Abstract
Seed yield and oil content are two important agricultural characteristics in oil crop breeding, and a lot of functional gene research is being concentrated on increasing these factors. In this study, by differential gene expression analyses between rapeseed lines (zy036 and 51070) which exhibit different levels of seed oil production, BnGRF2 (Brassica napus growth-regulating factor 2-like gene) was identified in the high oil-producing line zy036. To elucidate the possible roles of BnGRF2 in seed oil production, the cDNA sequences of the rapeseed GRF2 gene were isolated. The Blastn result showed that rapeseed contained BnGRF2a/2b which were located in the A genome (A1 and A3) and C genome (C1 and C6), respectively, and the dominantly expressed gene BnGRF2a was chosen for transgenic research. Analysis of 35S-BnGRF2a transgenic Arabidopsis showed that overexpressed BnGRF2a resulted in an increase in seed oil production of >50%. Moreover, BnGRF2a also induced a >20% enlargement in extended leaves and >40% improvement in photosynthetic efficiency because of an increase in the chlorophyll content. Furthermore, transcriptome analyses indicated that some genes associated with cell proliferation, photosynthesis, and oil synthesis were up-regulated, which revealed that cell number and plant photosynthesis contributed to the increased seed weight and oil content. Because of less efficient self-fertilization induced by the longer pistil in the 35S-BnGRF2a transgenic line, Napin-BnGRF2a transgenic lines were further used to identify the function of BnGRF2, and the results showed that seed oil production also could increase >40% compared with the wild-type control. The results suggest that improvement to economically important characteristics in oil crops may be achieved by manipulation of the GRF2 expression level.
Keywords: BnGRF2, chlorophyll, leaf morphology, oil production, seed size
Introduction
Vegetable oil is an important edible product and its use as an industrial resource is increasing, particularly as a source for biodiesel (Graef et al., 2009; Lu et al., 2011; Rogalski and Carrer, 2011). With the recent population growth and the developing global economy, the demand for vegetable oil has risen sharply (Lu et al., 2011). Over 22 Mt of rapeseed (Brassica napus) oil was produced globally during 2009–2010 and it represented the third largest source of vegetable oil (Tan et al., 2010). Therefore, increasing rapeseed oil production is of paramount importance for the supply of vegetable oil for food and non-food applications (Weselake et al., 2009). The amount of oil production per seed is determined by seed weight and oil content. Therefore, most genetic studies and breeding programmes on oilseed crops focus on elevating seed weight and/or oil content.
Seed weight, also characterized as seed mass or seed size generally, has been widely accepted as a complex trait controlled by polygenes. In plants, organ morphogenesis and size are the result of coordinated patterns of cell proliferation, expansion, and differentiation (Mizukami and Fischer, 2000; Horiguchi et al., 2005; Kozuka et al., 2005; Dupuy et al., 2010). After extensive studies over a long time, many genes which influence cell proliferation and expansion in seed development have been identified. In Arabidopsis, knowledge of most genes controlling seed mass development came from analyses of mutants. Several factors regulating seed size have been identified (Berger et al., 2006). Seed size is increased in ap2 or arf2/mnt mutants (Jofuku et al., 2005; Ohto et al., 2005; Schruff et al., 2006) and reduced in ttg2 mutants (Johnson et al., 2002; Garcia et al., 2005), and this effect appears to take place in the integument surrounding the seed. In addition to the integument, endosperm size has been found to play a major role in regulation of seed size (Berger et al., 2006). Loss-of-function of the WRKY transcription factor gene MINI3 and the leucine-rich repeat (LRR) kinase gene IKU2 (Garcia et al., 2003; Luo et al., 2005), which reduce seed mass, stop the nuclear divisions associated with cellularization. Some genes from quantitative trait loci (QTL) that influenced seed weight have been cloned in a number of crop plants, for example GS3 (Fan et al., 2006; Takano-Kai et al., 2009; Mao et al., 2010), GW2 (Song et al., 2007), qSW5 or GW5 (Shomura et al., 2008; Weng et al., 2008), and GIF1 (Wang et al., 2008) in rice, and SW4.1 in tomato (Orsi et al., 2009). In oilseed crops, although many QTLs that influence seed weight have been mapped, the specific genes associated with these QTLs have not been identified so far (Teng et al., 2009; Chen et al., 2011).
In oilseed plants, seed storage oils are composed primarily of triacylglycerol (TAG), which is synthesized from glycerol-3-phosphate and fatty acids (FAs) (Ohlrogge and Browse, 1995; Voelker and Kinney, 2001). The pathways of biosynthesis of FAs and TAG have been well characterized (Ohlrogge and Browse, 1995; Harwood, 1996; Beisson et al., 2003). Over the past 20 years, the model plant Arabidopsis thaliana and the oil crop rapeseed have been used as part of a concerted research effort centred on genes related directly to seed oil biosynthesis, including those involved in energy metabolism (PDHK and G6PDH), FA biosynthesis (ACCase and G3PDH), and the TAG synthesis pathway (GPAT, LPAAT, and DGAT) (Baud and Lepiniec, 2009; Weselake et al., 2009). However, since oil synthesis is a highly coordinated process that involves carbon metabolism, FA synthesis, and TAG synthesis pathways, manipulation of a single gene may not be an efficient approach for elevating seed oil content significantly (Thelen and Ohlrogge, 2002; Cahoon et al., 2007; Weselake et al., 2009). In recent years, candidate genes encoding seed-specific transcription factors such as GL2, WRI1, LEC1, and LEC2 in Arabidopsis and BnWRI1, BnLEC1, and BnL1L in rapeseed have been shown to play important roles in regulation of FA biosynthesis (Shen et al., 2006; Baud et al., 2007; Mu et al., 2008; Santos-Mendoza et al., 2008; Liu et al., 2010; Tan et al., 2010). Therefore, oil synthesis-related transcription factors might represent promising targets for producing a higher oil yield in oil crops through genetic engineering (Ekman et al., 2008).
GRF (growth-regulating factor) was first reported to perform a regulatory role in rice stem growth, and sequence analyses indicated that it might function as a transcription factor (Van der Knaap et al., 2000). Investigation of GRF-interacting factors (GIFs) suggests that GRFs may act as transcriptional activators (Kim and Kende, 2004). The conspicuous function of Arabidopsis GRFs was proved in development of leaves, cotyledons, and floral organs (Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Liu et al., 2009). Some researchers showed that GRFs functioned in Arabidopsis female reproductive development and ovule formation (Wynn et al., 2011) and were also expressed in rice embryo (Ye et al., 2004) and maize ear (Zhang et al., 2008), which suggested the possible roles of GRFs in seed development. In the present study, from microarray analyses of 20 d ovules in rapeseed lines with high and low levels of oil production (zy036 and 51070, respectively), a high expression level of a GRF2-like gene was observed in zy036, which suggested that this gene might have effects in controlling seed oil production. By transgenic work, it was proved that overexpression of BnGRF2 could increase oil production by inceasing the seed mass and oil content. Furthermore, by analysing its downstream genes, it was possible to explore how the gene acts on seed oil production.
Materials and methods
Plant growth conditions
Wild-type (WT) Col-2 and BnGRF2 transgenic Arabidopsis plants were grown in pots with compost soil. Seeds were pre-incubated in the dark for 3 d at 4 °C before transferring to a growth room with a continuous artificial light period of 16 h (24 °C) and a dark period of 8 h (22 °C) at a photon flux density of 100–120 μmol m−2 s−1. WT Arabidopsis and BnGRF2 transgenic plants were grown side by side in the same container to minimize variables that arise from differences in the microenvironment of the growth room.
