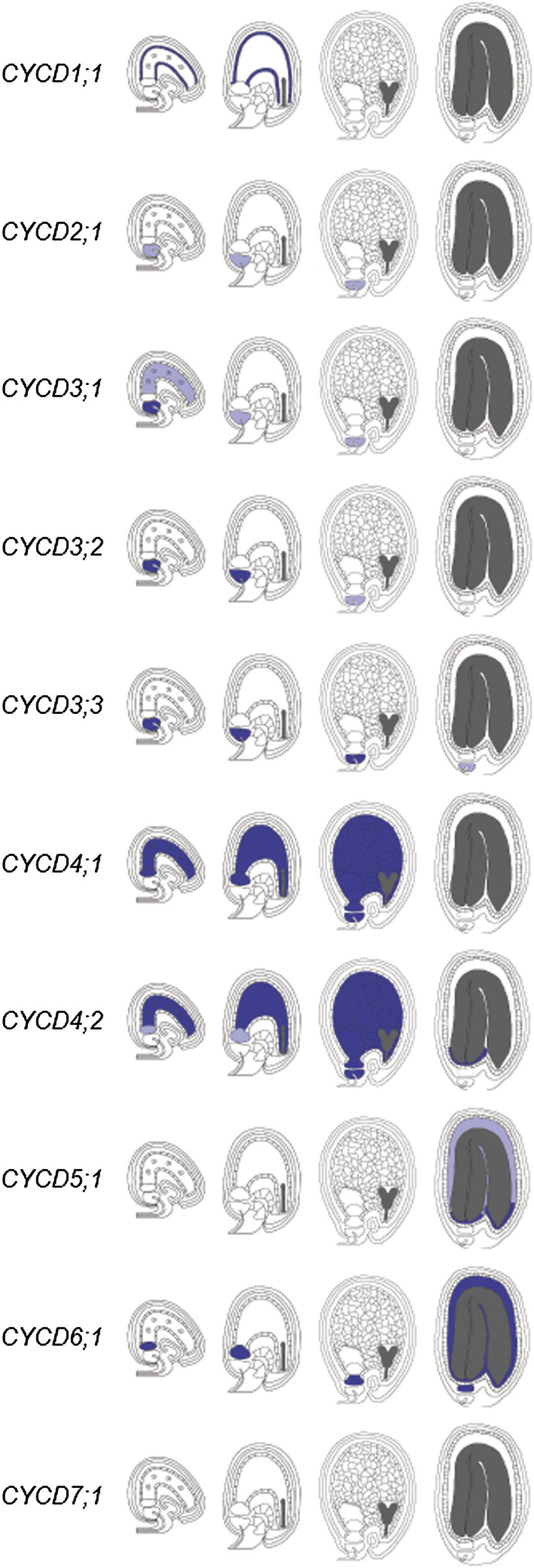
Fig. 4.
Summary of CYCD gene expression patterns during endosperm and peripheral seed tissue development using GUS reporters. Representative stages shown (from left to right): early syncytial, mid-syncytial, cellularized, and mature. Relative strengths of expression are represented as dark blue (strong), light blue (weak), and white (none).
Fig. 5.
Summary of CYCD gene expression patterns during embryogenesis using GUS reporters. Representative stages shown (left to right): one-cell zygote, globular, heart, and torpedo. Relative strengths of expression are represented as dark blue (strong), light blue (weak), and white (none).
To validate these GUS reporter analyses, the Genevestigator microarray expression database (Hruz et al., 2008) was examined to determine expression profiles for CYCDs in seed tissues (Fig. 6). Data was obtained for all CYCDs except CYCD7;1, since the ATH1 array lacks probe sets for this gene. All nine CYCDs were expressed in all tissues examined and, based on similarity of expression profiles, could be separated into four broadly distinct clusters which reflected phylogenetic subgroup structure (Fig. 6). Notably, CYCD3;1, CYCD3;2, and CYCD3;3 shared similar profiles with high embryo expression relative to the endosperm, with equal or lower expression in the suspensor. The Genevestigator data broadly agreed with results obtained using GUS reporters. Together, the results reveal CYCDs to have discrete and overlapping tissue-specific expression patterns in the seed, suggesting both distinct and redundant subgroup-specific functions for CYCDs in seed development.
Fig. 6.
Genevestigator expression profiles of CYCD genes in seed tissues. Data are shown as relative expression levels across different tissues. Sample points are joined for clarity. Note the scales in each panel are slightly different. Genes sharing similar profiles are organized into groups: (A) CYCD1;1 (red) and CYCD2;1 (dark blue); (B) CYCD3;1 (light green), CYCD3;2 (orange), and CYCD3;3 (purple); (C) CYCD4;1 (yellow) and CYCD4;2 (brown); (D) CYCD5;1 (dark green) and CYCD6;1 (light blue). CZE, chalazal endosperm; MCE, micropylar endosperm; PEN, peripheral endosperm.
Characterization of seed developmental progression in CYCD loss-of-function insertion mutant lines
In a systematic approach to investigate the functional significance of CYCDs in seed development, available loss-of-function insertion mutant combinations representing eight CYCDs were analysed for effects on seed developmental progression. Initial observations in 10 dap seeds showed that, in contrast to wild-type and all other cycd mutants, the cycd3;1cycd3;2cycd3;3 line had a significant increase in seed abortion at a frequency of 9.1%, compared to 0.2% in the wild type (Table 1).
Table 1.
