Abstract
Mitogen-activated protein kinase (MAPK) cascades are involved in various processes from plant growth and development to biotic and abiotic stress responses. MAPK kinases (MAPKKs), which link MAPKs and MAPKK kinases (MAPKKKs), play crucial roles in MAPK cascades to mediate a variety of stress responses in plants. However, few MAPKKs have been functionally characterized in cotton (Gossypium hirsutum). In this study, a novel gene, GhMKK5, from cotton belonging to the group C MAPKKs was isolated and characterized. The expression of GhMKK5 can be induced by pathogen infection, abiotic stresses, and multiple defence-related signal molecules. The overexpression of GhMKK5 in Nicotiana benthamiana enhanced the plants’ resistance to the bacterial pathogen Ralstonia solanacearum by elevating the expression of pathogen resistance (PR) genes, including PR1a, PR2, PR4, PR5, and NPR1, but increased the plants’ sensitivity to the oomycete pathogen Phytophthora parasitica var. nicotianae Tucker. Importantly, GhMKK5-overexpressing plants displayed markedly elevated expression of reactive oxygen species-related and cell death marker genes, such as NtRbohA and NtCDM, and resulted in hypersensitive response (HR)-like cell death characterized by the accumulation of H2O2. Furthermore, it was demonstrated that GhMKK5 overexpression in plants reduced their tolerance to salt and drought stresses, as determined by statistical analysis of seed germination, root length, leaf water loss, and survival rate. Drought obviously accelerated the cell death phenomenon in GhMKK5-overexpressing plants. These results suggest that GhMKK5 may play an important role in pathogen infection and the regulation of the salt and drought stress responses in plants.
Keywords: Abiotic stress tolerance, cell death, cotton, disease resistance, mitogen-activated protein kinase kinase
Introduction
Plants are constantly exposed to diverse stress stimuli including pathogen infection, drought, and high salinity throughout their life cycles. To survive, plants have developed a variety of mechanisms to perceive external signals and to protect themselves from environmental stress through a series of physiological and morphological changes (Tuteja, 2007; Ning et al., 2010). The perception and transduction of stimuli is regulated by an array of elaborate and intricate signalling networks. One of the universal signalling pathways involved in the responses to various biotic and abiotic stresses is the mitogen-activated protein kinase (MAPK) cascade (Rodriguez et al., 2010).
A typical MAPK cascade consists of three sequentially activated kinases, a MAPK kinase kinase (MAPKKK), a MAPK kinase (MAPKK), and a MAPK (MAPK Group, 2002). In the MAPK signal transduction cascade, a MAPK is activated by a MAPKK, which itself is activated by a MAPKKK. Each of the three tiers of kinases in a cell contains multiple members, which contributes to the specificity of the transmitted signal (Zhang and Klessig, 2001). In Arabidopsis, there are fewer MAPKKs (10) than MAPKs (20) and MAPKKKs (60) (MAPK Group, 2002). This fact suggests that various signal transduction pathways may converge at the MAPKK level in the MAPK cascades and that one type of MAPKKs is likely to be involved in multiple MAPK cascades and to carry out diverse biological functions (Xu et al., 2008; Andreasson and Ellis, 2010; Heinrich et al., 2011).
An increasing amount of evidence indicates that MAPKKs play important roles in plant biotic and abiotic stress responses. In Arabidopsis, the overexpression of AtMKK3 increases the expression of several pathogen reistance (PR) genes and enhanced the resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 (Doczi et al., 2007). The ectopic expression of AtMKK7 in local tissues induces PR gene expression and resistance to Pseudomonas syringae pv. maculicola (Psm) ES4326 in systemic tissues (Zhang et al., 2007). AtMKK1 can activate AtMPK3 to transmit drought, salt, and cold signals in Arabidopsis (Mizoguchi et al., 1998). Constitutively activated AtMKK9 can increase the sensitivity of transgenic plants to salt tolerance (Xu et al., 2008). In tobacco plants, SIMKK can activate SIMK to regulate salt stress signals (Kiegerl et al., 2000; Jonak et al., 2002). NtMEK2 serves as the upstream activator of SIPK and WIPK to regulate osmotic stress signals (Mikolajczyk et al., 2000). Intriguingly, some MAPKKs function in both biotic and abiotic stress responses. For example, AtMKK2-overexpressing transgenic plants exhibit markedly enhanced sensitivity to the fungal necrotroph Alternaria brassicicola, whereas these plants are more resistant to infection by PstDC3000 and Erwinia carotovora subsp. carotovora (Brader et al., 2007). At the same time, AtMKK2 is specifically activated by salt and cold, while the mkk2 mutant is hypersensitive to salt and cold stress, a combination that indicates that AtMKK2 positively regulates salt and cold stress (Teige et al., 2004). Although these reports provide us with a good overview of the functions of MAPKKs, the roles of MAPKKs in response to infection with different pathogens and to abiotic stress need to be further explored.
Biotic and abiotic stresses are typically associated with the rapid production of reactive oxygen species (ROS), including hydrogen peroxide (H2O2), superoxide anion (O2 –), and hydroxyl radicals (Kovtun et al., 2000). Moderate accumulation of ROS plays a central role in the regulation of biological processes, whereas high concentrations of ROS can result in oxidative stress and cause irreversible damage and hypersensitive response (HR)-like cell death (Kovtun et al., 2000). The HR is a typical sign of infection and is characterized by rapid cell death and often accompanies the formation of necrotic lesions. Emerging evidence has revealed that MAPK cascades are involved in the regulation of ROS homeostasis (Pitzschke and Hirt, 2009). Xing et al. (2007, 2008) found that AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signalling in Arabidopsis. Constitutive expression of AtMKK5 caused O3-induced ROS accumulation and cell death (Ren et al., 2002), whereas inhibiting the expression of AtMKK5 increased the O3 sensitivity and the H2O2 accumulation (Ahlfors et al., 2004; Miles et al., 2009). Recently in Nicotiana benthamiana, MEK2–SIPK and NPK1–MEK1–NTF6 cascades were reported to elevate ROS accumulation through the induced expression of the NbRbohb gene, leading to the increased tolerance to environmental stresses (Asai and Yoshioka, 2008). Furthermore, overexpression of constitutively active NtMEK2 activates SIPK/WIPK and results in HR-like cell death (Zhang et al., 1998; Yang et al., 2001). The cross-talk between the MAPK cascade and the ROS in the signal transduction network is very complicated, and many mechanisms remain largely unknown.
MAPKKs can be divided into four groups (A, B, C, and D) based on the phylogenetic analyses of amino acid sequences and phosphorylation motifs (MAPK Group, 2002; Hamel et al., 2006). Previous studies have mainly focused on MAPKKs in groups A, B, and D (Brader et al., 2007; Doczi et al., 2007; Zhang et al., 2007), whereas the information on group C MAPKKs is relatively limited, with published research primarily focusing on AtMKK4 and AtMKK5. It has been demonstrated that Arabidopsis AtMKK4 and AtMKK5 can activate both AtMPK3 and AtMPK6 to participate in flagellin perception and innate immunity, as well as in the regulation of the biosynthesis of camalexin (Asai et al., 2002; Ren et al., 2008). Recently, a novel group C MAPKK in maize, ZmMKK4, was reported to enhance salt and cold tolerance in transgenic Arabidopsis plants (Kong et al., 2011). These results imply that group C MAPKKs may be involved in various signalling processes and have multiple biological functions.
Cotton (Gossypium hirsutum) is one of the oldest and most important fibre and oil crops, and its growth and yield are severely inhibited under various biotic and abiotic stress conditions. Despite the important roles of MAPKK in biotic and abiotic stresses in plants, however, functional information on MAPKKs is scarce, especially in cotton. To address these concerns, the research details of the identification of a novel group C MAPKK gene, GhMKK5, from cotton were described. The results indicate that the expression of GhMKK5 is induced by chemical and biological stimuli. Ectopic expression of GhMKK5 in N. benthamiana altered the plants’ resistance to bacterial and oomycete pathogens and reduced salt and drought tolerance. In addition, GhMKK5-overexpressing plants exhibited a significant increase in H2O2 accumulation and H2O2-induced HR-like cell death. These results suggest that GhMKK5 may participate in multiple mechanisms by which plants respond to biotic and abiotic stresses. The findings further broaden our knowledge of the role of MAPKKs in signal transduction.
Materials and methods
Plant material, growth conditions, and treatments
Cotton (Gossypium hirsutum L. cv. lumian 22) seeds were placed in wet carbasus to accelerate germination, and the seedlings were transplanted into hydroponic culture under greenhouse conditions at 26±1 °C with a 16 h light/8 h dark cycle (light intensity of ∼200 μmol m−2 s−1 and relative humidity of 60–75%). Nicotiana benthamiana seeds were surface sterilized and planted on Murashige and Skoog (MS) medium for germination under greenhouse conditions. Two- or three-leaf stage N. benthamiana seedlings were transplanted into soil and maintained under greenhouse conditions. For the temperature treatment, uniformly developed cotton seedlings were transferred to 4 °C for given time periods. For other treatments, uniformly developed seedlings were cultured in solutions containing the indicated concentrations of NaCl, methyl viologen (MV), salicylic acid (SA), H2O2, methyl jasmonate (MeJA), abscisic acid (ABA), or ethephon for given time periods. For the pathogen treatment, 7-day-old cotton seedlings were inoculated with Fusarium oxysporum f. sp. vasinfectum conidial suspensions (105 conidia ml−1) using the root dip method. The treated cotyledons were collected for RNA extraction. Each treatment was repeated at least twice.