Vector construction and plant transformation
BLAST was used to compare the Arabidopsis GRF2 sequence against Brassica rapa and Brassica oleracea genome databases, and two homologues of GRF2 were identified in each species. Fragments encoding BnGRF2a and BnGRF2b were amplified from zy036 with the primers designed against the Brassica coding sequences using an RT-PCR kit (Qiagen, Düsseldorf, Germany). PCR amplifications were carried out with 35 cycles of 94 °C for 30 s, 58 °C for 90 s, and 72 °C for 90 s. The amplicons were cloned into the Gateway entry vector pCR/GW/TOPO (Invitrogen, Carlsbad, CA, USA) using TA overhangs. Proper orientation and integrity were confirmed by sequencing with the BnGRF2aR and M13 primers.
To construct the expression plasmid pEarleyGate100-BnGRF2a, the BnGRF2a coding region was transferred from pCR/GW/TOPO to the pEarleyGate100 (Invitrogen) using the Gateway™ LR Clonase™ Enzyme Mix (Invitrogen). The construct was confirmed by PCR with the Cauliflower mosaic virus (CaMV) 35S promoter primer pCaMVp and BnGRF2aR.
For construction of the napin promoter vector, the BnGRF2a coding region was obtained with the primers BnGRF2aSmaI and BnGRF2aBamHI. PCR amplification was carried out with 30 cycles of 94 °C for 30 s, 60 °C for 90 s, and 72 °C for 90 s, followed by digestion with SmaI and BamHI, and ligation into the pBI vector (reconstructed with the napin promoter). The constructed vector was confirmed by PCR with primers NapinP and BnGRF2aR. Arabidopsis was transformed with Agrobacterium tumefaciens strain GV3101 using the floral dip method (Clough and Bent, 1998). Transformants expressing the bar resistance gene were selected and confirmed by PCR with pCaMV, NapinP, and BnGRF2R primers. Homozygous lines overexpressing BnGRF2a were selected for phenotypic and microarray analyses. Primers used for gene isolation and expression vector confirmation are listed in Supplementary Table S1 available at JXB online (the underlined bases indicate additional restriction sites for SmaI and BamHI).
Real-time RT-PCR
Total RNA was extracted from rapeseed tissues and Arabidopsis tissues (young leaf of 20 d after germinating, fully expanded adult leaf and silique of 10 d after flowering) using the Plant Mini RNeasy kit (Qiagen). The reverse transcription reaction was performed using the First Strand cDNA Synthesis Kit for RT-PCR (Takara, Dalian, China). Primers (BnGRF2aRTf, BnGRF2aRTr, BnGRF2bRTf, and BnGRF2bRTr) were designed to detect expression of BnGRF2a and BnGRF2b in rapeseed. Rapeseed β-actin2 and β-actin3 (Gao et al., 2004; Weng et al., 2005) served as endogenous reference genes and were amplified using the primers Bnactin2-F, Bnactin2-R, and Bnactin3F, Bnactin3R. Primers for BnGRF2a (BnGRF2aRTf and BnGRF2aRTr) and AtGRF2 (AtGRF2RTf and AtGRF2RTr) were used to detect BnGRF2a expression levels in transgenic Arabidopsis. Arabidopsis β-actin1 (At2g37620) and UBQ10 (At4g05320) served as an endogenous reference genes and were amplified using the primers AtactinF1, AtactinR, and AtUBQ10F, AtUBQ10R (Czechowski et al., 2005; Rus et al., 2006). The real-time PCR contained 1 μl of 10-fold diluted first-strand cDNA, 1 μl of SYBR Green (Applied Biosystems, Carlsbad, CA, USA), 0.2 mM dNTP, 1× LA buffer, 0.5 U of LA Taq (Takara) and 10 μM of each primer. Initial denaturion time was 10 min, followed by 40 cycles of 95 °C for 15 s, 58 °C for 30 s, and 72 °C for 30 s. A melting curve was performed after PCR cycles to verify that only one PCR product was amplified. Experiments were performed in triplicate and data were obtained from three independent experiments with similar results. The absolute slope values of the curve of log cDNA dilution versus ΔC(T) were assessed and the efficiencies of primers for target and reference genes were considered equal if the absolute slopes were <0.1. Primers used for real-time PCR are listed in Supplementary Table S1 at JXB online.
Insertion site analyses of transgenic lines
Leaf DNA from two transgenic lines, 35S-BnGRF2a-2 and 35S-BnGRF2a-10, was extracted using the Plant DNA extraction kit (Qiagen). Random primers AD1 and AD2 as well as vector primers LBR1, LBR2, and LBR3 were used for TAIL (thermal asymmetric interlaced) PCR. Cycle settings in PCR amplifications were carried out according to the descriptions in Liu et al. (1995).
Pleiotropic phenotypes of transgenic Arabidopsis leaves and seeds
Leaves were excised from WT Col-2, 35SBnGRF2a-2, and 35SBnGRF2a-10 plants using a razor blade and scanned to acquire digital images for determination of the surface area. As described previously (Tsuge et al., 1996), the leaves were fixed with FAA and cleared with chloral hydrate solution (200 g of chloral hydrate, 20 g of glycerol, and 50 ml of dH2O). Leaf cells were observed by differential interference contrast (DIC) microscopy (DM6000B; Leica Microsystems, Japan). Seeds from the WT and the Napin-BnGRF2a-3 line were photographed under a dissection microscope, weighed, and measured. Mature embryos were imbibed for 1 h and dissected under a microscope according to the description in Ohto et al. (2005) with slight modification, and photographed with an Olympus compound microscope. Seed oil contents were detected by NMR PQ001 (Niumag, China).
Chlorophyll content and photosynthesis detection
Tissue samples (1.0 g) were obtained from fully expanded leaves and 15 d siliques after flowering. At least three replicates were taken from different plants of each line. After freezing in liquid nitrogen, the samples were ground to a power in 50 ml of extraction solution (2:1 acetone:95% alcohol) and incubated in the dark at 4 °C overnight to ensure complete extraction of chlorophyll. The cell debris was pelleted by centrifugation for 1 min at 15 000 g, and absorbance of the supernatant was measured at 646.6, 663.6, and 750 nm, as described previously (Porra et al., 1989). Chlorophyll content was expressed in micrograms. The photosynthetic parameters were measured with a portable photosynthesis system LI-6400XT (LI-COR Biosciences, Lincoln, NE, USA) in a growth room. The measurement conditions were as follows: 24 °C, a photon flux density of 100–120 μmol m−2 s−1, and 40–60% humidity. All data were determined as the means of five plants.
Microarray analyses
Microarray analyses were performed with WT and 35S-BnGRF2-overexpressing plants by the Agilent Arabidopsis Oligo DNA microarray. Young leaves were harvested from 20-day-old Arabidopsis plants after germination and frozen immediately in liquid nitrogen. Total RNA was extracted using the RNeasy plant mini kit (Qiagen). Spectrophotometric analyses of the isolated total RNA were carried out at 260 nm and 280 nm to determine sample concentration and purity. Regulated genes were identified with a stringent significance threshold, namely a mean >1.5-fold change (transgenic relative to WT control samples) and a P value ≤0.01, based on at least three out four replicates. Primers used in real-time PCR experiments of six genes are listed in Supplementary Table S1 at JXB online.