Seed abortion in wild-type and cycd mutants
| Genotype | Normal seeds (%) | Aborted seeds (%) | Seeds scored (n) |
| Wild type | 99.8 | 0.2 | 360 |
| cycd1;1 | 100 | 0 | 552 |
| cycd2;1cycd4;1 | 99.5 | 0.5 | 439 |
| cycd3;1cycd3;2cycd3;3 | 90.9 | 9.1 | 671 |
| cycd6;1 | 99.4 | 0.6 | 586 |
| cycd7;1 | 100 | 0 | 401 |
Microscopic observations of staged seeds were performed at 2, 3, 4, 5, 7, and 9 dap (Table 2). In siliques of wild-type and all cycd mutant lines except for cycd3;1cycd3;2cycd3;3, progression of seed development was generally synchronous, with the majority reaching maturity by 9 dap, and mutants were phenotypically indistinguishable from the wild type, indicating that the respective genes are not essential for seed development. No additional phenotypes were observed in cycd6;1 other than those described of delayed divisions in cortical/endodermal root precursors (Sozzani et al., 2010).
Table 2.
Seed developmental progression in wild-type and cycd mutants Values are percentages. dap, days after pollination.
| dap | Genotype | 2-Cell | Quadrant/octant | Dermatogen | Globular | Heart | Torpedo | Bent cotyledon to Mature |
| 2 | Wild type | 35 | 65 | |||||
| cycd1;1 | 32 | 68 | ||||||
| cycd2;1cycd4;1 | 30 | 70 | ||||||
| cycd3;1cycd3;2cycd3;3 | 56 | 44 | ||||||
| cycd6;1 | 44 | 56 | ||||||
| cycd7;1 | 28 | 72 | ||||||
| 3 | Wild type | 15 | 30 | 55 | ||||
| cycd1;1 | 5 | 15 | 30 | 50 | ||||
| cycd2;1cycd4;1 | 17 | 29 | 54 | |||||
| cycd3;1cycd3;2cycd3;3 | 3 | 24 | 51 | 22 | ||||
| cycd6;1 | 9 | 39 | 52 | |||||
| cycd7;1 | 10 | 28 | 62 | |||||
| 4 | Wild type | 11 | 89 | |||||
| cycd1;1 | 10 | 90 | ||||||
| cycd2;1cycd4;1 | 21 | 79 | ||||||
| cycd3;1cycd3;2cycd3;3 | 12 | 65 | 23 | |||||
| cycd6;1 | 19 | 81 | ||||||
| cycd7;1 | 15 | 85 | ||||||
| 5 | Wild type | 25 | 75 | |||||
| cycd1;1 | 33 | 67 | ||||||
| cycd2;1cycd4;1 | 35 | 65 | ||||||
| cycd3;1cycd3;2cycd3;3 | 14 | 79 | 7 | |||||
| cycd6;1 | 27 | 73 | ||||||
| cycd7;1 | 26 | 74 | ||||||
| 7 | Wild type | 14 | 86 | |||||
| cycd1;1 | 17 | 83 | ||||||
| cycd2;1cycd4;1 | 14 | 86 | ||||||
| cycd3;1cycd3;2cycd3;3 | 7 | 17 | 41 | 35 | ||||
| cycd6;1 | 15 | 85 | ||||||
| cycd7;1 | 18 | 82 | ||||||
| 9 | Wild type | 100 | ||||||
| cycd1;1 | 100 | |||||||
| cycd2;1cycd4;1 | 100 | |||||||
| cycd3;1cycd3;2cycd3;3 | 2 | 7 | 11 | 80 | ||||
| cycd6;1 | 100 | |||||||
| cycd7;1 | 100 |
Progression of seed development in cycd3;1cycd3;2cycd3;3 was severely delayed and less synchronous (Table 2). At 3 dap, the first delay compared to the wild type were observed. At 9 dap, retardation in development was most pronounced with 80% of mutant seeds reaching mature stages and 20% at the globular, transition, or torpedo stages, compared to 100% of wild-type seeds that had reached seed maturity. No abnormalities were seen outside the embryo. These results suggest that CYCD3 genes play important roles in embryo development and that their loss reduces the rate of embryo progression.
Generation of CYCD-overexpressing lines
To gain further insight into the role of CYCD3 in controlling embryonic cell divisions and to explore the functional relevance of D-type cyclins in seed development more generally, two genes, CYCD3;1 and a previously uncharacterized cyclin, CYCD7;1, were overexpressed in specific seed domains using the mGAL4:VP16 / UAS two-component gene expression system (Haseloff, 1999). An ACT driver line based on RIBOSOMAL PROTEIN S5A (RPS5A) promoter (ACT RPS5A) (Weijers et al., 2003) was chosen as it drives strong GAL4 expression in the embryo from early stages, with transient activity in the proliferating endosperm. A further ACT driver line was generated based on the FWA promoter (ACT FWA), which is active exclusively in the proliferating endosperm as early as the central cell stage (Kinoshita et al., 2004). Effector (EF) lines were generated that harboured CYCD3;1 and CYCD7;1 coding sequences under control of the GAL4-responsive UAS promoter, and contained an associated GAL4-responsive GUS reporter gene to confirm transgene expression. No GUS activity was detected in all ACT and EF lines prior to transactivation and all were phenotypically normal compared to the wild type (Table 3) confirming that the UAS promoter was inactive in the absence of the GAL4 protein. To confirm that ACT RPS5A and ACT FWA lines are suitable for driving transgene expression in the desired embryo and endosperm tissues, both lines were crossed to an EF line harbouring a UAS promoter-driven GFP-GUS fusion reporter gene (EF pUAS:GGi) (Weijers et al., 2003) and GUS activity was monitored during seed development in F1 progeny. Both lines showed the expected pattern of activity for the RPS5A and FWA promoters, confirming that the ACT lines were suitable for further analysis (Supplementary Fig. S1, available at JXB online).
Table 3.