Disease resistance analysis
The bacterial pathogen Ralstonia solanacearum was cultured overnight at 37 °C in Luria–Bertani (LB) broth, harvested by centrifugation, and resuspended in sterile tap water. The oomycete pathogen Phytophthora parasitica var. nicotianae Tucker was cultured on potato dextrose agar (PDA) medium at 28 °C for 2 weeks, and the zoospores were then suspended in 1% glucose. For the disease resistance analysis, 8-week-old T3 generation overexpression (OE) and vector control (Vec) plants were inoculated with R. solanacearum bacterial suspensions [108 colony-forming units (cfu) ml−1] or P. parasitica var. nicotianae Tucker zoospore suspensions (105 zoospores ml−1) using the root dip method. Alternatively, the inoculation was also performed using the detached leaves of 8-week-old OE and Vec plants. The inoculated plants were kept in a moist chamber. Bacterial growth was monitored by performing serial dilutions onto King’s B agar medium containing 100 μg ml−1 rifampicin. The disease resistance analysis was repeated three times.
Salt and drought stress analysis
For the salt treatment, T3 generation OE and Vec seeds were surface-sterilized and sown on MS medium supplemented with different concentrations of NaCl (0, 100, and 200 mM). The germination percentage was measured daily after sowing. To examine the post-germination response, the seeds sown on MS medium for 3 d with radicle emergence were transferred onto MS medium with different concentrations of NaCl (0, 100, and 200 mM), and root elongation was determined. In addition, leaf discs, 1.3 cm in diameter, were detached from healthy, fully expanded leaves of OE and Vec plants of the same age. The discs were floated in solutions of different concentrations of salt (0, 100, and 200 mM) for 72 h and then immersed in 80% acetone for 48 h to extract the chlorophyll. Chlorophylls a and b were then subjected to spectrophotometric measurement. For the drought treatment, the seed germination percentage on MS medium with different concentrations of mannitol (0, 50, and 100 mM) was measured by the method above, and the post-germination root elongation was observed. Additionally, water was withheld completely from 8-week-old OE and Vec plants sown in soil for 10 d, after which the plants were watered for 2 d to allow them to recover, and the survival rate (the number of surviving plants relative to the total number of treated plants) was recorded. For the transpirational water loss assay, leaves of OE and Vec plants were detached and placed on an electronic balance at room temperature, and the changes in fresh weight were recorded over time. The rate of water loss was assumed to be equivalent to the loss of fresh weight of the samples. Salt and drought stress analysis was repeated at least three times.
Cloning of the GhMKK5 gene
Cloning was performed as described previously (Shi et al., 2011), and the primer sequences are provided in Supplementary Table S1 available at JXB online.
Vector construction and genetic transformation
The GhMKK5 cDNA (GenBank accession no. HQ637469) was inserted into the binary vector pBI121 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter via the XbaI and SalI sites. The recombinant plasmid was electroporated into Agrobacterium tumefaciens (strain LBA4404) for N. benthamiana transformation using the leaf disc method (Horsch et al., 1985), and transformants were screened for kanamycin (100 mg l−1) resistance and further confirmed by PCR. The plants transformed with vector pBI121 were used as the controls.
3,3-Diaminobenzidine (DAB) and trypan blue staining assays
For DAB staining, leaves were incubated in a DAB solution (1 mg ml−1, pH 3.8) for 12 h at 25 °C in the dark. After staining, the leaves were soaked in 95% ethanol overnight to remove the chlorophyll. For trypan blue staining, trypan blue was dissolved in lactophenol (0.33 mg l−1) and mixed with ethanol at a proportion of 1:1 (v/v). Leaves were incubated in the solution, boiled for 5 min, and soaked in chloral hydrate (2.5 g ml−1) overnight to remove the chlorophyll.
Northern blot and semi-quantitative RT-PCR analyses
A 20 μg aliquot of total RNA that had been extracted using the RNeasy Mini Kit (Qiagen, USA) according to the manufacturer’s instructions was separated on 1% agarose–formaldehyde gels and transferred onto Hybond-N+ membranes. The northern blot hybridizations were performed as described previously (Wang et al., 2007). The gene-specific primers TPF/TPR were used in the hybridization analyses. To analyse gene expression in transgenic plants by RT-PCR, total RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol and used for the first-strand cDNA synthesis using the reverse transcriptase system (TransGen Biotech, Beijing, China). The amplifications were performed at 94 °C for 5 min, followed by 25–30 cycles of amplification (94 °C for 50 s, 50–53 °C for 50 s, and 72 °C for 50 s). The PCR products were separated on a 1.8% agarose gel and visualized after ethidium bromide staining. The N. benthamiana β-actin gene was used as a loading control in each reaction.
Subcellular localization of GhMKK5
The open reading frame (ORF) of GhMKK5 was obtained by PCR amplification using the specific primers ZPF and ZPR. The fragments were fused onto the N-terminus of the green fluorescent protein (GFP) gene controlled by the CaMV 35S promoter. For transient expression, the plasmid DNA was transformed into onion epidermal cells using the particle bombardment method as described by Shi et al. (2010). Then, tissues were incubated on MS agar medium under dark condition at 25 °C for 12 h. The fluorescence was observed using a confocal laser scanning microscope (LSM 510 META, ZEISS, Germany). The 35S::GFP was used as a control.
Results
Isolation and sequence analysis of GhMKK5
A fragment was isolated from the cDNA of cotton cotyledons using the degenerate primers MP1 and MP2. Based on the internal sequence, the specific primers 5P1/5P2, 3P1/3P2, and TPF/TPR were used for 5′ and 3′ RACE (rapid amplification of cDNA ends)-PCR, and the amplification of the full-length sequence, respectively. The full-length cDNA sequence consisted of 1483 nucleotides, containing a 1053 bp ORF, a 225 bp 5′-untranslated region (UTR), and a 205 bp 3′ UTR. This clone exhibits a high level of sequence similarity to AtMKK5 in Arabidopsis; therefore, this gene was designated as GhMKK5 (GenBank accession no. HQ637469).
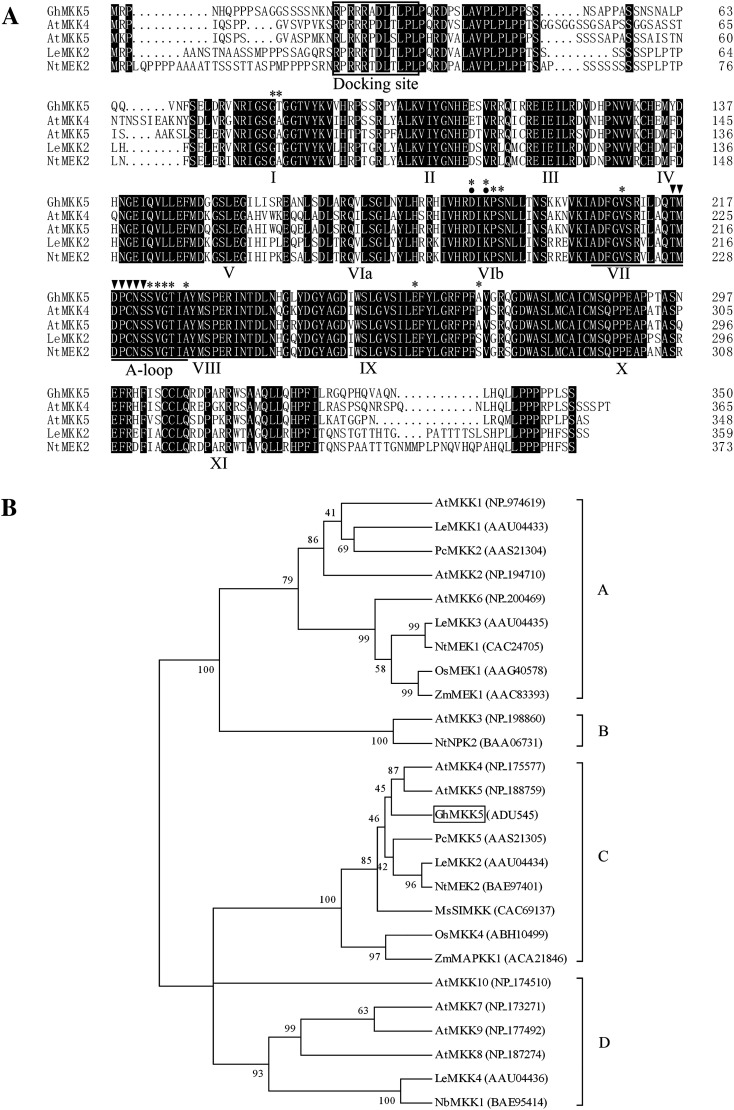
The predicted protein encoded by GhMKK5 consisted of 360 amino acid residues with a putative molecular weight of 38.90 kDa and an isoelectric point of 8.99. Multiple sequence alignments against MAPKKs from various plants were performed using the DNAMAN software. Consistent with other plant MAPKKs, the GhMKK5 protein exhibits the same family signature, including 11 conserved subdomains, a conserved S/TXXXXXS/T motif, a catalytic loop (activation-loop), and a docking site. Furthermore, GhMKK5 has a homology of 74.86% with AtMKK4, 75.21% with AtMKK5 from Arabidopsis thaliana, 73.46% with NtMEK2 from Nicotiana tabacum, and 73.63% with Solanum lycopersicum (previously named Lycopersicon esculentum) (Fig. 1A). Additionally, the conserved aspartate and lysine residues within the active site motif, -D(L/I/V)K-, and putative substrate binding sites were predicted using a bioinformatics tool available on the NCBI sever.
Fig. 1.