Results
Sequence analyses of BnGRF2 from the rapeseed line zy036
The amount of oil per seed from rapeseed lines zy036 and 51070 (2.14 mg and 1.38 mg per seed, respectively) has been reported (Hua et al., 2012). Gene expression analyses of 20 d ovules from zy036 and 51070 showed differential expression of BnGRF2 (homologous to Arabidopsis AtGRF2, At4g37740) (Supplementary Table S2 at JXB online). To isolate the gene for functional analyses, the Arabidopsis GRF2 cDNA sequence was compared with the Brassica genomic databases: B. rapa (Wang et al., 2011) and B. oleracea (scaffold sequences, unpublished) using BLASTN. The results indicated that rapeseed contained two GRF2 homologues named GRF2a/2b which are located in the A genome (A1 and A3 chromosome) and the C genome (C1 and C6 chromosome), respectively. With primers designed against Brassica GRF2a/2b, the coding region sequences of BnGRF2a (GenBank accession nos JN831660 and JN831661) and BnGRF2b (GenBank accession nos JN831662 and JN831663) were isolated from 20 d ovules of zy036. As shown in Fig. 1, the predicted polypeptides encoded by BnGRF2a/2b were homologous to AtGRF2, with 65% and 69% identity, respectively. Similar to the other members of the AtGRF family, BnGRF2 contains highly conserved QLQ and WRC domains, with the cysteine (C) and histidine (H) residues of the C3H motif, as well as the TQL and GGPL motifs (Kim et al., 2003).
Fig. 1.
Sequence alignment of AtGRF2 and BnGRF2. Black boxes indicate the QLQ and WRC domains. The cysteine (C) and histidine (H) residues of the C3H motif in the WRC domain are shown with a black dot. The TQL and GGPL motifs are highlighted with ellipses. (This figure is available in colour at JXB online.)
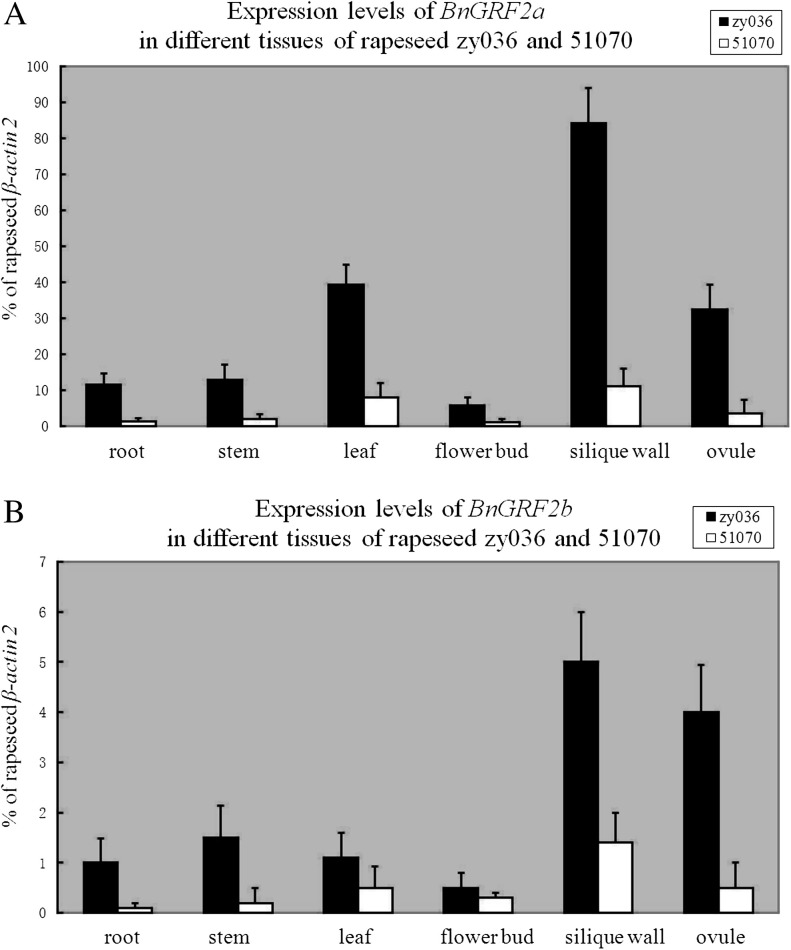
To investigate the BnGRF2 expression pattern in detail, BnGRF2a/2b transcript levels were analysed in different rapeseed tissues (root, stem, leaf, flower bud, silique wall, and ovule) in zy036 and 51070. Real-time PCR results confirmed differential expression of the BnGRF2 gene in the ovule, with the expression level of BnGRF2a/2b >8-fold higher in zy036 than in 51070. Differential BnGRF2a/2b expression levels were also observed in the other five tissues, with the highest and lowest levels being found in the silique wall and flower bud, respectively (Fig. 2; Supplementary Fig. S1 at JXB online). Moreover, there were 10- to 30-fold higher expression levels of BnGRF2a than BnGRF2b in different tissues, which suggested that BnGRF2a was the dominant expressed gene compared with BnGRF2b and should be chosen for further research.
Fig. 2.
Real-time PCR analyses of the BnGRF2 gene in different tissues (root, stem, leaf, flower bud, silique wall, and ovule) between rapeseed lines zy036 and 51070. (A) Expression analyses of BnGRF2a. (B) Expression analyses of BnGRF2b. Data expression was normalized to rapeseed β-actin2, and expression levels of BnGRF2 were compared with that of β-actin2. Data presented are mean values of three biological replicates, and error bars represent standard deviations.
Transformation of Arabidopsis with 35S-BnGRF2a and identification of transgenic lines
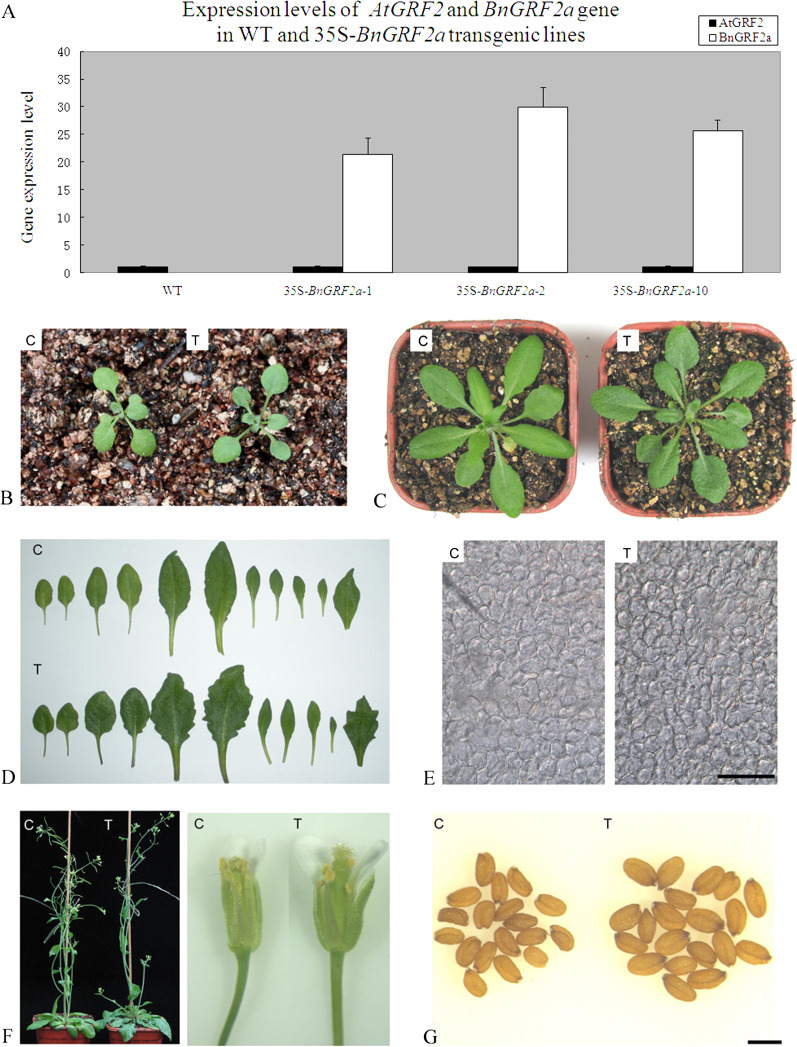
With the aim of studying the possible function of BnGRF2, Arabidopsis plants were transfected with BnGRF2a under the control of the CaMV 35S promoter. After three rounds of Basta selection, 21 independent homogenous T3 transgenic lines were obtained. Total RNA from young leaves of independent transgenic Arabidopsis lines was extracted for quantitative RT-PCR. The results showed that BnGRF2a was highly expressed in all the selected lines and the transgenic plants with phenotypes different from that of the WT exhibited higher expression levels of BnGRF2 (Fig. 3A). Insertion sites of the transgenic lines were identified to confirm the successful transformation of BnGRF2a. Sequence analyses of insertion sites indicated that the vector sequence inserted into different Arabidopsis chromosomes. For example, the BnGRF2a gene was inserted in chromosomes 1 and 2 for 35S-BnGRF2a-2 and 35S-BnGRF2a-10, respectively (Supplementary Fig. S2 at JXB online).