Seed abortion in wild-type, CYCD overexpression and control lines
| Genotype | Normal seeds (%) | Aborted seeds (%) | Seeds scored (n) |
| Wild type | 99.8 | 0.2 | 360 |
| ACT RPS5A | 99.7 | 0.3 | 652 |
| ACT FWA | 99.8 | 0.2 | 764 |
| EF CYCD3;1 | 100 | 0 | 324 |
| EF CYCD7;1 | 99.7 | 0.3 | 581 |
| PRPS5A>>CYCD3;1 | 91.1 | 8.9 | 737 |
| PFWA>>CYCD3;1 | 99.6 | 0.4 | 689 |
| PRPS5A>>CYCD7;1 | 84.3 | 15.7 | 980 |
| PFWA>>CYCD7;1 | 99.3 | 0.7 | 559 |
CYCD3;1 overexpression stimulates embryonic and suspensor cell proliferation and delays embryogenesis
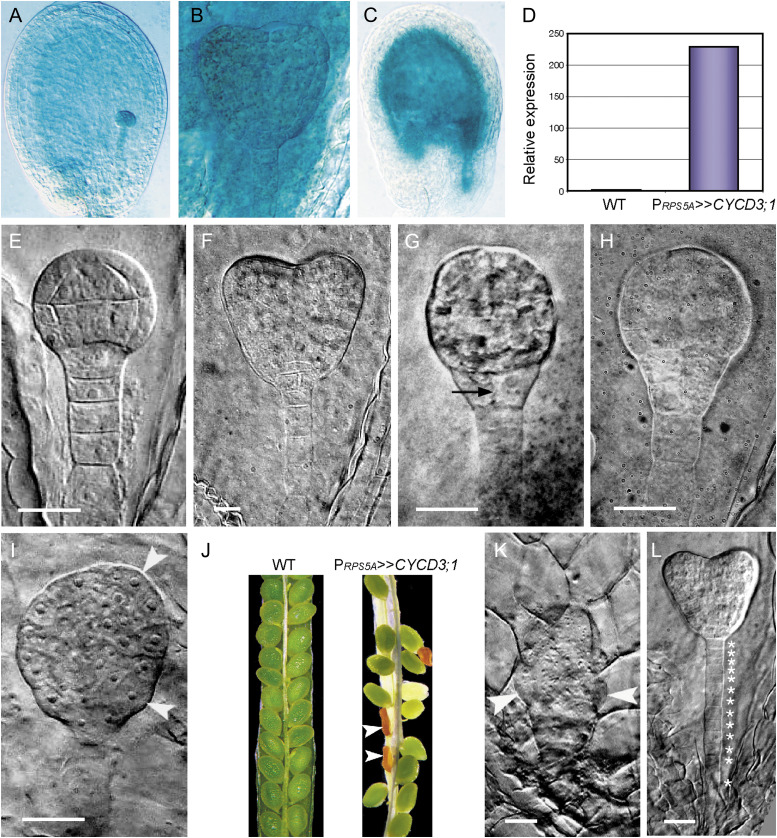
Overexpression of CYCD3;1 in the embryo and endosperm was achieved by crossing EF CYCD3;1 with the ACT RPS5A and ACT FWA lines. Transactivation was confirmed by the expected pattern of GUS activity of the RPS5A and FWA promoters (Fig. 7A–C; Weijers et al., 2003; Kinoshita et al., 2004) and the approximately 230-fold higher transcript levels in PRPS5A>>CYCD3;1 siliques (Fig. 7D), with similar levels achieved in PFWA>>CYCD3;1 seeds (data not shown). Seeds of the PRPS5A>>CYCD3;1 line exhibited a severe retardation in developmental progression associated with cell-division-induced embryonic defects, which was most pronounced at 9 dap (Table 4 and Supplementary Table S1). In contrast, progression of seed development in PFWA>>CYCD3;1 was normal and seeds were indistinguishable from the wild type (Tables 3 and 4).
Fig. 7.
Transactivation of CYCD3;1 causes ectopic cell divisions in the embryo and suspensor. (A–D) Cell type-specific transactivation of CYCD3;1 in PRPS5A>>CYCD3;1 (A, B, D) and PFWA>>CYCD3;1 (C). (A) Globular-stage seed with strong global GUS activity in the embryo, suspensor, and endosperm. (B) Global GUS activity in a heart-stage embryo with staining in the suspensor and endosperm. (C) Strong GUS staining throughout all domains of the endosperm. (D) CYCD3;1 transcript levels in siliques containing globular-stage seeds: relative transcript abundance is scaled to expression in the wild type (1-fold expression). (E–L) Wild-type embryo development (E, F) and phenotypes of PRPS5A>>CYCD3;1 embryos (G–L). (E) Wild-type globular-stage embryo. (F) Wild-type heart-stage embryo. (G) Globular-stage embryo showing premature division of the hypophysis (arrow). (H) Globular-stage embryo showing extra divisions in the hypophysis and lower tier. (I) Overproliferated globular-stage embryo with poor cell alignment and protuberances in the protoderm (arrowheads). (J) Typical seed set in the wild type (WT) and PRPS5A>>CYCD3;1 siliques at 10 dap showing elevated levels of seed abortion in PRPS5A>>CYCD3;1 line (arrowheads). (K) Overproliferated globular-stage embryo with large outgrowths (arrowheads). (L) Heart-stage embryo showing extra divisions in the suspensor (individual cells indicated by asterisks). Embryos from GUS-stained and unstained seeds were visualized during development. Bars = 25 μm (E, G–L), 10 μm (F).
Table 4.