Comparison of the deduced amino acid sequences of GhMKK5 and closely related plant MAPKKs. (A) Alignment of the amino acid sequence of GhMKK5 (ADU545) with those of AtMKK4 (NP_175577), AtMKK5 (NP_188759), LeMKK2 (AAU04434), and NtMEK2 (BAE97401). Identical amino acids are shaded in black. The protein kinase subdomains are shown with numerals (I–XI) on the bottom of the sequences, and the activation loop (A-loop) is underlined. The serine and/or threonine residues in the conserved consensus motif S/TXXXXXS/T between subdomains VII and VIII of MAPKKs are marked with filled inverted triangles above. The conserved aspartate and lysine residues within the active site motif, -D(L/I/V)K-, are indicated by filled circles. The docking site is boxed. The asterisks above the sequences represent the putative substrate-binding sites. (B) The phylogenetic relationships between GhMKK5 and other plant MAPKK proteins. The Neighbor–Joining phylogenetic tree was created with Clustal W using MEGA 4.1. The numbers above or below branches indicate the bootstrap values (>50%) from 500 replicates. The gene name is followed by the protein ID. The species of origin of the MAPKKs are indicated by the abbreviation before the gene names: At, A. thaliana; Gh, Gossypium hirsutum; Le, Lycopersicon esculentum (Solanum lycopersicum); Ms, Medicago sativa; Nb, Nicotiana benthamiana; Nt, Nicotiana tabacum; Os, Oryza sativa; Pc, Petroselinum crispum, and Zm, Zea mays.
To investigate the evolutionary relationship among MAPKKs from different species, phylogenetic analysis was performed based on the amino acid sequences by the Neighbor–Joining method using the software MEGA version 4.1. As shown in Fig. 1B, GhMKK5 exhibited high similarity to the members of MAPKK group C, such as AtMKK4, AtMKK5, NtMEK2, and PcMKK5. These results suggest that GhMKK5 is a member of MAPKK group C.
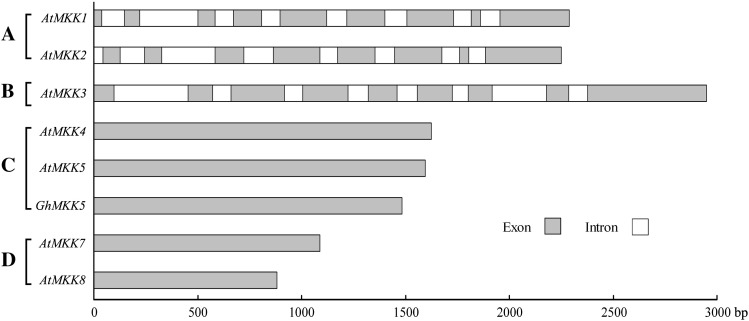
To investigate the genomic structure of GhMKK5, the specific primers TPF and TPR were used to amplify the sequence of the GhMKK5 from genomic DNA. Interestingly, sequence comparison revealed that GhMKK5 has no intron structure, which is similar to the characteristics of group C and D MAPKKs (Fig. 2). This result further indicates that GhMKK5 is a member of MAPKK group C.
Fig. 2.
Comparison of the genomic DNA sequences of GhMKK5 and several MAPKK genes of Arabidopsis available in GenBank. The white boxes indicate the introns, and the grey boxes represent the exons. The scale indicates the length of the sequence. A, B, C, and D indicate the groups of MAPKKs.
Subcellular localization of GhMKK5
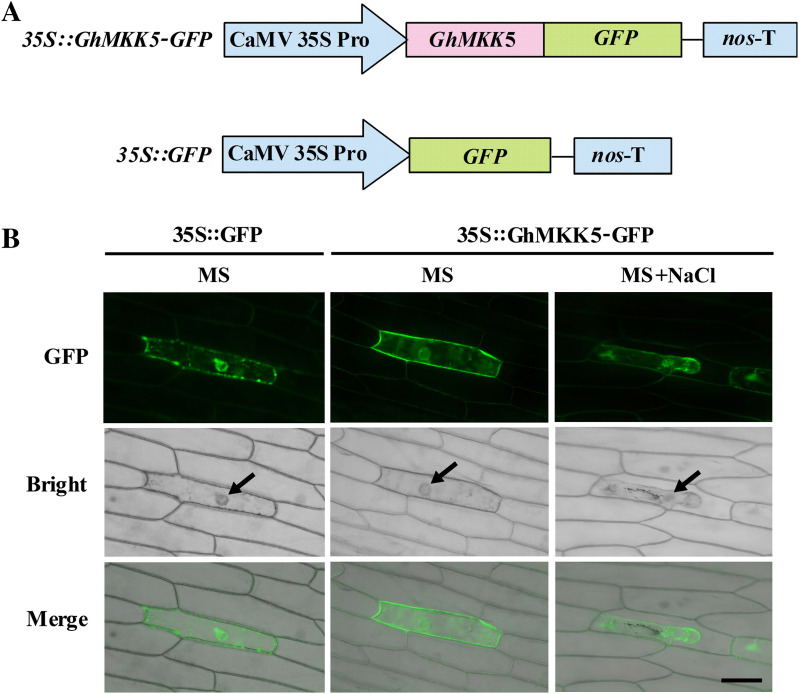
Bioinformatics analysis using the Plant-mPLoc program predicted that GhMKK5 is localized in both the nucleus and the cytoplasm. However, the CELLO version 2 program predicted that GhMKK5 is localized in the nucleus. To test these predictions, the overexpression vector 35S::GhMKK5–GFP was constructed to investigate the localization of GhMKK5 (Fig. 3A). The 35S::GFP construct was also produced as a control. The 35S::GhMKK5–GFP and 35S::GFP constructs were introduced into onion epidermal cells using particle bombardment. As shown in Fig. 3B, fluorescent signals from 35S::GhMKK5–GFP and 35S::GFP were detected in both the cytoplasm and the nucleus. There may be alternative protein localizations when cells are exposed to environmental stress. Therefore, to confirm further the localization of GhMKK5, the transiently expressing onion epidermises were transferred to MS medium containing 200 mM NaCl. Following a subculture until the occurrence of plasmolysis, the GhMKK5::GFP fusion protein still emitted green fluorescence in both the cytoplasm and the nucleus (Fig. 3B). These results suggest that GhMKK5 may function in the cytoplasm and the nucleus.
Fig. 3.
Subcellular localization of the GhMKK5 protein in onion epidermal cells. (A) Schematic diagram of the 35S::GhMKK5–GFP fusion construct and the 35S::GFP construct. (B) Transient expression of 35S::GhMKK5–GFP fusion and 35S::GFP constructs in onion epidermal cells. Green fluorescence was observed 12 h after particle bombardment using a confocal microscope. The arrow indicates the nuclei. Bar=200 μm.
Expression patterns of GhMKK5 in different tissues and under diverse stress conditions
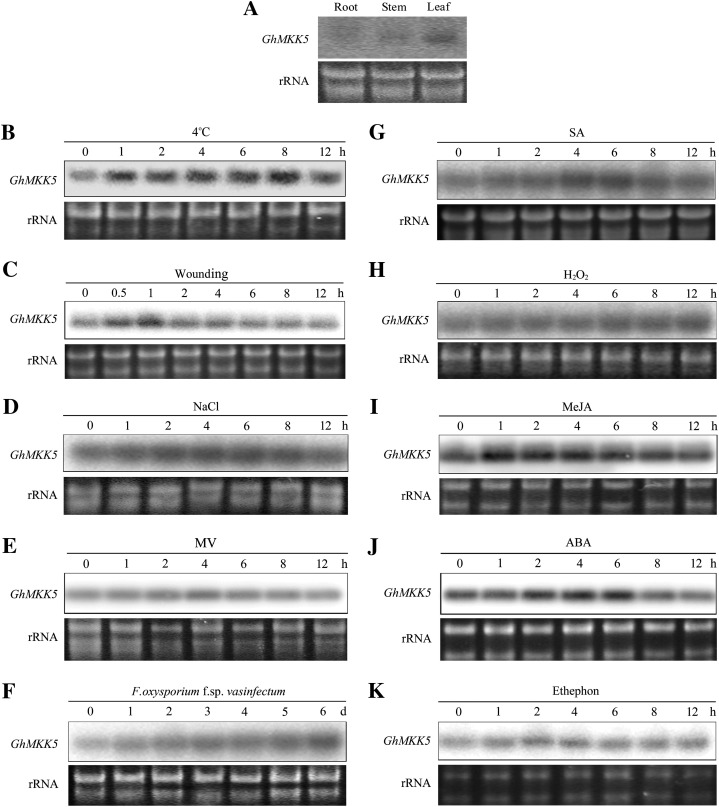
To investigate the expression patterns of GhMKK5, the RNA of 7-day-old cotton seedlings obtained from hydroponic culture was extracted for northern blot. As shown in Fig. 4A, the transcripts of GhMKK5 were primarily observed in cotyledons, as opposed to roots and stems, which indicated that the expression of GhMKK5 was tissue specific.
Fig. 4.
Expression profiles of GhMKK5 in different tissues and under different stress conditions. (A) Tissue-specific expression of GhMKK5 analysed by northern blot using the total RNA extracted from roots, stems, and cotyledon leaves of 7-day-old cotton seedlings. For stress treatments, 7-day-old cotton seedlings obtained from hydroponic culture were treated with low temperature (4 °C) (B), wounding (C), 200 mM NaCl (D), 10 μM MV (E), F. oxysporum f. sp. vasinfectum (F), 10 mM SA (G), 100 μM H2O2 (H), 100 μM MeJA (I), 100 μM ABA (J), and 5 mM ET released from ethephon (K). Total RNA was isolated at the indicated times after the treatments and was subjected to northern blot analysis using α-32P-labelled GhMKK5 cDNA fragments as a probe. The ethidium bromide (EB)-stained rRNA is included as a loading control. This experiment was repeated at least twice.