Fig. 3.
Phenotype analyses induced by overexpression of BnGRF2a in Arabidopsis. (A) Real-time PCR analyses of BnGRF2a expression levels in the adult leaves of 35S-BnGRF2a transgenic Arabidopsis lines. Data expression was normalized to Arabidopsis β-actin1 (At2g37620) and relative to the expression of AtGRF2. Data presented are mean values of three biological replicates and error bars represent standard deviations. (B–D) Leaf phenotypes at different developmental stages (B, 2 weeks after germination; C, bolting time; D, flowering time). (E) Paradermal view of palisade cells in the subepidermal layer of adult leaves in the wild type and the 35S-BnGRF2a-2 transgenic line. Bar, 100 μm. (F) The longer pistil induced near sterility in 35S-BnGRF2a-2 transgenic Arabidopsis. (G) A BnGRF2a transgenic plant produces large seeds (shown are mature dried seeds from the wild type and the 35S-BnGRF2a-2 transgenic line). C, wild-type control; T, transgenic plant. Bar, 500 μm. (This figure is available in colour at JXB online.)
Pleiotropic phenotypes in 35S-BnGRF2a transgenic Arabidopsis
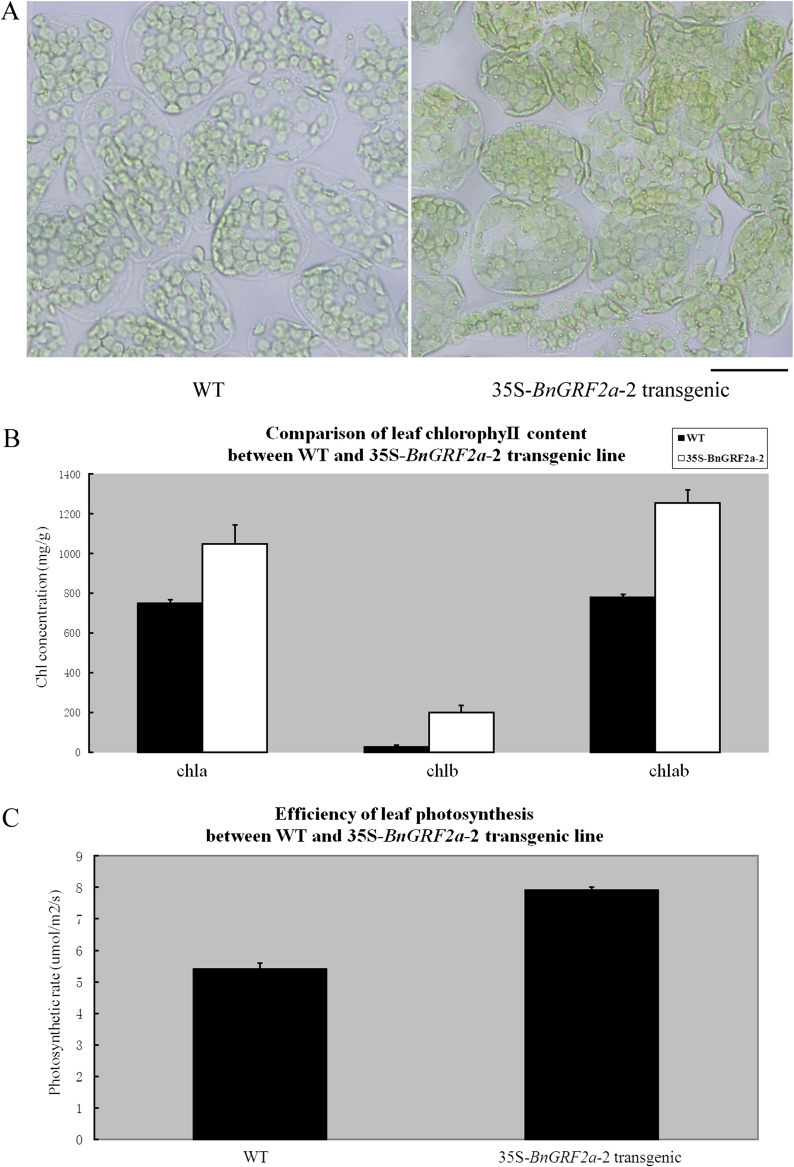
The 35S-BnGRF2a transgenic lines showed clear morphological differentiation enabled them to be distinguished from the WT at ∼2 weeks after germination, with transgenic cotyledons exhibiting a longer petiole (Fig. 3B, C). At the flowering stage, the leaf area of transgenic plants was 20–30% larger than that of the WT control (Fig. 3D, Table 1). Leaf width and tooth morphology were also affected, while the lengths of blades and petioles similar to those of the control (Fig. 3D). A DIC microscope was used to compare palisade cells in the subepidermal layers of mature leaves of WT control and transgenic plants. Since no significant difference in epidermal cell size was observed, it was likely that the enlarged leaf area of BnGRF2a-overexpressing plants was mediated directly by an increase in cell number (Fig. 3E). In addition, the fully expanded leaves of transgenic lines contained more chloroplasts and higher chlorophyll contents than those of the WT control (Fig. 4A, B). Consistent with this result, it was not surprising to find that the photosynthetic rate of transgenic leaves was >40% higher than that of the WT control (Fig. 4C).
Table 1.
Phenotypic parameters including bolting time, flowering time, leaf number, leaf area, and seed characters in wild-type control and 35S-BnGRF2a transgenic lines
| WT | 35S-BnGRF2a−2 | 35S-BnGRF2a−10 | |
| Bolting time (d) | 20.9±0.5 | 25.7±0.8 | 27.8±0.6 |
| Flowering time (d) | 25.1±0.3 | 30.3±0.5 | 32.8±0.9 |
| No. of leaves before flowering | 10.2±0.6 | 11.3±0.2 | 10.0±0.3 |
| Adult leaf area (cm2) | 3.79±0.26 | 4.63±0.12 | 5.17±0.11 |
| Seed weight (mg/100 seeds) | 1.9±0.25 | 2.9±0.21 | 3.0±0.30 |
| No. of cells in a leaf | 134 100±9200 | 163 800±4200 | 182 900±3900 |
| Seed oil production (mg/100 seeds) | 0.59±0.07 | 0.91±0.09 | 1.01±0.12 |
Fig. 4.
The detection of chloroplast (A), chlorophyll content (B), and photosynthetic efficiency (C) in fully expanded leaves of 4-week-old plants of the wild-type (WT) control and the 35S-BnGRF2a-2 line. Bar, 20 μm. Data presented in B and C are mean values of three biological replicates and error bars represent standard deviations. (This figure is available in colour at JXB online.)