Seed developmental progression in wild-type and CYCD overexpressors Values are percentages and values in boldface are percentages of seeds with abnormal characteristics. dap, days after pollination. The details of the phenotypes observed in the PRPS5A >>CYCD3;1 and PRPS5A >>CYCD7;1 lines are described in Supplementary Table S1 and Supplementary Table S2, respectively.
| dap | Genotype | 2-Cell | Quadrant/octant | Dermatogen | Globular | Heart | Torpedo | Bent cotyledon to Mature |
| 2 | Wild type | 35 | 65 | |||||
| PRPS5A>>CYCD3;1 | 52 | 48 | ||||||
| PRPS5A>>CYCD7;1 | 61 | 39 | ||||||
| PFWA>>CYCD3;1 | 25 | 75 | ||||||
| PFWA>>CYCD7;1 | 30 | 70 | ||||||
| 3 | Wild type | 15 | 30 | 55 | ||||
| PRPS5A>>CYCD3;1 | 28 | 49 | 23 | |||||
| PRPS5A>>CYCD7;1 | 6 | 19 | 48 | 27 | ||||
| PFWA>>CYCD3;1 | 8 | 31 | 61 | |||||
| PFWA>>CYCD7;1 | 6 | 29 | 65 | |||||
| 4 | Wild type | 11 | 89 | |||||
| PRPS5A>>CYCD3;1 | 25+53 | 7+15 | ||||||
| PRPS5A>>CYCD7;1 | 2 | 10 | 53 | 35 | ||||
| PFWA>>CYCD3;1 | 14 | 86 | ||||||
| PFWA>>CYCD7;1 | 6 | 94 | ||||||
| 5 | Wild type | 25 | 75 | |||||
| PRPS5A>>CYCD3;1 | 2+9 | 16+47 | 26 | |||||
| PRPS5A>>CYCD7;1 | 2 | 3 | 13+13 | 19+30 | 20 | |||
| PFWA>>CYCD3;1 | 47 | 53 | ||||||
| PFWA>>CYCD7;1 | 38 | 62 | ||||||
| 7 | Wild type | 14 | 86 | |||||
| PRPS5A>>CYCD3;1 | 6 | 8+22 | 18+6 | 40 | ||||
| PRPS5A>>CYCD7;1 | 1 | 11 | 12+13 | 39 | 11+13 | |||
| PFWA>>CYCD3;1 | 17 | 83 | ||||||
| PFWA>>CYCD7;1 | 20 | 80 | ||||||
| 9 | Wild type | 100 | ||||||
| PRPS5A>>CYCD3;1 | 8 | 1+6 | 3+7 | 75 | ||||
| PRPS5A>>CYCD7;1 | 9 | 11 | 7+12 | 22+39 | ||||
| PFWA>>CYCD3;1 | 100 | |||||||
| PFWA>>CYCD7;1 | 100 |
During the transition from the globular to the heart stage in wild-type seeds, the uppermost suspensor cell, the hypophysis, undergoes stereotyped asymmetric divisions giving rise to the quiescent centre of the root meristem and the central root cap (Fig. 7E, F). However, in some PRPS5A>>CYCD3;1 seeds, embryos underwent premature division of the hypophysis (Fig. 7G). In stronger phenotypes, extra rounds of proliferation with irregular division planes in the hypophysis and lower tier of the inner cell layer led to a highly disorganized basal region, causing it to bulge out at the periphery (Fig. 7H), whereas in other cases, uncontrolled divisions in both upper and lower tier cell files led to protuberances in the protoderm and an overall increase in embryo size (Fig. 7I). There was a significantly higher incidence of seed abortion in the PRPS5A>>CYCD3;1 line (8.9% compared to 0.2% in the wild type) (Table 3 and Fig. 7J). Inspection of aborted seed revealed predominantly globular-stage embryos that had undergone substantial uncontrolled proliferation, causing distinct outgrowths (Fig. 7K). In a related phenotype, CYCD3;1 overexpression induced divisions in the suspensor, resulting in a more filamentous structure composed of 10–13 smaller cells, compared to the 7–9 cells present in wild-type suspensors (compare Fig. 7F with Fig. 7L; Table 5). Importantly, the extra cell divisions that were stimulated upon overexpression of CYCD3;1 correlated with the domains of expression determined for all three CYCD3 genes (Figs. 2–6), supporting a dose-responsive role for CYCD3 in controlling embryonic cell cycle activity. It is concluded that specific embryonic tissues show particular sensitivity and responses to elevated CYCD levels.
Table 5.
Frequencies of suspensor cell numbers in PRPS5A>>CYCD3;1 seeds Approximately 150 seeds were analysed per line at each time point. PRPS5A>>CYCD3;1 embryos were from ACT RPS5A plants pollinated using EF CYCD3;1 plants as pollen parents.