To study the effect of stresses on the expression of GhMKK5, the cotton seedlings were exposed to diverse environmental stresses. As shown in Fig. 4B–E, the expression of GhMKK5 could be induced by low temperature and NaCl. Wounding treatment rapidly and transiently enhanced the expression level of GhMKK5, and there was a mild accumulation of GhMKK5 transcripts upon MV treatment. In addition, F. oxysporum f. sp. vasinfectum could also activate the expression of GhMKK5, although the induction process was slow (Fig. 4F). These results indicated that GhMKK5 is stress responsive and may play crucial roles in the plant stress response.
To explore the signal transduction mechanism of GhMKK5 in response to environmental stresses, the responsiveness of GhMKK5 to diverse signalling molecules was also examined. As shown in Fig. 4G–K, the transcripts of GhMKK5 accumulated significantly under SA, MeJA, and ABA treatments. In contrast, H2O2 exerted a negligible effect on the expression of GhMKK5. In addition, although the induction was slight, ethephon caused an increase in GhMKK5 transcripts 2 h after treatment. These results indicated that GhMKK5 is responsive to multiple defence-related signal molecules, suggesting a role for GhMKK5 in multiple plant defence signal transduction pathways.
Enhanced resistance to bacterial pathogen in GhMKK5-overexpressing plants
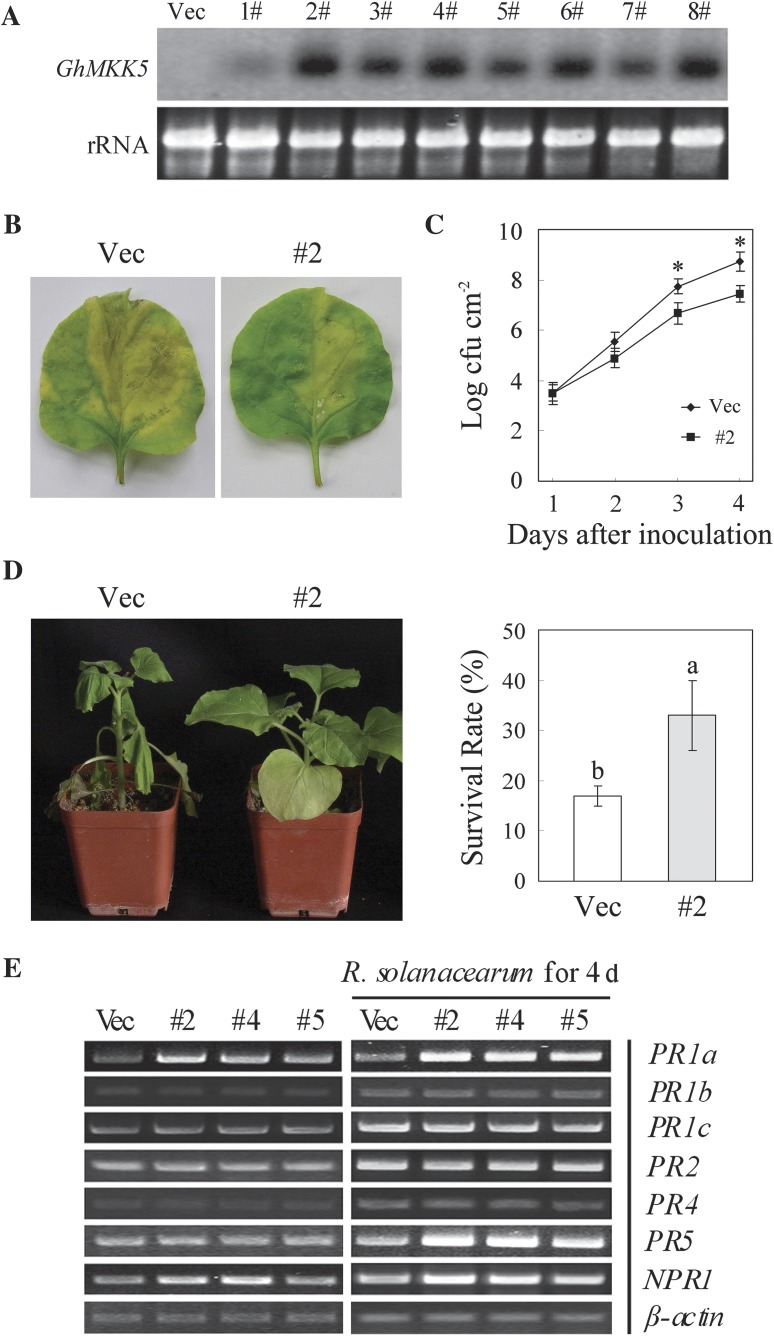
To test the function of GhMKK5, transgenic N. benthamiana plants overexpressing GhMKK5 were produced. A total of 22 independent OE lines were obtained by kanamycin resistance selection and confirmed by PCR detection (data not shown). Eight T1 progeny from OE lines were randomly selected for detecting GhMKK5 expression levels in different lines using northern blot analysis (Fig. 5A). Three representative lines (#2, #4, and #5) exhibiting different expression levels were selected and their T3 progeny transgenic plants were used for functional analysis.
Fig. 5.
Overexpression of GhMKK5 conferred on transgenic plants enhanced resistance to bacterial pathogens. (A) Northern blot analysis of GhMKK5 expression in T1 OE and Vec plants. (B) Signs of disease of the detached leaves from OE (#2) and Vec plants inoculated with 20 μl of a suspension (OD600=0.6–0.8) of R. solanacearum. The inoculated leaves were maintained under greenhouse conditions with 80% relative humidity for 1 week, and photographs taken at day 4 are presented. (C) Bacterial growth in the leaves of Vec and #2 plants at different time points after infiltration with R. solanacearum. Data are the means ±SE of three independent experiments (n=6). Asterisks (*) above lines indicate significant differences (P < 0.05) according to Duncan’s multiple range test. (D) Disease signs and survival rates of the OE (#2) and Vec plants inoculated with R. solanacearum using the trickle irrigation method. Data are the means ±SE of three independent experiments (n ≥ 30). Different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test. (E) RT-PCR analysis of the expression of PR genes in OE (#2, #4, and #5) and Vec plants under normal and R. solanacearum inoculation conditions. OE, overexpression; Vec, vector control; #2, #4, #5, particular overexpressing lines.
To examine the role of GhMKK5 in the defence response to bacterial pathogens, the detached leaves from OE and Vec plants were inoculated with 20 μl of a suspension (OD600=0.6–0.8) of R. solanacearum. Four days after inoculation, the leaves of Vec plants showed chlorotic signs and enlarged water-soaked lesions. In contrast, the leaves of GhMKK5-overexpressing plants exhibited only mild disease signs 4 d after inoculation (Fig. 5B). To confirm the disease signs, bacterial growth in the leaves of OE and Vec plants was measured after inoculation. Four days after inoculation, transgenic line #2 showed an almost 10-fold reduction in bacterial growth (Fig. 5C). When the plants were inoculated with R. solanacearum using the trickle irrigation method, Vec plants displayed marked wilting, whereas OE plants were relatively less affected, with a survival rate approximately twice that of the Vec plants (Fig. 5D). These results indicated that the overexpression of GhMKK5 enhanced the resistance to bacterial pathogens in transgenic plants.
Altered expression of defence-related genes in GhMKK5-overexpressing plants
To elucidate the possible mechanisms of enhanced bacterial resistance in OE plants, the expression levels of several defence-related genes in N. benthamiana, including PR1a, PR1b, PR1c, PR2, PR4, PR5, and NPR1, were analysed using RT-PCR. As shown in Fig. 5E, the expression levels of PR1a and NPR1 were constitutively elevated without any stress treatment, whereas the expression levels of PR1b, PR1c, PR2, PR4, and PR5 without any stress treatment were not obviously altered. However, when treated with R. solanacearum, the expression levels of PR1a, PR5, and NPR1 were more strongly induced in OE plants than in Vec plants. Although the mRNA levels of PR1b, PR1c, and PR2 were also increased by the infection, there was no significant difference between the OE and Vec plants. In contrast, the induction of PR4 in OE plants was relatively milder than in Vec plants after infection. Notably, PR1a, PR5, and NPR1 are the marker genes for SA signalling, and PR4 is the marker gene for JA signalling. Therefore, it was proposed that the GhMKK5-dependent activation of the defence-related genes plays a key role in the enhanced bacterial resistance observed in OE plants, which might be related to SA-dependent and JA-dependent signalling pathways.