The bolting and flowering times of transgenic lines were delayed by ∼5 d (Fig. 3E, Table 1). The flower buds of transgenic Arabidopsis had a longer pistil than the WT, but developed a normal stamen, which resulted in less efficient self-fertilization and thus they were nearly sterile (Fig. 3F). Transgenic plants developed greater seed mass (obtained by artificial pollination) with weight increases of >40% and oil production increases of >50% compared with the WT control (Fig. 3G, Table 1). However, reduced fertility in 35S-BnGRF2 transgenic lines led to the question of whether the large seed mass phenotype could result simply from allocation of extra resources to the few seeds produced. To determine whether the larger seed size is caused by reduced siliques or a higher BnGRF2a expression level, all flowers on a WT plant were removed, with the exception of three flowers on each secondary inflorescence. Seeds from 35S-BnGRF2 transgenic plants were heavier (2.9±0.21 mg and 3.0±0.30 mg, respectively) than those from the deflowered WT control (2.3±0.29 mg), even though the seed number in the silique was comparable. Together, these results suggest that the effect of BnGRF2a on seed mass is not primarily due to its effect on fertility in 35S-BnGRF2 transgenic lines.
Seed analyses in Napin-BnGRF2a transgenic lines
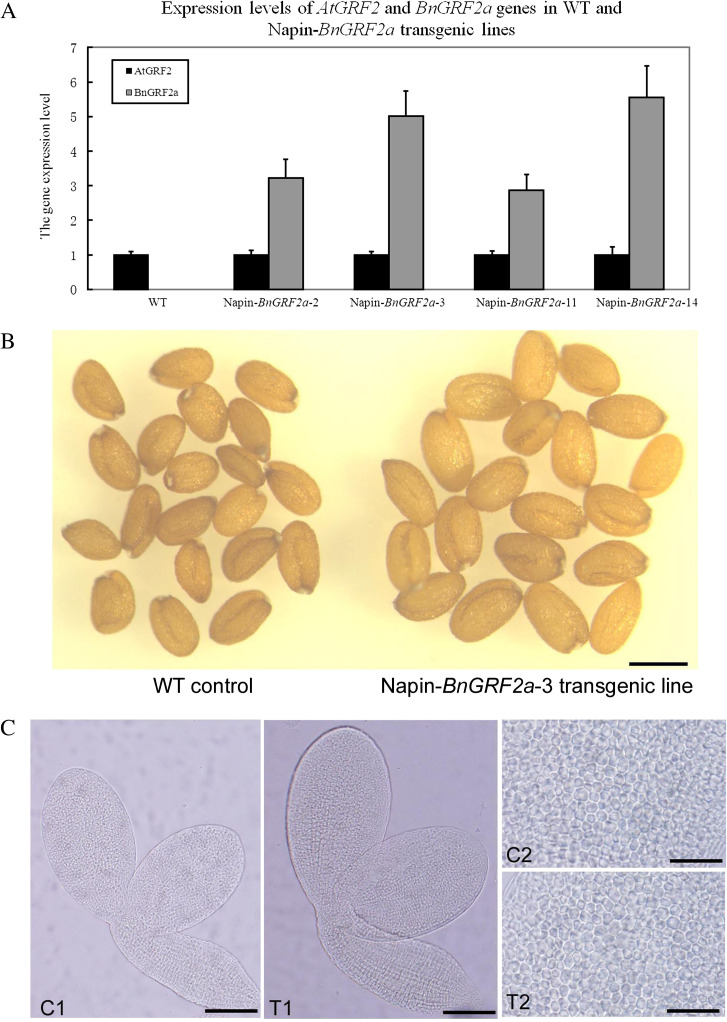
To confirm further the effect of BnGRF2a on seed mass, an overexpression vector driven by the napin promoter was constructed. Following transformation and basta selection of Arabidopsis, 31 independent homogenous transgenic lines were obtained. Among four lines, seed sizes were increased to different degrees, and quantitative RT-PCR analyses of the transgenic siliques (10 d after flowering) indicated that these lines exhibited high levels of BnGRF2a expression (Fig. 5A).
Fig. 5.
Analyses to BnGRF2a expression levels and seed phenotypes in Napin-BnGRF2a Arabidopsis lines. (A) Relative gene expression of BnGRF2a in the silique (10 d after flowering) of the transgenic Napin-BnGRF2a Arabidopsis line was identified by qRT-PCR. Data presented are mean values of three biological replicates, and error bars represent standard deviations. Data expression was normalized to β-actin1 (At2g37620) and relative to the expression of AtGRF2. (B) Mature seeds obtained from mature dried seeds of the wild type and the Napin-BnGRF2a-3 line. Bar, 500 μm. (C) Mature embryos obtained from mature dried seeds of the wild type (C1) and the Napin-BnGRF2a-3 line (T1). Bar, 240 μm. Epidermal cell layer in the central region of cotyledons from embryos of the wild type (C2) and the Napin-BnGRF2a-3 line (T2). Bar, 80 μm. (This figure is available in colour at JXB online.)
Phenotypic analyses showed the obviously increased widths of the cotyledons, and the hypocotyl generated larger seeds in the Napin-BnGRF2a-3 line, which increased the cotyledon area resulting in it being ∼1.4-fold larger than that of the WT (Fig. 5B, C, Table 2). Comparison of the epidermal cell layer from the central region of the cotyledon showed no significant difference in cell size between transgenic Arabidopsis and the WT (Fig. 5C, Table 2). Compared with the control seeds, the transgenic seed showed a >30% increase in weight without change in seed number per silique, which means seed yield could increase >30%. The chlorophyll content of 15 d ovules was >20% higher in the Napin-BnGRF2a-3 line and seed oil production in mature seed was >40% higher (Table 2).
Table 2.
Comparison of seed characteristics between wild-type Arabidopsis and Napin-BnGRF2a transgenic lines
| Genotype | Weight (mg/100 seeds) | Oil production (mg/100 seeds) | Dimensions (length×width, mm) | Area per cotyledon (mm2) | No. of cells in a cotyledon | Chlorophyll content (mg/g) |
| Col-2 WT | 1.9±0.24 | 0.61±0.08 | 0.40±0.020×0.23±0.011 | 0.09±0.01 | 717±80 | 0.29±0.04 |
| Napin-BnGRF2a−3 | 2.5±0.19 | 0.86±0.07 | 0.43±0.023×0.30±0.025 | 0.13±0.01 | 1035±80 | 0.35±0.06 |
| Napin-BnGRF2a−14 | 2.4±0.27 | 0.92±0.10 | 0.42±0.018×0.29±0.017 | 0.12±0.02 | 955±160 | 0.39±0.01 |
Microarray analyses of BnGRF2a-regulated pathways
To investigate the genome-wide effects on transcription resulting from BnGRF2a overexpression, microarray experiments were performed on three independent biological replicates of 35S-BnGRF2a and WT control plants. RNA from young leaves of 4-week-old plants was chosen for gene expression analyses. An overview of the differential gene expression between the two genotypes is presented in Supplementary Table S3 at JXB online. Genetic up-regulation of the BnGRF2a expression level in Arabidopsis resulted in increased expression of 1601 genes and decreased expression of 1234 genes.