| Stage | Genotype | Number of suspensor cells | ||||||||
| 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ≥13 | ||
| Globular | Wild type | 14.5 | 27.3 | 41.8 | 16.4 | |||||
| PRPS5A>>CYCD3;1 | 5 | 30 | 55 | 10 | ||||||
| Heart | Wild type | 33 | 55 | 12 | ||||||
| PRPS5A>>CYCD3;1 | 10 | 14 | 21 | 32 | 16 | 7 | ||||
Overexpression of CYCD7;1 induces cell proliferation and cell enlargement in the embryo and endosperm leading to overgrowth
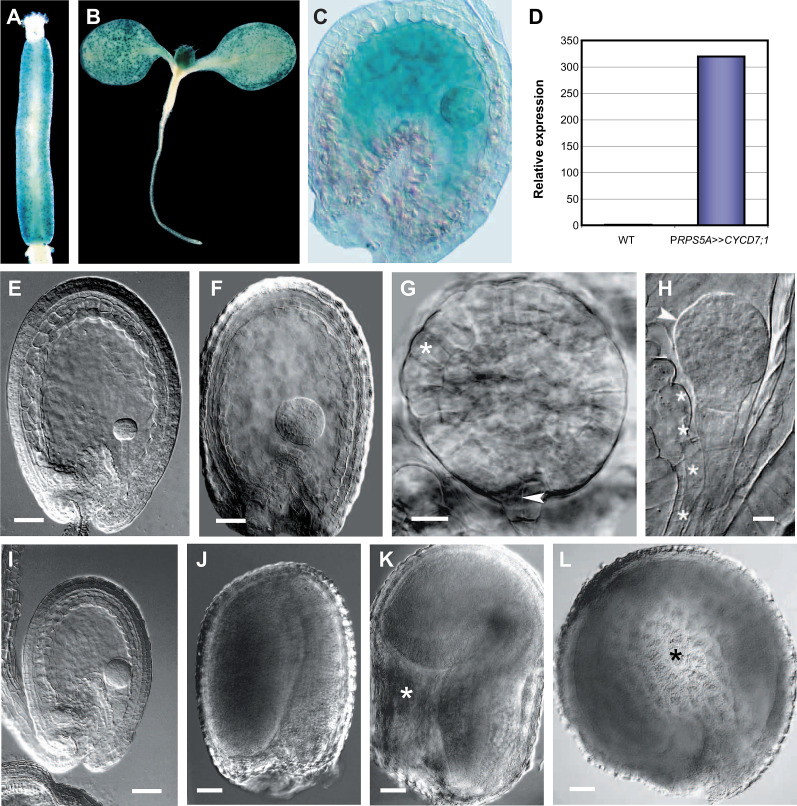
Whether the stimulation of embryonic cell divisions by CYCD3;1 overexpression was a general property of CYCDs was tested by ectopically expressing CYCD7;1. Based on reporter transgenic lines, CYCD7;1 is not expressed in the seed (Fig. 3) but is present in dividing cells of the stomatal lineage in leaf and floral tissues (Fig. 8A,B). Transactivation of CYCD7;1 was confirmed by GUS activity (Fig. 8C) and the detection of approximately 320-fold higher transcript levels in PRPS5A>>CYCD7;1 siliques (Fig. 8D). Similar results were obtained for PFWA>>CYCD7;1 (data not shown). Seed developmental progression in the PRPS5A>>CYCD7;1 line was highly retarded being most pronounced at 9 dap and presented a number of cell-division-induced embryonic defects (Table 4 and Supplementary Table S2). In contrast, progression of development in PFWA>>CYCD7;1 seeds was comparable to the wild type.
Fig. 8.
CYCD7;1 overexpression drives cell proliferation and cell growth in seed tissues. (A–D) Localization of GUS expression for pCYCD7;1:GUS (A, B) and cell-type-specific transactivation of CYCD7;1 in PRPS5A>>CYCD7;1 (C, D). (A) pCYCD7;1:GUS expression in stomatal cells lining the gynoecium. (B) pCYCD7;1:GUS expression in stomatal cells on the surface of developing leaves. (C) Global GUS activity in the embryo, suspensor, and endosperm of globular-stage seeds. (D) CYCD7;1 transcript levels in siliques containing globular-stage seeds: relative transcript abundance was scaled to expression in the wild type (1-fold expression). (E–L) Resulting phenotypic effects. (E) Wild-type globular seed. (F) PRPS5A>>CYCD7;1 seed containing an enlarged globular-stage embryo and endosperm. (G) Overproliferated PRPS5A>>CYCD7;1 globular-stage embryo showing enlarged protodermal cells (asterisk) and premature division of the hypophysis (arrowhead). (H) Overproliferated embryo with protuberances (arrowhead) and enlarged suspensor cells (asterisks). (I) Overproliferated embryo contained within a reduced endosperm. (J) Wild-type mature seed. (K) Enlarged PRPS5A>>CYCD7;1 seed with endosperm cavity (asterisk). (L) Enlarged PRPS5A>>CYCD7;1 seed with abnormal circular shape (cavity highlighted with asterisk). Bars = 50 μm (E, F, I–L), 20 μm (G, H).
Examination of PRPS5A>>CYCD7;1 seeds from 5 dap onwards revealed dramatic effects on both the embryo and endosperm. In wild-type seeds, the embryo and endosperm are tightly coordinated during seed development and undergo highly ordered nuclear and cellular divisions followed by cell enlargement, so that by the globular stage, embryos are surrounded by a late syncytial-stage endosperm of invariant size (Fig. 8E). However, in many PRPS5A>>CYCD7;1 seeds, the embryo and endosperm were significantly enlarged, which in the embryo was due to extra rounds of cell division followed by cell expansion (Fig. 8F–H). Ectopic embryo divisions were visible in the protoderm and inner tissue layers, followed by cell enlargement in a significant proportion of cells (Fig. 8G, compare with Fig. 7E). In a related phenotype, there was a marked growth of cells comprising the suspensor (Fig. 8H). In other cases, a reduction in endosperm was commonly observed (Fig. 8I). A significant proportion of mature seeds in the overexpressor line exhibited excessive endosperm and integument growth and were usually characterized by the presence of a persistent chalazal endosperm cavity (Fig. 8J, K), which in extreme cases led to large spherical seed morphologies (Fig. 8L). The PRPS5A>>CYCD7;1 line was associated with a significant increase in seed lethality (Table 3). These results indicate that while CYCD7;1 expression can drive cell proliferation in seed tissues, CYCD7;1 may have additional effects on cell growth, suggesting not all CYCDs are functionally equivalent.
Discussion
D-type cyclins have distinct and overlapping roles in seed development
The expression analyses performed here showed CYCDs to have discrete cell-specific expression patterns during seed development. This study has compared these with the general pattern of mitotic cycling cells, as revealed by the CYCB1;1 reporter fusion (Colón-Carmona et al., 1999). In general, their cumulative overall expression is strongly correlated with proliferating tissues in the embryo and endosperm, consistent with their proposed roles as key G1–S cell cycle regulators (Nieuwland et al., 2009a). However, distinctive patterns are seen for different CYCD genes. The expression patterns described for CYCD3;2, CYCD4;1 and CYCD6;1 are in agreement with previous partial expression studies using in situ hybridization and reporter transgenic lines (De Veylder et al., 1999; Swaminathan et al., 2000; Sozzani et al., 2010). There was substantial overlap in expression domains among the members of CYCD subgroups, which broadly reflected phylogenetic structure, implying that related genes coregulate the cell cycle in specific groups of cells. This suggests functional redundancy among various CYCD subtypes, which is consistent with the lack of embryonic phenotypes in single and double loss-of-function mutants representing CYCD1;1, CYCD2;1, and CYCD4;1 reported here, the delayed embryo development in cycd3;1cycd3;2cycd3;3 (discussed below), and the delayed formative ground tissue divisions in cycd6;1 (Sozzani et al., 2010). The data presented provides a valuable starting point for the identification of candidate CYCDs to target in higher-order loss-of-function insertion mutant combinations in order to further delineate functional roles for CYCDs in the development of specific seed tissues.