Enhanced susceptibility of GhMKK5-overexpressing plants to oomycete pathogen infection
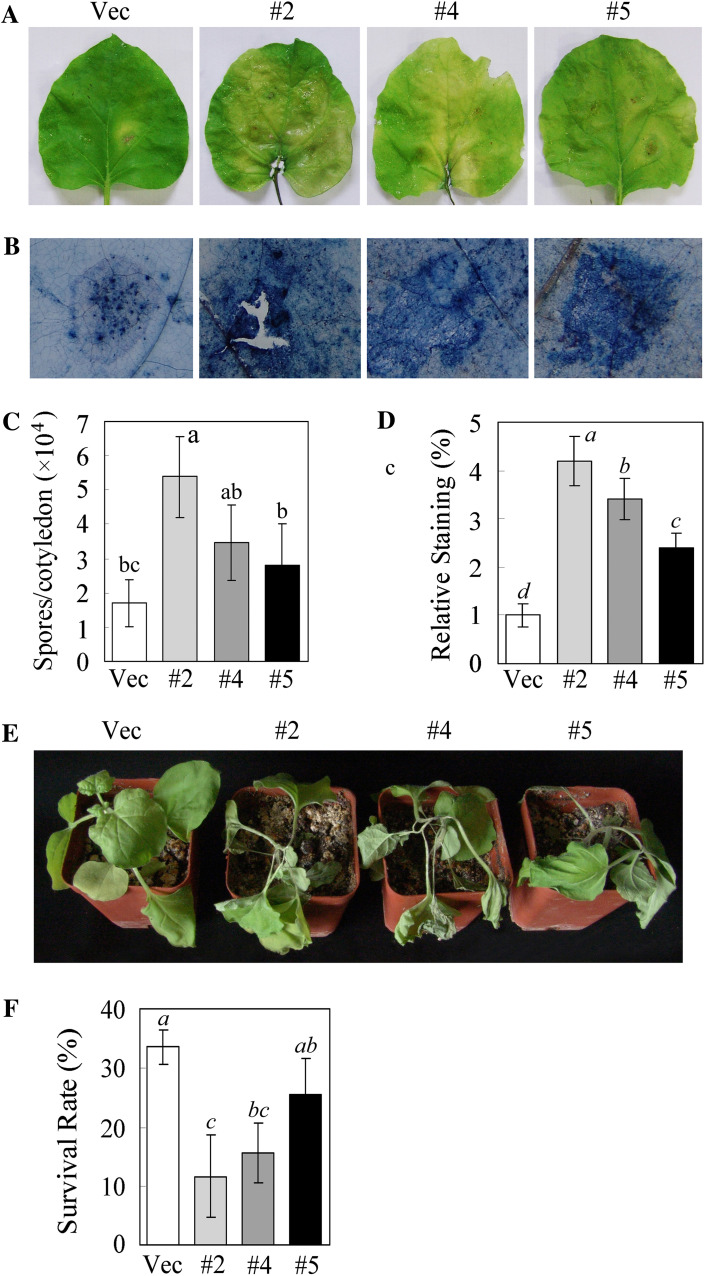
To determine whether the ectopic expression of GhMKK5 in N. benthamiana plants alters their resistance to oomycete pathogens, transgenic plants were challenged with the P. parasitica pathogen. Six days after inoculation, the leaves of OE plants exhibited wilting and yellowing with necrotic lesions, whereas the leaves of Vec plants showed less severe disease signs (Fig. 6A). Trypan blue staining of the inoculated leaves indicated that leaves of OE plants showed relatively more staining than those of Vec plants (Fig. 6B, D), which indicated that the overexpression of GhMKK5 increased the level of P. parasitica-induced cell death. Spore counts allow accurate quantification of pathogen reproduction. As shown in Fig. 6C, both OE and Vec plants suffered heavy sporulation, but the zoospore production by OE lines was greater, by ∼1.6- to 3.1-fold, than that by Vec plants. When the plants were inoculated with P. parasitica using the trickle irrigation method, both OE and Vec plants displayed wilting and leaf etiolation, but OE plants were more significantly affected, with a 10–20% survival rate reduction compared with Vec plants (Fig. 6E, F). These results indicated that the overexpression of GhMKK5 enhanced the susceptibility to oomycete pathogen infection in transgenic plants.
Fig. 6.
Overexpression of GhMKK5 conferred on transgenic plants enhanced susceptibility to P. parasitica infection. (A) Disease signs on the detached leaves of OE and Vec plants 6 d after inoculation. (B) Trypan blue staining for the pathogen infection. The average number of spores per cotyledon at 6 d after P. parasitica infection and the relative staining of trypan blue are shown in (C) and (D), respectively. Each experiment contained the spore counts from 10 inoculated cotyledons of OE and Vec plants. Data are the means ±SE of three independent experiments. Different letters above the columns indicate significant differences (P < 0.05), and different italic letters indicate highly significant differences (P < 0.01) according to Duncan’s multiple range test. (E) Disease signs of the transgenic plants inoculated with the P. parasitica pathogen using the trickle irrigation method. These plants’ survival rates are given in (F). Data are the means ±SE of three independent experiments (n ≥ 30). Different italic letters indicate highly significant differences (P < 0.01) according to Duncan’s multiple range test. OE, overexpression; Vec, vector control; #2, #4, #5, particular overexpressing lines.
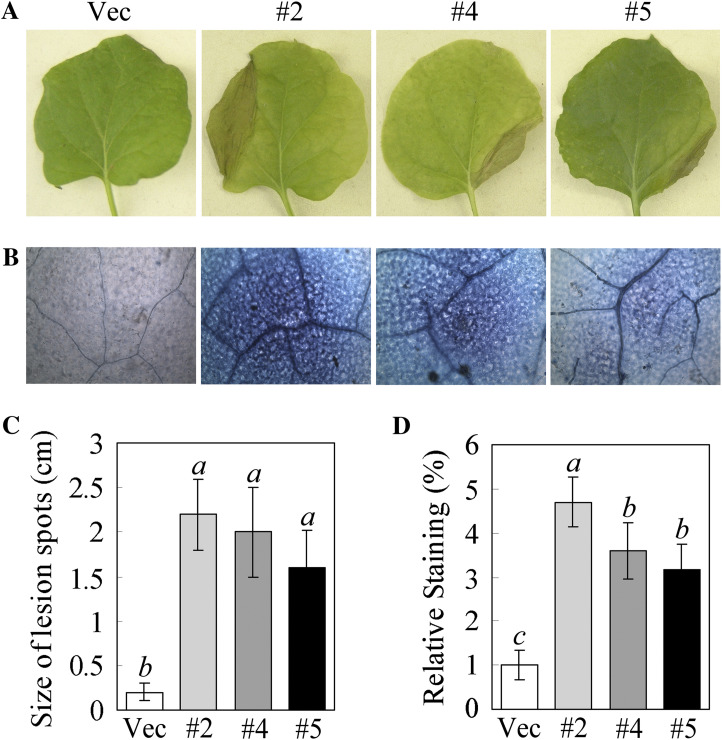
Overexpression of GhMKK5 in transgenic plants results in HR-like cell death
The T3 progeny transgenic plants were sown in soil and maintained routinely. Interestingly, the bottom leaves of OE plants first showed a HR-like cell death phenomenon ∼2 months after germination under greenhouse conditions (Fig. 7A), and the lesion size in the leaves of OE plants was much larger than that in the Vec plants (Fig. 7C). There was more trypan blue staining in the OE plants than in the Vec plants, which indicated that the leaves of OE plants showed relatively more cell death (Fig. 7B, D). These results indicated that the overexpression of GhMKK5 resulted in HR-like cell death in transgenic plants.
Fig. 7.
The HR-like cell death in transgenic plants induced by the overexpression of GhMKK5. (A) HR-like cell death signs in the leaves of OE plants. (B) Trypan blue staining for cell death in the leaves of transgenic plants. (C) The size of lesions in the leaves with HR-like cell death. (D) The relative levels of staining for cell death in the leaves of transgenic plants. OE, overexpression. Data are the means ±SE of three independent experiments (n=15). Different italic letters indicate highly significant differences (P < 0.01) according to Duncan’s multiple range test. Vec, vector control; #2, #4, #5, particular overexpressing lines.
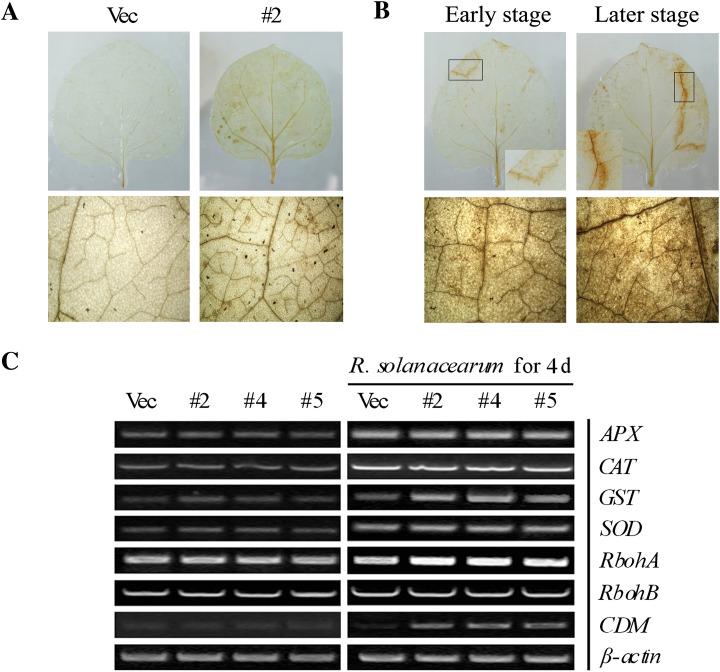
GhMKK5 regulates the accumulation of H2O2
To determine the reason for the HR-like cell death in OE plants, the H2O2 accumulation in the leaves was analysed by DAB staining. As shown in Fig. 8A, OE plants accumulated relatively higher amounts of H2O2 under normal conditions. In addition, the accumulation of H2O2 in leaves that exhibited HR-like cell death was detected. Notably, DAB staining showed that H2O2 was primarily found surrounding the necrotic lesions. Along with the aggravation of cell death, the accumulation of H2O2 was significantly elevated (Fig. 8B). However, there was no significant variation in the level of H2O2 in the Vec plants (data not shown).
Fig. 8.
H2O2 generation induces cell death mediated by the overexpression of GhMKK5. (A) The accumulation of H2O2 in transgenic plants visualized by DAB staining. (B) The significant accumulation of H2O2 surrounding the necrotic lesions in #2 plants. The black box indicates the region containing the necrotic lesions. (C) The expression of ROS and cell death-related genes in transgenic plants under normal and R. solanacearum infection conditions. OE, overexpression; Vec, vector control. #2, #4, #5, particular overexpressing lines.