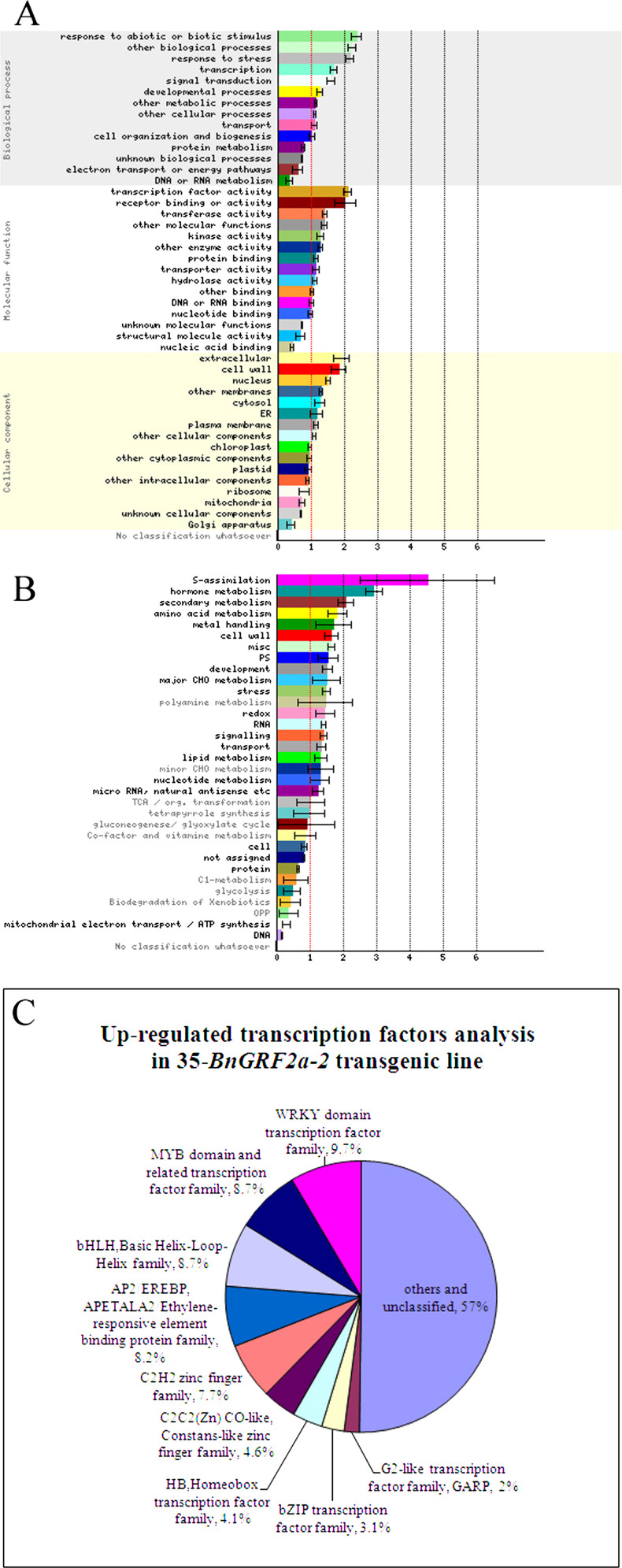
To classify and analyse the genes affected by BnGRF2a, the differentially expressed genes were submitted to an enrichment analyses (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi). When the genes were classified according to Gene Ontology (GO) terms relating to biological process, cellular compartment, and molecular function, transcription factor-related terms were highly represented and significantly enriched (Supplementary Table S4 at JXB online; Fig. 6A). When classified with MapMan, 313 of 345 differentially expressed transcription factor genes were classified into 49 gene families (Fig. 6B). Among the up-regulated transcription factors, 42.9% (84/196) belonged to five main families, namely the WRKY domain family, the MYB domain and related transcription factor family, the basic helix–loop–helix (bHLH) family, the AP2/EREBP family, and the C2H2 zinc finger family (Fig. 6C). In addition to transcription factors, the regulated genes were mainly involved in several different metabolic processes relating to hormones, secondary substances, carbohydrates, lipids, and amino acids (Fig. 6B).
Fig. 6.
Analyses of dfferentially expressed genes induced by the BnGRF2a gene in Arabidopsis. (A) Gene Ontology (GO) analyses of differentially expressed genes. (B) Differentially expressed genes classified with MapMan. (C) Analyses of transcription factors up-regulated in the BnGRF2 transgenic line. (This figure is available in colour at JXB online.)
Up-regulated genes in possible BnGRF2-regulated pathways
Corresponding to the pleiotropic phenotypes of the transgenic lines, there was up-regulation of a number of genes that were thought to be involved in regulatory pathways (Supplementary Table S5 at JXB online; Table 3). In 35S-BnGRF2a and Napin-BnGRF2a transgenic lines, overexpression of BnGRF2a resulted in an enlarged leaf and cotyledon area, respectively, which were induced by increasing cell numbers. According to microarray analyses, some genes related to cell division [PHB3 (At5g40770), RCC1 (At3g53830, At1g69710), and MAPKKK16 (At4g26890)] and the cell cycle [CYCH;1 (At5g27620), CYCA2;1 (At5g25380), CYCA2;3 (At1g15570), CYCD4;2 (At5g10440), CYCD6;1 (At4g03270), and CYCP4;1 (At2g44740)] were up-regulated in the BnGRF2a transgenic lines (Table 3).
Table 3.
List of up-regulated genes related to pleiotropic phenotypes (cell number, photosynthesis, and lipid and fatty acid synthesis) in the 35S-BnGRF2a-2 transgenic Arabidopsis line
| Gene ID | Short discription | GO function | Subcellar localization | Transcript level |
| Cell division and cycle | ||||
| At1g15570 | CYCA2;3, cyclin-dependent protein kinase regulator | Cell cycle regulation | Nucleus | 1.68 |
| At1g69710 | Putative regulator of chromosome condensation (RCC1) family protein | Chromatin binding | Cellular component | 2.03 |
| At2g44740 | CYCP4;1, cyclin-dependent protein kinase | Cell cycle regulation | Nucleus | 1.82 |
| At3g53830 | Regulator of chromosome condensation (RCC1) family protein/UVB-resistance protein-related | Chromatin binding | Cellular component | 1.61 |
| At4g03270 | CYCD6;1, cyclin-dependent protein kinase | Cell cycle regulation | Nucleus | 1.74 |
| At4g26890 | MAPKKK16 (mitogen-activated protein kinase kinase kinase 16); kinase | Protein tyrosine kinase activity | 1.87 | |
| At5g10440 | CYCD4;2, cyclin-dependent protein kinase | Cell cycle regulation | Nucleus | 1.75 |
| At5g25380 | CYCA2;1, cyclin-dependent protein kinase regulator | Cell cycle regulation | Nucleus | 2.58 |
| At5g27620 | CYCH;1, cyclin-dependent protein kinase/protein binding/protein kinase | Cell cycle regulation | Nucleus | 1.80 |
| At5g40770 | ATPHB3 (prohibitin 3) | Cell division | 1.60 | |
| Photosynthesis | ||||
| At1g15820 | LHCB6 (light-harvesting complex PSII) | Photosynthesis | Plastoglobule | 1.61 |
| At1g20780 | armadillo/beta-catenin repeat protein-related/U-box domain-containing protein | Regulation of chlorophyll biosynthetic process | Plasma membrane | 1.64 |
| At1g27730 | STZ (salt tolerance zinc finger); nucleic acid binding/transcription factor/zinc ion binding | Photosynthesis | Nucleus | 5.50 |
| At1g44446 | CH1 (chlorophyll b biosynthesis); chlorophyllide a oxygenase | Chlorophyll biosynthetic process | Thylakoid membrane | 1.68 |
| At1g68190 | Zinc finger (B-box type) family protein | Transcription regulation | Intracellular | 1.64 |
| At2g05100 | LHCB2.1 (photosystem II light-harvesting complex gene) | Photosynthesis | Chloroplast thylakoid membrane | 1.80 |
| At2g28000 | CPN60A (chloroplast/60 kDa chaperonin alpha subunit) | Chloroplast organization | Chloroplast envelope | 1.59 |
| At3g09490 | Chloroplast lumen common family protein | Photosynthesis, light reaction | Chloroplast thylakoid lumen | 1.67 |
| At3g27690 | LHCB2:4 (photosystem II light-harvesting complex gene) | Photosynthesis | Chloroplast envelope | 1.59 |
| At3g47470 | LHCA4 (photosystem I light-harvesting complex gene) | Photosynthesis, light harvesting | Chloroplast thylakoid membrane | 1.71 |
| At4g01330 | Protein kinase family protein | Protein amino acid phosphorylation | Plasma membrane | 1.99 |
| At5g44190 | GLK2 (GOLDEN2-LIKE 2); DNA binding/transcription factor | Regulation of chlorophyll biosynthetic process | Nucleus | 2.23 |