To date, there is limited information about how the cell cycle is fine tuned and integrated with patterning programmes to control the timing and location of specific cell divisions in the seed, although spatiotemporal control of the expression of regulatory proteins is presumably a major determinant (Menges et al., 2005; Sozzani et al., 2010). However, since both the main kinase partner of CYCD, CDKA, and the target of CDK-CYCD activity, RBR, show indistinct, non-cell-type-specific expression in the seed (Hemerly et al., 1993; Wildwater et al., 2005; Johnston et al., 2010), and many other core cell cycle genes generally have highly overlapping expression domains (Menges et al., 2005; Engler et al., 2009), other factors must be involved in governing localized cell division patterns. The data presented here suggests that CYCDs could provide a major contribution to conferring spatiotemporal specificity to the CYCD-RBR-E2F pathway and that correct patterning of seed tissues is achieved through developmental-stage- and cell-type-specific expression of distinct subsets of CYCDs. In support of this, precise spatiotemporal regulation of CYCD6;1 expression was recently shown to be required for a specific formative cell division in the embryonic ground tissue (Sozzani et al., 2010). The present study shows that perturbation of CYCD levels through overexpression or loss-of-function affects embryo development. CYCD subgroups defined in Arabidopsis have counterparts across the angiosperms including poplar (Populus trichocarpa) and rice (Oryza sativa) (Menges et al., 2007), consistent with conserved subgroup-specific roles for CYCDs in cell cycle regulation among higher plants.
CYCD3 is required for normal rate of embryonic development
Retarded developmental progression was observed in the cycd3;1cycd3;2cycd3;3 mutant but not in the other single and double mutant combinations tested. Except for CYCD4;2 and CYCD5;1, these represented each of the single-member CYCD subgroups present in Arabidopsis and, in addition, the cycd2;1 cycd4;1 combination. It should be noted that among higher plants, the CYCD2/CYCD4 subgroups are not separable and this study did not test a loss of function of all three members of these combined groups. However, overall the results suggest either a greater degree of redundancy in the function of the other CYCD gene subgroups and/or a particularly significant role for the CYCD3 subgroup, which may correlate with the partially overlapping expression domains of all three CYCD3 genes. Furthermore, embryo but not endosperm tissue was observed to be responsive to overexpression of CYCD3;1, which was able to stimulate extra cell divisions largely in domains of the embryo where CYCD3 genes were shown to be active. These divisions led in some embryos to perturbed patterning and lethality. These observations suggest that a correct level of CYCD3 activity is necessary for critical cell divisions required for normal patterning and morphogenesis of the embryo.
A necessity for strict regulation of the cell cycle for normal embryo development has been demonstrated in previous studies where the expression of cell cycle regulatory genes were manipulated, including CDKA (Hemerly et al., 2000), CYCA3;2 (Yu et al., 2003), DNA polymerase ϵ (Jenik et al., 2005), and CYCD6;1 (Sozzani et al., 2010). Embryonic delays seen in cycd3;1cycd3;2cycd3;3 mutant and/or the overexpressor resemble those seen in cdka and antisense-induced cyca3;2 mutants (Hemerly et al., 2000; Yu et al., 2003). In all mutants, a lack of sufficient cell divisions contributes to the inhibited growth of the embryonic shoot and root systems, despite the initial establishment of organ primordia and apical–basal axiality. Indeed, since all three CYCD3 proteins appear to bind CDKA exclusively (Van Leene et al., 2010) and CDKA is constitutively expressed during the cell cycle (Hemerly et al., 1993; Menges et al., 2005), it is reasonable to expect that the embryo effects observed in cycd3;1cycd3;2cycd3;3 are likely to be at least partially attributable to the lack of functional CDKA-CYCD3 complex formation. Nevertheless, the degree of overlap in CYCD expression patterns and the lack of complete penetrance in cycd3;1cycd3;2cycd3;3, suggest that even CYCD3 acts redundantly with other cyclins in regulating these cell divisions. However, no effects were observed in embryos mutant in single or double mutant combinations of other CYCD groups, suggesting that the CYCD3 group plays the most significant role. Strikingly, the CYCD3 subgroup is conserved in both mono- and dicotyledonous plants and, unlike any other CYCD classes, has the distinctive feature of having the cyclin box encoded in a single exon (Menges et al., 2007), suggesting that genes of the CYCD3 type have an ancient origin in higher plants and likely evolutionarily conserved functions.
Previous analysis has shown that CYCD3 genes are regulated by cytokinin (Riou-Khamlichi et al., 1999) and are required for normal cytokinin responses by shoot tissue (Dewitte et al., 2007). The correct specification of cell fate decisions in the cells that will give rise to the embryonic root requires an interplay between the hormones auxin and cytokinin (Müller and Sheen, 2008), and the TCS cytokinin reporter is highly expressed in the globular-stage embryo suspensor and its apical cell, the hypophysis. After division of the hypophysis, cytokinin signalling remains high in the suspensor, a tissue that is clearly responsive to increased CYCD3;1 expression revealed by ectopic cell divisions, and also in the lens-shaped (upper) daughter of the hypophysis which goes on to form the centre of the developing root meristem (Müller and Sheen, 2008). The present study observed that the CYCD3;1-overexpressing embryos showing lethality were predominantly globular-stage embryos with substantial uncontrolled proliferation and that, both in these lethal cases and in others that apparently recovered, irregular division planes were observed in a highly disorganized basal region. Current studies are investigating whether a hyperactive cell division response to cytokinin in overexpressers and a defect in the mutants could explain the phenotypes observed in the developing embryo, as previously observed in post-embryonic growth (Dewitte et al., 2007).