To investigate the possible mechanisms underlying the elevated H2O2 accumulation in OE plants, the expression levels of genes that encode ROS-scavenging enzymes, such as manganese superoxide dismutase (MnSOD), catalase 1 (CAT1), ascorbate peroxidase (APX), and glutathione S-transferase (GST), and the ROS producers, respiratory burst oxidase homologues (RbohA and RbohB), were determined by RT-PCR. As shown in Fig. 8C, there was no visible difference in the expression levels of the ROS-scavenging and ROS-producing genes between the OE and Vec plants under normal conditions. However, when the plants were inoculated with R. solanacearum, the expression of GST and RbohA was strongly enhanced in OE plants (Fig. 8C). Although the transcript accumulation of APX, CAT, and SOD was markedly elevated by the R. solanacearum infection, there was still no visible difference in the expression of these genes between the OE and Vec plants. High concentrations of ROS can result in HR cell death. Interestingly, the overexpression of GhMKK5 enhanced the expression of the cell death marker gene CDM in the transgenic plants (Fig. 8C). These results suggest that the HR-like cell death in GhMKK5-overexpressing plants is conferred by the RbohA-mediated ROS production and that GhMKK5 may be involved in the regulation of ROS signalling.
Overexpression of GhMKK5 reduced the salt tolerance in transgenic plants
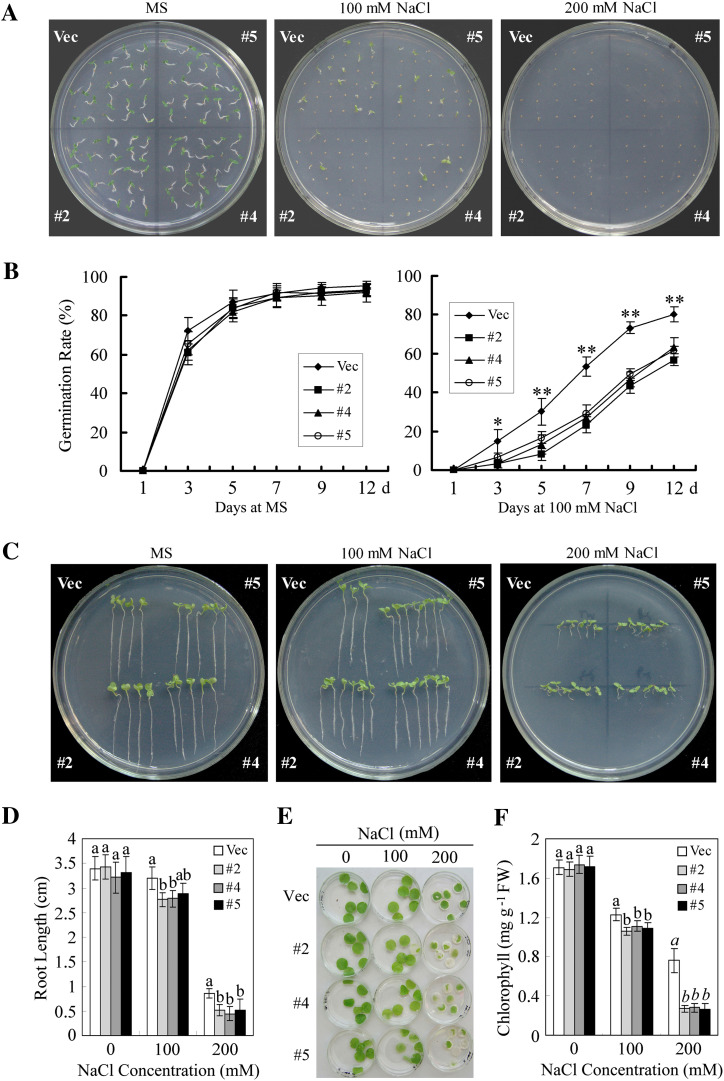
To determine the possible effects of GhMKK5 on salt tolerance, the transgenic seed germination capacity on MS agar medium containing different concentrations of NaCl was analysed. As shown in Fig. 9A, no significant difference in seed germination was observed between the OE and Vec lines without NaCl treatment. When the NaCl concentration was increased to 100 mM, a drastic decrease in the germination rates of both OE and Vec seeds was observed, whereas the OE seeds exhibited a germination rate ∼20% lower than that of the Vec seeds 7 d after sowing (Fig. 9B). As the concentration was further increased to 200 mM, both OE and Vec seeds failed to germinate at the seventh day after sowing (Fig. 9A). To confirm the reduced tolerance to salt, the post-germination growth of the transgenic plants was also tested. The seeds of OE and Vec plants were allowed to germinate on normal MS medium for 3 d and were then transferred to medium containing various NaCl concentrations, ranging from 0 mM to 200 mM. The seedlings’ growth was monitored by measuring their root length. As shown in Fig. 9C and D, the roots of the OE seedlings were much shorter than those of the Vec plant in the presence of 100 mM and 200 mM NaCl.
Fig. 9.
Reduced salt tolerance in transgenic plants overexpressing GhMKK5. (A) Seed germination in the presence of the indicated NaCl concentrations. (B) Germination rates of the OE and Vec lines under normal and NaCl treatment conditions. Germination was scored daily, and the result on the MS medium with 100 mM NaCl is presented. Data are the means ±SE of three independent experiments (n=3). Asterisks (* or **) above lines indicate (highly) significant differences (*P < 0.05; **P < 0.01) according to Duncan’s multiple range test. (C) The post-germination seedling development of OE and Vec lines on MS medium containing different concentrations of NaCl. The seeds sown on MS medium for 3 d with radicle emergence were transferred onto MS medium with different concentrations of NaCl. The plates were held erect, and a photograph was taken 14 d after transfer. (D) Root length of the seedlings 14 d after transfer onto NaCl-containing plates. Data are the means ±SE of three independent experiments (n=6). Different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test. (E) Leaf discs from OE and Vec plants were infiltrated with different concentrations of NaCl (0, 100, and 200 mM). (F) Relative chlorophyll content in the leaf disks 4 d after NaCl treatments. Data are the means ±SE of three independent experiments (n=6). Different letters above the columns indicate significant differences (P < 0.05), and different italic letters indicate a highly significant difference (P < 0.01) according to Duncan’s multiple range test. OE, overexpression; Vec, vector control. #2, #4, #5, particular overexpressing lines. (This figure is available in colour at JXB online.)
To determine whether the reduced salt tolerance conferred by the overexpression of GhMKK5 is also effective at the vegetable growth stage, leaf discs from 8-week-old transgenic plants were soaked in the solutions containing various NaCl concentrations, ranging from 0 mM to 200 mM. As shown in Fig. 9E, the discs incubated in water without NaCl showed no abnormalities. After incubation in different concentrations of NaCl for 72 h, signs of bleaching or chlorosis appeared in the leaf discs from both OE and Vec plants. However, NaCl treatment led to more severe damage in the leaf discs of OE plants. This result was further confirmed by measuring the chlorophyll content in the leaf discs after NaCl treatment (Fig. 9F). These results indicated that the overexpression of GhMKK5 confers reduced tolerance to salt stress in transgenic plants.
Overexpression of GhMKK5 reduced the drought tolerance in transgenic plants
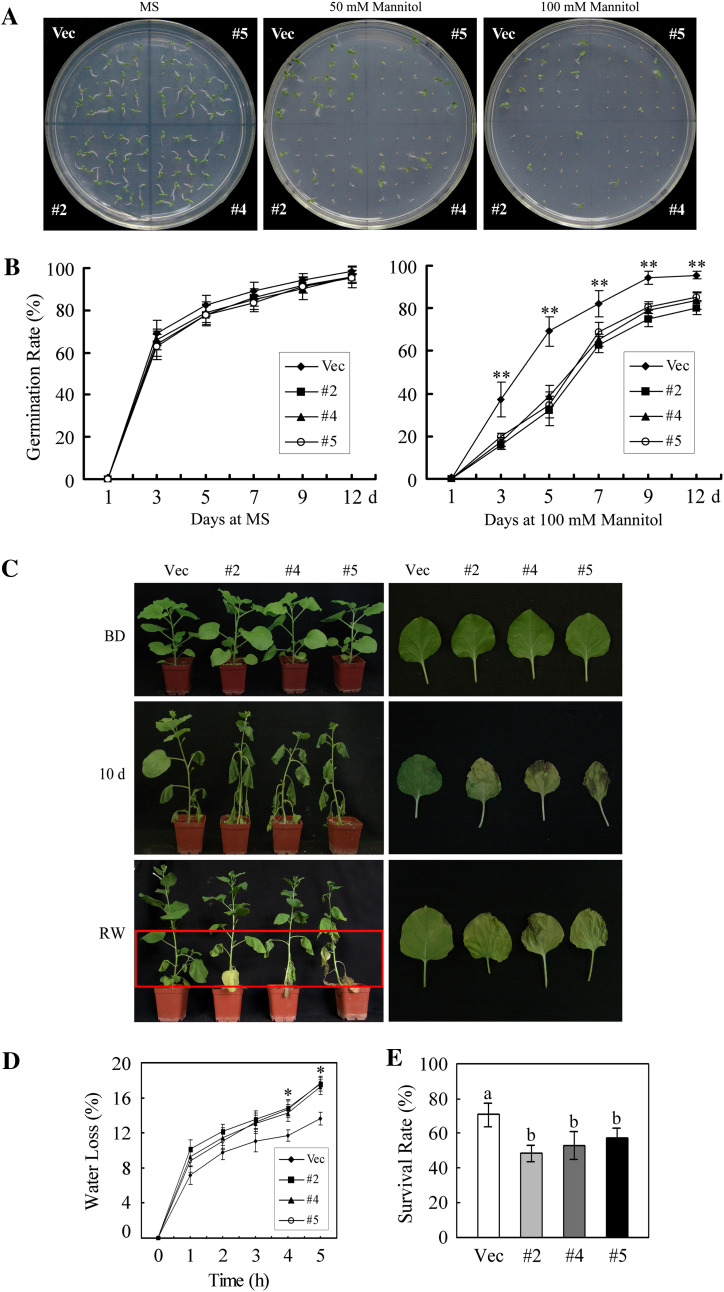
To investigate the response of GhMKK5-overexpressing plants to drought stresses, seeds of OE and Vec plants were exposed to MS medium containing different concentrations of mannitol (0, 50, and 100 mM) to mimic drought stress. As shown in Fig. 10A and B, treatment with mannitol significantly inhibited the germination of OE and Vec seeds, resulting in ∼14% and 19% reductions in the germination rate in the presence of 100 mM mannitol 7 d after sowing, respectively.