| At5g48490 | Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | Lipid transport | Endomembrane system | 2.53 |
| At5g54190 | PORA; oxidoreductase/protochlorophyllide reductase | Chlorophyll biosynthetic process | Chloroplast | 2.50 |
| At5g54270 | LHCB3 (light-harvesting chlorophyll-binding protein) | Photosynthesis | Chloroplast thylakoid membrane | 1.90 |
| Lipid and fatty acid synthesis | ||||
| At1g01120 | KCS1 (3-ketoacyl-CoA synthase 1); acyltransferase | Fatty acid elongase activity | Cytosolic ribosome | 1.91 |
| At2g15090 | Fatty acid elongase, putative | Fatty acid elongation | Endoplasmic reticulum | 3.48 |
| At2g28630 | beta-Ketoacyl-CoA synthase family protein | Acyltransferase activity | Endoplasmic reticulum | 1.89 |
| At2g46720 | HIC (high carbon dioxide); acyltransferase | Fatty acid elongation | Endomembrane system | 1.67 |
| At3g10280 | Fatty acid elongase 3-ketoacyl-CoA synthase, putative | Fatty acid elongation | Endomembrane system | 1.55 |
| At4g01950 | ATGPAT3/GPAT3 (glycerol-3-phosphate acyltransferase 3) | Acyltransferase activity | 2.81 | |
| At4g22753 | SMO1-3 (sterol 4-alpha methyl oxidase); catalytic | Fatty acid biosynthetic process | Endoplasmic reticulum | 1.57 |
| At4g33110 | Coclaurine N-methyltransferase, putative | Lipid biosynthetic process | Plasma membrane | 1.54 |
| At4g34250 | Fatty acid elongase, putative | Transferase activity | Endomembrane system | 2.62 |
| At5g04530 | beta-Ketoacyl-CoA synthase family protein | Transferase activity | Endomembrane system | 3.81 |
| At5g08030 | Glycerophosphoryl diester phosphodiesterase family protein | Glycerol metabolic process | Endomembrane system | 3.18 |
| At5g38020 | S-Adenosyl-L-methionine:carboxyl methyltransferase family protein | Fatty acid biosynthetic process | Cellular component | 1.71 |
| At5g56100 | Glycine-rich protein/oleosin | Lipid storage | Membrane | 2.01 |
| At5g61610 | Glycine-rich protein/oleosin | Lipid storage | Membrane | 4.79 |
Transcript level, transcript level ratio between the transgenic sample and the WT control
As might be expected from the increased chlorophyll content and photosynthetic efficiency caused by BnGRF2, some genes relating to light harvesting and chlorophyll biosynthesis were up-regulated. GLK2 (golden2-like) (At5g44190), which was up-regulated in the transgenic lines, is known to stimulate expression of genes related to light harvesting and chlorophyll biosynthesis (Waters et al., 2008, 2009). Partially up-regulated genes such as lhcb2.4 (At3g27690), CHI (At1g44446), lhcb3 (At5g54270), lhcb6 (At1g15820), and LTP (At5g48490) were also found to be coincident with the up-regulated genes induced by GLK2. Also, some genes related to light harvesting and chlorophyll biosynthesis such as PUB44 (At1g20780), CPN60A (At2g28000), PORA (At5g54190), lhcb2.1 (At2g05100), and lhca4 (At3g47470) were up-regulated in the transgenic lines (Table 3).
In addition, some up-regulated genes such as KCS16 (At4g34250), GPAT (at4g01950), F4I10.40 (at4g33110), GDPD6 (at5g08030), SAMT (AT5G38020), SMO1-3 (AT4G22753), and oleosin proteins (at5g56100 and at5g61610) were classified as being involved in lipid and FA biosynthetic and storage processes by GO category (Table 3).
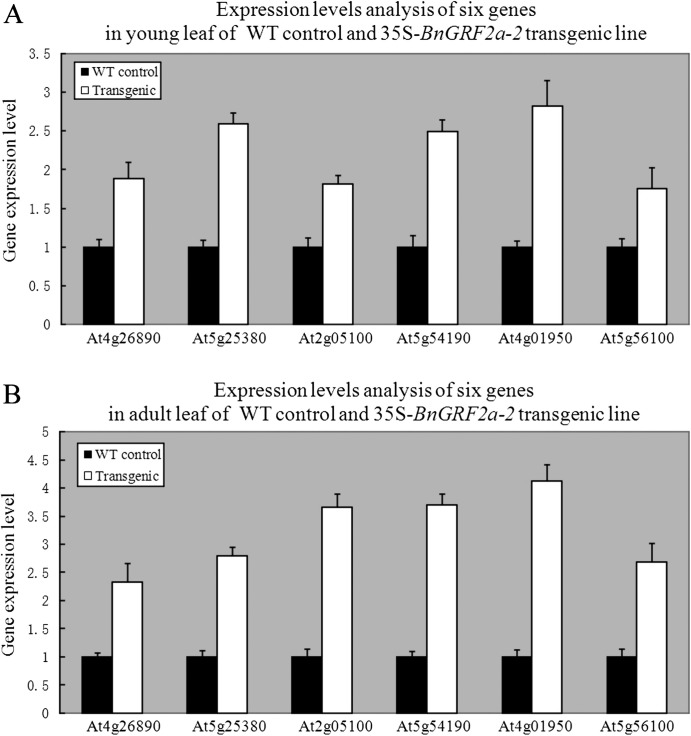
According to the leaf phenotypes of the 35S-BnGRF2a transgenic line, although the expression level of most up-regulated genes in transgenic young leaves was <2-fold compared with the WT control (Table 3), they might be more highly expressed in the adult leaf. To confirm this, six genes, MAPKKK16, CYCA2;1, lhcb2.1, PORA, GPAT, and oleosin, were chosen to detect their expression levels in adult leaf of the 35S-BnGRF2a transgenic lines. The results showed that most genes were expressed more highly in adult leaf than in young leaf (Fig. 7). The up-regulation of these genes was further verified in seed of the Napin-BnGRF2a-3 line (Supplementary Fig. S3 at JXB online).
Fig. 7.
Comparison of the expression level of up-regulated genes in young leaf and adult leaf between the wild-type (WT) control and the 35S-BnGRF2a-2 transgenic line. (A) Data from microarray analyses. (B) Data from real-time PCR analyses. Data expression was normalized to β-actin1 (At2g37620) and relative to the expression of genes in the WT control. Data presented are mean values of three biological replicates, and error bars represent standard deviations.
Discussion
The present study began with gene expression analyses of 20 d ovules from the zy036 and 51070 rapeseed lines, which exhibited different levels of seed oil production. The differentially expressed gene BnGRF2 was chosen for further examination to elucidate its function in oil production. BLAST analyses of the Brassica sequence databases indicated that two BnGRF2 homologues (BnGRF2a and BnGRF2b) are located in the rapeseed A or C genome. Real-time PCR showed that the two homologues were expressed differentially in six rapeseed tissues, and BnGRF2a is expressed dominantly compared with BnGRF2b. In B. rapa and B. oleracea, quite a lot of homologous genes showed differences in expression levels (unpublished), which is a ubiquitous phenomenon during polyploid genome evolution, and suggested the importance of dominantly expressed genes in plant development. Therefore, BnGRF2a was chosen for further functional research.