Ectopic CYCD7;1 expression alters cell proliferation and seed development
Critical cell cycle events during the development of the seed, including the onset and progression of proliferation of the syncytial and cellular endosperm and the integument layer and the timing of cellularization, are known to be regulated by the RBR-E2F pathway (Sabelli and Larkins, 2009; Sun et al., 2010). Manipulation of these processes, for example in seeds with an excess of paternal genomes from interploidy crosses (Scott et al., 1998), or overexpression of SHORT HYPOCOTYL UNDER BLUE 1 (SHB1) (Zhou et al., 2009) promotes endosperm proliferation and a delay in cellularization.
In contrast to the effects seen with ectopic CYCD3;1 expression, the results presented here demonstrate that ectopic CYCD7;1 expression can drive growth of both the embryo and endosperm, with lethal consequences in a significant proportion of seeds. Although the specific effects of CYCD7;1 overexpression on endosperm proliferation was not investigated, CYCD7;1 could act by promoting nuclear divisions in the syncytial endosperm prior to cellularization in a manner reminiscent of seeds with a paternal genome excess (Scott et al., 1998). This proposal is supported by the promotive effects that ectopic CYCD7;1 expression had on cell divisions and growth of the embryo and is consistent with the proposed role of CYCDs as positive regulators of cell cycle activity (Nieuwland et al., 2009a). Importantly, the spatiotemporal domain of activity of the RPS5A promoter ensured that high levels of CYCD7;1 was expressed at key phases in the early development of the endosperm during which active nuclear and cell proliferation events occur (Weijers et al., 2003; Sun et al., 2010). Therefore, it is not unreasonable to expect that extra rounds of divisions, and perhaps a delay in cellularization, are likely to have been the main drivers of the overgrowth of the endosperm observed. However, a direct contribution by the enlarged embryo in endosperm growth cannot be ruled out, since embryo development is known to have an influence on these processes (Hutchison et al., 2006; Nowack et al., 2007; Kondou et al., 2008). In this regard, it was interesting to note the difference in phenotypes in embryos and associated suspensor tissues overexpressing CYCD3;1 and CYCD7;1, the former increasing the number of cells and the latter the size of contributing cells. The apparent promotion of cell growth by CYCD7;1 in the embryo suggests that CYCDs are not all functionally equivalent and indicates a potential novel role for CYCD7;1. Intriguingly, a role in promoting cell growth was proposed for CYCD genes in Drosophila (Emmerich et al., 2004), suggesting this could be a function conserved in certain CYCD plant subgroups. In this interpretation, the effect of CYCD7;1 on division in some tissues could be a consequence of the promotion of growth.
In post-embryonic development, most examples of alteration of cell division rates do not cause phenotypes that affect pattern or organ size, but rather alter the cellular composition of tissues (Harashima and Schnittger, 2010). Increased expression of CYCD3;1 results in organs with an increased number of contributing cells (Dewitte et al., 2003), whereas cycd3;1cycd3;2cycd3;3 mutants have a reduced number of larger cells with higher levels of endoreduplication (Dewitte et al., 2007). In contrast, the present data suggest that correct spatiotemporal regulation of CYCD expression and cell division play an important role in the normal pattern and rate of growth from the two-cell stage of development.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Activity of the RPS5A and FWA activator lines during seed development.
Supplementary Table S1. Frequencies of phenotypic classes observed in PRPS5A >>CYCD3;1 seeds.
Supplementary Table S2. Frequencies of phenotypic classes observed in PRPS5A>>CYCD7;1 seeds.
Acknowledgments
The authors wish to thank Bart den Boer for valuable advice, Susan Howroyd for excellent technical assistance, Jim Haseloff (Cambridge, UK) for plasmid pBIN-Gal4–mGFP5er, Tetsu Kinoshita (Japan) for plasmid pBCH2-PFWA:▵FWA-GFP, Plant Systems Biology (Belgium) for plasmid pKGWFS7, and Remko Offringa (Netherlands) for plasmid pSDM7021 and the ACT pRPS5A:GAL4 and EF pUAS:GFP-GUS-intron lines. This work was supported by a Cooperative Award in Science and Engineering (CASE) studentship from the Biotechnology and Biological Sciences Research Council (ref: BBS/S/L/2003/10186).