Fig. 10.
Reduced drought tolerance in GhMKK5-overexpressing plants. (A) Seed germination on MS medium in the presence of the indicated mannitol concentrations. (B) Germination rates of the OE and Vec lines under normal and 100 mM mannitol conditions. Data are the means ±SE of three independent experiments (n=3). Asterisks (**) above lines indicate highly significant differences (**P < 0.01) according to Duncan’s multiple range test. (C) Phenotype of plants submitted to drought stress at the vegetable stage. Water was withheld from 8-week-old plants for 10 d, and then the plants were watered for 2 d to allow them to recover. The leaves with drought-induced necrotic spots are shown on the right. BD, before drought treatment; RW, rewatering. (D) Water loss from detached leaves of OE and Vec plants. The rate of water loss was calculated by the loss in fresh weight of the samples. Data are the means ±SE of three independent experiments (n=6). Asterisks (*) above lines indicate significant differences (*P < 0.05) according to Duncan’s multiple range test. (E) The survival rate of the transgenic plants under drought conditions. Data are the means ±SE of three independent experiments (n ≥ 30). Different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test. OE, overexpression; Vec, vector control; #2, #4, #5, particular overexpressing lines. (This figure is available in colour at JXB online.)
To confirm further the drought tolerance of transgenic plants at the vegetable growth stage, water was withheld from the OE and Vec plants for 10 d. After 10 d of drought treatment, OE plants completely wilted, whereas Vec plants were less affected (Fig. 10C). Moreover, under drought stress, OE plants showed a mild decrease in growth over the Vec plants. When they were watered after the drought treatment, the Vec plants recovered faster than the OE plants, although several leaves did not recover completely (Fig. 10C). Interestingly, drought stress drove the Vec plants to develop pale green leaves, whereas OE leaves showed marked etiolation and stress-induced necrotic spots on the leaves. In addition, the rate of water loss from detached leaves of OE plants was higher than that of Vec plants under drought conditions (Fig. 10D), and the OE plants displayed a greater decrease in the survival rate (Fig. 10E).
Discussion
The tolerance mechanisms of plants are sophisticated, among which MAPK cascades play crucial roles. As the nodal point of MAPK cascades, the role and significance of MAPKKs in response to biotic and abiotic stresses have only recently begun to emerge. In the present study, the cotton group C MAPKK gene GhMKK5 was characterized. To the authors’ knowledge, this is the first time that a cotton group C MAPKK gene has been identified. Although several reports have indicated that MAPKKs are involved in stress signal transduction, the negative regulation mechanism is rarely reported, and the responsiveness of the MAPKK-mediated signalling pathway to different pathogens is not well documented. However, cotton transformation is very difficult and the whole process is time consuming (Li et al., 2004). Considering the easy transformation and convenience of studying the resistance to pathogens, such as bacteria and oomycetes, functional analysis was firstly performed by heterologous ectopic expression in N. benthamiana. In this study, the results suggested that the overexpression of GhMKK5 enhanced the resistance to bacterial pathogens but reduced the resistance to oomycete pathogens and the tolerance to salt and drought stress. Notably, transgenic plants overexpressing GhMKK5 showed marked H2O2 accumulation and H2O2-induced HR-like cell death. These findings not only extend the knowledge of the biological function of the group C MAPKKs but also provide new clues for further exploration of the role of GhMKK5 in the regulation of plant defence responses.
The investigation of MAPKK protein subcellular localization is very important for exploring the roles of MAPK cascades in signal transduction. Takahashi et al. (2007) have reported that a group D MAPKK in N. benthamiana, NbMKK1, was localized in the nucleus. Similarly, the constitutively active AtMKK9 is also localized in the nucleus (Yoo et al., 2008). Recently, a group C MAPKK in Zea mays, ZmMKK4, was also reported to be localized in the nucleus (Kong et al., 2011). In contrast, in the present study, the results indicated that GhMKK5 was localized in both the cytoplasm and the nucleus (Fig. 3). Several lines of evidence have suggested that MAPKKs may be located not only in the cytoplasm, but also in the nucleus, and that the proteins may have altered localization under different environmental conditions. The human extracellular signal-regulated kinase (ERK) 1/2 cascade components ERK1/2 (MAPK), MEK1/2 (MAPKK), and Raf1 (MAPKKK) were demonstrated to be localized in the cytoplasm in quiescent cells (Kondoh et al., 2005; Takahashi et al., 2007). However, the stimulation of cells caused a rapid translocation of Raf1 to the plasma membrane and the translocation of MEK1/2 and ERK1/2 to the nuclei. In contrast, parsley PcMKK5 was retained in the cytoplasm after phytophthora-derived (Pep-13) elicitor treatment (Lee et al., 2004). In this study, the localization of GhMKK5 showed negligible alteration after salt stress treatment (Fig. 3), which was in agreement with the pattern of PcMKK5, although the localizations of the two proteins were different. In contrast, human MEK5, in addition to its downstream MAPK, ERK5, is always localized to the nucleus irrespective of cellular conditions (Raviv et al., 2004). Furthermore, MAPKKs in Arabidopsis, including AtMKK4 and AtMKK5, contain transport peptide, and AtMKK4 is localized in chloroplasts (Samuel et al., 2008). No obvious nuclear localization signal (NLS) was found in the GhMKK5 amino acid sequence (data not shown). The mechanisms of nuclear localization of GhMKK5 and the regulation of its interaction with its downstream targets should be addressed in future work.
The inducibility of the expression of GhMKK5 by diverse signalling molecules and pathogens suggested that GhMKK5 might be involved in defence responses. To test the role of GhMKK5 in pathogen infection, transgenic plants were inoculated with bacterial and oomycete pathogens, respectively. As shown in Fig. 6, the overexpression of GhMKK5 elevated the resistance to the bacterial pathogen R. solanacearum. In contrast, overexpressing plants displayed increased sensitivity to P. parasitica var. nicotianae Tucker. Similarly, the overexpression of constitutively active AtMKK2 enhanced the resistance to the bacterial pathogen PstDC3000 and the fungal pathogen Erwinia carotovora subsp. carotovora, but OE plants exhibited increased sensitivity to the fungal necrotroph Alternaria brassicicola (Brader et al., 2007). Moreover, AtMKK1 can be activated by pathogen elicitors and shows a role in enhanced disease resistance (Teige et al., 2004). Recently, Zhang et al. (2007) have indicated that the overexpression of Arabidopsis AtMKK7 leads to the activation of plant basal and systemic acquired resistance (SAR). Furthermore, the homologue of AtMKK7 in tomato, LeMKK4, is involved in Pto-mediated immune responses (Pedley and Martin, 2004), and the homologue in N. benthamiana, NbMKK1, mediates INF1-induced cell death signalling and non-host resistance to Pseudomonas cichorii (Takahashi et al., 2007). In this study, the overexpression of GhMKK5 resulted in the increased accumulation of H2O2 followed by H2O2-induced HR-like cell death in transgenic plants. Consistent with this result, the overexpression of constitutively active AtMKK4 or AtMKK5 results in oxidative burst and cell death characterized by the accumulation of H2O2 in the leaves and the formation of necrotic lesions (Ren et al., 2002; Asai and Yoshioka, 2008). In tobacco, the overexpression of the homologue of AtMKK4 and AtMKK5, NtMEK2, also caused the formation of hypersensitive lesions (Jin et al., 2003). Recently, it was reported that the overexpression of NbMKK1 triggered a rapid generation of H2O2, which was followed by HR-like cell death in N. benthamiana leaves (Takahashi et al., 2007). These results suggest that plant MAPKKs have pleiotropic functions, including roles in regulating defence responses either positively or negatively.
It has been previously established that the signalling molecules SA, ET, and JA play important roles in the regulation of complex defence mechanisms (Spoel et al., 2007). SA is an essential signalling molecule that induces SAR and is implicated in resistance to biotrophic pathogens (Spoel et al., 2007; Spoel and Dong, 2008; Leon-Reyes et al., 2009). ET and JA are typically associated with the defence responses to necrotrophic pathogens and herbivorous insects (Spoel et al., 2007; Spoel and Dong, 2008). In this study, GhMKK5 could be significantly induced by SA, MeJA, and ethephon, which suggested that GhMKK5 might be involved in the SA or JA/ET signalling pathways. In addition, the expression of the marker genes for various pathways (PR1a and PR5 for SA signalling; PR4 for JA signalling) was greatly elevated in GhMKK5-overexpressing plants (Fig. 5). Furthermore, another SA signalling pathway marker gene, NPR1, which is very important for plant SAR, was also significantly increased. In Arabidopsis, although the levels of SA, JA, and ET were not significantly affected, the constitutively active AtMKK2 increases the expression levels of genes that encode enzymes in ET and JA signalling. However, upon PstDC3000 infection, transgenic plants depress the levels of increase of JA and SA, which suggests that AtMKK2 affected hormone levels during pathogen infection (Brader et al., 2007). Therefore, whether the levels of SA, JA, and ET were affected in GhMKK5-overexpressing plants should be further determined.
The tolerance of transgenic plants to abiotic stress, including salt and drought stress, was also determined. Interestingly, transgenic plants overexpressing GhMKK5 showed reduced salt and drought tolerance with decreased seed germination and other indicators of reduced tolerance (Figs 5, 6). In line with this observation, Arabidopsis AtMKK9 can activate AtMPK3/AtMPK6 to participate in the negative response to salt stress (Xu et al., 2008). In addition, AtMPK4 functions as a negative regulator of SAR, and the activation of AtMPK4 is regulated by AtMKK1, suggesting that AtMKK1 may be involved in the AtMPK4-related negative response to environmental stresses (Petersen et al., 2000; Xing et al., 2008). In contrast, Arabidopsis AtMKK2 can also be induced by salt stress, but it can enhance transgenic plants’ tolerance to salt stress (Teige et al., 2004). Recently, it was reported that the overexpression of ZmMKK4 increased transgenic plants’ cold and salt stress tolerance (Kong et al., 2011). In this study, the results indicated that the overexpression of GhMKK5 reduced salt stress tolerance, which suggests that the regulatory mechanisms of the response to abiotic stress involving GhMKK5 significantly differ from those involving AtMKK2. In addition, comparison of the MAPKKs in plants reported previously indicated that MAPKKs function differently across species and even in the same species. Further physiological and genetic experiments are necessary to elucidate the potential functions of GhMKK5.
In conclusion, the results of the present study strongly suggest that GhMKK5 is responsive to a variety of stress and signal molecule treatments and that the ectopic expression of GhMKK5 enhanced transgenic plants’ bacterial pathogen resistance while also increasing their oomycete pathogen sensitivity. Furthermore, GhMKK5-overexpressing plants exhibited reduced salt and drought stress tolerance. Although the complex regulation mechanisms involving GhMKK5 are still unclear, this work provides further insight into the mechanisms by which MAPKK regulates the stress response. As we learn more about MAPK cascades and their regulation, potential applications in genetic improvement should become possible.
Supplementary data
Supplementary data are available at JXB online.
Table S1. The primers used in this study.
Acknowledgments
This work was financially supported by the Genetically Modified Organisms Breeding Major Projects of China (2009ZX08009-092B) and the National Natural Science Foundation of China (grant no. 31171837).
References
- Ahlfors R, Macioszek V, Rudd J, Brosche M, Schlichting R, Scheel D, Kangasjarvi J. Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. The Plant Journal. 2004;40:512–522. doi: 10.1111/j.1365-313X.2004.02229.x. [DOI] [PubMed] [Google Scholar]
- Andreasson E, Ellis B. Convergence and specificity in the Arabidopsis MAPK nexus. Trends in Plant Science. 2010;15:106–113. doi: 10.1016/j.tplants.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Asai S, Yoshioka H. The role of radical burst via MAPK signaling in plant immunity. Plant Signaling Behavior. 2008;3:920–922. doi: 10.4161/psb.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Brader G, Djamei A, Teige M, Palva ET, Hirt H. The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis . Molecular Plant-Microbe Interactions. 2007;20:589–596. doi: 10.1094/MPMI-20-5-0589. [DOI] [PubMed] [Google Scholar]
- Doczi R, Brader G, Pettko-Szandtner A, Rajh I, Djamei A, Pitzschke A, Teige M, Hirt H. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. The Plant Cell. 2007;19:3266–3279. doi: 10.1105/tpc.106.050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends in Plant Sciences. 2006;11:192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Heinrich M, Baldwin IT, Wu J. Two mitogen-activated protein kinase kinases, MKK1 and MEK2, are involved in wounding- and specialist lepidopteran herbivore Manduca sexta-induced responses in Nicotiana attenuata . Journal of Experimental Botany. 2011;62:4355–4365. doi: 10.1093/jxb/err162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Rogers SG, Fraley RT. Transgenic plants. Cold Spring Harbor Symposia on Quantitative Biology. 1985;50:433–437. doi: 10.1101/sqb.1985.050.01.054. [DOI] [PubMed] [Google Scholar]
- Jin H, Liu Y, Yang KY, Kim CY, Baker B, Zhang S. Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. The Plant Journal. 2003;33:719–731. doi: 10.1046/j.1365-313x.2003.01664.x. [DOI] [PubMed] [Google Scholar]
- Jonak C, Okresz L, Bogre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Current Opinion in Plant Biology. 2002;5:415–424. doi: 10.1016/s1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- Kiegerl S, Cardinale F, Siligan C, et al. SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. The Plant Cell. 2000;12:2247–2258. doi: 10.1105/tpc.12.11.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh K, Torii S, Nishida E. Control of MAP kinase signaling to the nucleus. Chromosoma. 2005;114:86–91. doi: 10.1007/s00412-005-0341-9. [DOI] [PubMed] [Google Scholar]
- Kong X, Pan J, Zhang M, Xing X, Zhou Y, Liu Y, Li D. ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis . Plant, Cell and Environment. 2011;34:1291–1303. doi: 10.1111/j.1365-3040.2011.02329.x. [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proceedings of the National Academy of Sciences, USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Rudd JJ, Macioszek VK, Scheel D. Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. Journal of Biological Chemistry. 2004;279:22440–22448. doi: 10.1074/jbc.M401099200. [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RA, Ritsema T, Pieterse CM. Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiology. 2009;149:1797–1809. doi: 10.1104/pp.108.133926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang XD, Zhao X, Dutt Y. Improvement of cotton fiber quality by transforming the acsA and acsB genes into Gossypium hirsutum L. by means of vacuum infiltration. Plant Cell Reports. 2004;22:691–697. doi: 10.1007/s00299-003-0751-1. [DOI] [PubMed] [Google Scholar]
- MAPK Group. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends in Plant Science. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk M, Awotunde OS, Muszynska G, Klessig DF, Dobrowolska G. Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. The Plant Cell. 2000;12:165–178. [PMC free article] [PubMed] [Google Scholar]
- Miles GP, Samuel MA, Ellis BE. Suppression of MKK5 reduces ozone-induced signal transmission to both MPK3 and MPK6 and confers increased ozone sensitivity in Arabidopsis thaliana . Plant Signaling Behavior. 2009;4:687–692. doi: 10.4161/psb.4.8.9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Ichimura K, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K. Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Letters. 1998;437:56–60. doi: 10.1016/s0014-5793(98)01197-1. [DOI] [PubMed] [Google Scholar]
- Ning J, Li X, Hicks LM, Xiong L. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiology. 2010;152:876–890. doi: 10.1104/pp.109.149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley KF, Martin GB. Identification of MAPKs and their possible MAPK kinase activators involved in the Pto-mediated defense response of tomato. Journal of Biological Chemistry. 2004;279:49229–49235. doi: 10.1074/jbc.M410323200. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H. Disentangling the complexity of mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiology. 2009;149:606–615. doi: 10.1104/pp.108.131557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviv Z, Kalie E, Seger R. MEK5 and ERK5 are localized in the nuclei of resting as well as stimulated cells, while MEKK2 translocates from the cytosol to the nucleus upon stimulation. Journal of Cell Science. 2004;117:1773–1784. doi: 10.1242/jcs.01040. [DOI] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S. A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis . Proceedings of the National Academy of Sciences, USA. 2008;105:5638–5643. doi: 10.1073/pnas.0711301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis . Journal of Biological Chemistry. 2002;277:559–565. doi: 10.1074/jbc.M109495200. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annual Review of Plant Biology. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Samuel MA, Chaal BK, Lampard G, Green BR, Ellis BE. Surviving the passage: non-canonical stromal targeting of an Arabidopsis mitogen-activated protein kinase kinase. Plant Signaling Behavior. 2008;3:6–12. doi: 10.4161/psb.3.1.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, An HL, Zhang L, Gao Z, Guo XQ. GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Molecular Biology. 2010;74:1–17. doi: 10.1007/s11103-010-9661-0. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhang L, An H, Wu C, Guo X. GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Molecular Biology. 2011;12:22. doi: 10.1186/1471-2199-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host and Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proceedings of the National Academy of Sciences, USA. 2007;104:18842–18847. doi: 10.1073/pnas.0708139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Nasir KH, Ito A, Kanzaki H, Matsumura H, Saitoh H, Fujisawa S, Kamoun S, Terauchi R. A high-throughput screen of cell-death-inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1-induced cell death signaling and non-host resistance to Pseudomonas cichorii . The Plant Journal. 2007;49:1030–1040. doi: 10.1111/j.1365-313X.2006.03022.x. [DOI] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis . Molecular Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Tuteja N. Abscisic acid and abiotic stress signaling. Plant Signaling Behavior. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhang Y, Wang J, Wu X, Guo X. A novel MAP kinase gene in cotton (Gossypium hirsutum L.), GhMAPK, is involved in response to diverse environmental stresses. Journal of Biochemistry and Molecular Biology. 2007;40:325–332. doi: 10.5483/bmbrep.2007.40.3.325. [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J. AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis . Journal of Experimental Botany. 2007;58:2969–2981. doi: 10.1093/jxb/erm144. [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis . The Plant Journal. 2008;54:440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis . Journal of Biological Chemistry. 2008;283:26996–27006. doi: 10.1074/jbc.M801392200. [DOI] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S. Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proceedings of the National Academy of Sciences, USA. 2001;98:741–746. doi: 10.1073/pnas.98.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Du H, Klessig DF. Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. The Plant Cell. 1998;10:435–450. doi: 10.1105/tpc.10.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF. MAPK cascades in plant defense signaling. Trends in Plant Science. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dai Y, Xiong Y, DeFraia C, Li J, Dong X, Mou Z. Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. The Plant Journal. 2007;52:1066–1079. doi: 10.1111/j.1365-313X.2007.03294.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.