References
- Clough S, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cockcroft CE. Expression analysis of Arabidopsis thaliana cyclin CycD2. 1998 PhD thesis, University of Cambridge, United Kingdom. [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. The Plant Journal. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- De Veylder L, De Almeida Engler J, Burssens S, Manevski A, Lescure B, Van Montagu M, Engler G, Inzé D. A new D-type cyclin of Arabidopsis thaliana expressed during lateral root primordia formation. Planta. 1999;208:453–462. doi: 10.1007/s004250050582. [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy J, Jacqmard A, Kilby N, Murray JAH. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. The Plant Cell. 2003;15:79–92. doi: 10.1105/tpc.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proceedings of the National Academy of Sciences, USA. 2007;104:14537–14542. doi: 10.1073/pnas.0704166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich J, Meyer CA, de la Cruz AF, Edgar BA, Lehner CF. Cyclin D does not provide essential Cdk4-independent functions in Drosophila . Genetics. 2004;168:867–875. doi: 10.1534/genetics.104.027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler Jde A, De Veylder L, De Groodt R, et al. Systematic analysis of cell-cycle gene expression during Arabidopsis development. The Plant Journal. 2009;59:645–660. doi: 10.1111/j.1365-313X.2009.03893.x. [DOI] [PubMed] [Google Scholar]
- Gruissem W. Function of the Retinoblastoma-related protein in plants. In: Inzé D, editor. Cell Cycle Control and Plant Development. Oxford, UK: Blackwell Publishing; 2007. pp. 164–186. [Google Scholar]
- Harashima H, Schnittger A. The integration of cell division, growth and differentiation. Current Opinion in Plant Biology. 2010;13:66–74. doi: 10.1016/j.pbi.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Haseloff J. GFP variants for multispectral imaging of living cells. Methods in Cell Biology. 1999;58:139–151. doi: 10.1016/s0091-679x(08)61953-6. [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira PCG, Van Montagu M, Engler G, Inzé D. Cell division events are essential for embryo patterning and morphogenesis: studies on dominant-negative cdc2aAt mutants of Arabidopsis . The Plant Journal. 2000;23:123–130. doi: 10.1046/j.1365-313x.2000.00800.x. [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, De Almeida Engler J, Van Montagu M, Engler G, Inzé D. Cdc2a expression in Arabidopsis is linked with competence for cell division. The Plant Cell. 1993;5:1711–1723. doi: 10.1105/tpc.5.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics. 2008 doi: 10.1155/2008/420747. ID 420747 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. The Plant Cell. 2006;18:3073–3087. doi: 10.1105/tpc.106.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Jurkuta REJ, Barton KM. Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase ϵ. The Plant Cell. 2005;17:3362–3377. doi: 10.1105/tpc.105.036889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Kirioukhova O, Barrell PJ, Rutten T, Moore JM, Baskar R, Grossniklaus U, Gruissem W. Dosage-sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genetics. 2010;6 doi: 10.1371/journal.pgen.1000988. e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Mayer U. Arabidopsis. In: Bard JBL, editor. Embryos, Colour Atlas of Development. London, UK: Wolfe Publishing; 1994. pp. 7–21. [Google Scholar]
- Karimi M, Inzé D, Depicker A. Gateway vectors for Agrobacterium-mediated plant transformation. Trends in Plant Sciences. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- Kondou Y, Nakazawa M, Kawashima M, et al. RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiology. 2008;147:1924–1935. doi: 10.1104/pp.108.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono A, Umeda-Hara C, Adachi S, Nagata N, Konomi M, Nakagawa T, Uchimiya H, Umeda M. The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. The Plant Cell. 2007;19:1265–1277. doi: 10.1105/tpc.106.046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masubelele NH, Dewitte W, Menges M, Maughan S, Collins C, Huntley R, Nieuwland J, Scofield S, Murray JAH. D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis . Proceedings of the National Academy of Sciences, USA. 2005;102:15694–15699. doi: 10.1073/pnas.0507581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, De Jager SM, Gruissem W, Murray JAH. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. The Plant Journal. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- Menges M, Pavesi G, Morandini P, Bögre L, Murray JAH. Genomic organisation and evolutionary conservation of plant D-type cyclins. Plant Physiology. 2007;145:1558–1576. doi: 10.1104/pp.107.104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lukowitz W, Bayer M. Talk global, act local – patterning the Arabidopsis embryo. Current Opinion in Plant Biology. 2008;11:28–33. doi: 10.1016/j.pbi.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Maughan S, Dewitte W, Scofield S, Sanz L, Murray JAH. The D-type cyclin CYCD4;1 modulates lateral root density in Arabidopsis by affecting the basal meristem region. Proceedings of the National Academy of Sciences, USA. 2009b;106:22528–22533. doi: 10.1073/pnas.0906354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland J, Scofield S, Murray JAH. Control of division and differentiation of plant stem cells and their derivatives. Seminars in Cell and Developmental Biology. 2009a;20:1134–1142. doi: 10.1016/j.semcdb.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Nowack MK, Shirzadi R, Dissmeyer N, Dolf A, Endl E, Grini PE, Schnittger A. Bypassing genomic imprinting allows seed development. Nature. 2007;447:312–315. doi: 10.1038/nature05770. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JM, Murray JAH. Sugar control of the plant cell cycle: Differential regulation of Arabidopsis D-type cyclin gene expression. Molecular and Cellular Biology. 2000;20:4513–4521. doi: 10.1128/mcb.20.13.4513-4521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins PA. The development of endosperm in grasses. Plant Physiology. 2009;149:14–26. doi: 10.1104/pp.108.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Dewitte W, Forzani C, et al. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. The Plant Cell. 2011;23:641–660. doi: 10.1105/tpc.110.080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana . Development. 1998;125:3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes and Development. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JAH, Benfey PN. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B, Salehian Z. An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Molecular Biology Reporter. 2002;20:107–114. [Google Scholar]
- Sun X, Shantharaj D, Kang X, Ni M. Transcriptional and hormonal signalling control of Arabidopsis seed development. Current Opinion in Plant Biology. 2010;13:1–10. doi: 10.1016/j.pbi.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Yang Y, Grotz N, Campisi L, Jack T. An enhancer trap line associated with a D-class cyclin gene in Arabidopsis . Plant Physiology. 2000;124:1658–1667. doi: 10.1104/pp.124.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inzé D. Genome-wide analysis of core cell cycle genes in Arabidopsis . The Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana . Molecular Systems Biology. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Van Hamburg J-P, Van Rijn E, Hooykaas PJJ, Offringa R. Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiology. 2003;133:1882–1892. doi: 10.1104/pp.103.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The retinoblastoma-related gene regulates stem cell maintenance in Arabidopsis root. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shen W- H. The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. The Plant Cell. 2003;15:2763–2777. doi: 10.1105/tpc.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Kang X, Zhao X, Zhang X, Ni M. SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. The Plant Cell. 2009;21:106–117. doi: 10.1105/tpc.108.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.