In a previous study, GRFs were believed to function as transcription activators (Kim and Kende 2004); therefore, it was not a surprise that BnGRF2 induced pleiotropic phenotypes in Arabidopsis. However, it must be noted that greater phenotypic differences were observed in transgenic overexpression of BnGRF2 (Supplementary Table S6 at JXB online), compared with previous reports regarding overexpression of AtGRF2 or other GRF genes (Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Liu et al., 2009). First, the increased cell number enlarged the leaf area in BnGRF2a transgenic lines. However, in Arabidopsis, overexpression of AtGRF2 resulted in larger leaves because of an increase in cell size. Secondly, the adult leaf colour of the transgenic line was dark green. Thirdly, because of the longer pistil and normal stamen, near sterility was induced compared with AtGRF2 transgenic plants. Finally, the seed phenotype was not referred to in previous studies. It is thought that this discrepancy is due to the difference in gene sequences. Although BnGRF2 and AtGRF2 show good conservation of key domains, they only exhibit 69% identity overall, and these sequence differences might be responsible for the phenotypic differences. Research on ZmGRF2 and ZmGIF3 in Arabidopsis which only showed delayed bolting of the inflorescence stem and acceleration of the elongation of the stem seemed to confirm this view point (Zhang et al., 2008).
To elucidate how the metabolic pathways participate in the phenotypic changes observed, genome-wide analyses of the BnGRF2a-responsive transcriptome were performed by a microarray method. Generally, genes regulate seed mass by changing the cell number (Song et al., 2007; Weng et al., 2008), cell size (Liu et al., 2010), or both (Ohto et al., 2005). In the present experiments, the cell number in the leaf and seed was found to be increased in BnGRF2a-overexpressing transgenic lines, which was consistent with the AtGRF5 gene in the leaf (Horiguchi et al., 2005), but different from AtGRF1 and AtGRF2 genes which increase the leaf cell size in Arabidopsis (Kim et al., 2003). With respect to the increased cell number, it was found that some genes related to cell division and the cell cycle were up-regulated (Inze and De Veylder, 2006; Meyerowitz, 1997).
Improvement of the photosynthetic rate in transgenic lines was found, which was supported by some up-regulated genes related to light harvesting and chlorophyll biosynthesis. Just like the phenotype of the leaf in the 35S-BnGRF2a transgenic lines, the chlorophyll content of developing seeds was also higher in transgenic Napin-BnGRF2a Arabidopsis, which coincided with differences in the leaf and ovule between zy036 and 51070 (data not shown). Interestingly, the transgenic seeds contained 10% more oil than WT seeds, possibly as a result of improved photosynthetic efficiency.
Light reactions of photosynthesis play an important role in defining plant fitness and productivity by influencing a seed’s carbon economy and how that carbon is stored (Goffman, 2005). Previous studies with developing rapeseed embryos had shown that lipid accumulation was stimulated by light (Aach and Heise, 1998; Ruuska et al., 2004; Schwender et al., 2004). This finding suggested that FA synthesis in the embryo might be dependent on the supply of reducing power (NADPH and NADH) and ATP provided by the light reactions of photosynthesis (Ruuska et al., 2004; Goffman et al., 2005; Schwender et al., 2006). To a certain extent, photosynthetic efficiency increases with an increasing concentration of light and, thus, light stimulates lipid accumulation by increasing photosynthetic efficiency (Li et al., 2006). Some genes relating to FA synthesis and oil storage were up-regulated in the transgenic lines. For example, KCS16 is a member of the 3-ketoacyl-CoA synthase family, which is involved in the biosynthesis of very long chain FAs. In Arabidopsis, a kcs16 mutant had a reduced eicosenoic acid content in seeds (Joubès et al., 2008). Modest seed oil content increases were also observed in recombinant expression studies using modified safflower GPAT cDNA and GPAT from Escherichia coli (Jain et al., 2000).
Of course, for oil crop breeding, what is of concern is the agricultural characteristics such as seed weight and oil content. At present, much research on improving oil production by transgenic technology is ongoing. However, previous studies focused on genes which changed only seed mass or oil content. Also, most of the genes encode the enzymes and transcription factors regulating carbon metabolism, FA synthesis, and TAG synthesis pathways (Baud and Lepiniec, 2009; Weselake et al., 2009). In the present study, it was found that BnGRF2a increases oil production by regulating the capacity of both the source (changing photosynthesis efficiency) and the sink (changing cell number), which is more significant for elevating oil production in oil crop breeding. Although overexpression of BnGRF2a in flower buds induced less efficient self-fertilization, such a drawback could be avoided using a tissue-specific promoter. Thus, this gene may represent a suitable candidate for genetic manipulation to improve the economic characteristics of seed weight or oil production in crops.
Supplementary data
Supplementary data are available at JXB online.
Figure S1 . Real-time PCR analyses of the BnGRF2a gene in different tissues of rapeseed lines zy036 and 51070.
Figure S2 . Localization of the insertion point of the vector sequence in the 35S-BnGRF2a-2 and 35S-BnGRF2a-10 transgenic lines.
Figure S3 . Expression level comparison of up-regulated genes between siliques (10 d after flowering) of WT control and the Napin-BnGRF2a-3 transgenic line.
Table S1 . Primers used for gene isolation, expression vector confirmation, and real-time PCR in this study.
Table S2 . Up-regulated genes of 20 d ovules in zy036 compared with 51070.
Table S3 . An overview of differential gene expression between the leaf of 35S-BnGRF2a-2 and WT control plants.
Table S4 . Transcription factors involved in differentially expressed genes between the leaf of 35S-BnGRF2a-2 and WT control plants.
Table S5 . Up-regulated genes that were thought to be involved in possible BnGRF2-regulated pathways.
Table S6 . Phenotype comparisons between transgenic Arabidopsis lines overexpressing BnGRF2a and AtGRF.
Acknowledgments
This study was supported by the National Key Basic Research Program of China (2011CB109300) and the National Transgenic Research Projects of China (2009ZX08009-018B).
References
- Aach H, Heise K. On the compartmentation of triacylglycerol synthesis in developing seeds of Brassica napus . Botanica Acta. 1998;111:123–129. [Google Scholar]
- Baud S, Lepiniec L. Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiology and Biochemistry. 2009;47:448–455. doi: 10.1016/j.plaphy.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. The Plant Journal. 2007;50:825–838. doi: 10.1111/j.1365-313X.2007.03092.x. [DOI] [PubMed] [Google Scholar]
- Berger F, Grini PE, Schnittger A. Endosperm: an integrator of seed growth and development. Current Opinion in Biotechnology. 2006;9:664–670. doi: 10.1016/j.pbi.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, et al. Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiology. 2003;132:681–697. doi: 10.1104/pp.103.022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Shockey JM, Dietrich CR, Gidda SK, Mullen RT, Dyer JM. Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Current Opinion in Plant Biology. 2007;10:236–244. doi: 10.1016/j.pbi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Chen W, Zhang YS, Yao JB, Ma CZ, Tu JX, Fu TD. Quantitative trait loci mapping for two seed yield component traits in an oilseed rape (Brassica napus) cross. Plant Breeding. 2011;130:640–646. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genomewide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy L, Mackenzie J, Haseloff J. Coordination of plant cell division and expansion in a simple morphogenetic system. Proceedings of the National Academy of Sciences, USA. 2010;107:2711–2716. doi: 10.1073/pnas.0906322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman A, Hayden DM, Dehesh K, Bulow L, Stymne S. Carbon partitioning between oil and carbohydrates in developing oat (Avena sativa L.) seeds. Journal of Experimental Botany. 2008;59:4247–4257. doi: 10.1093/jxb/ern266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theoretical and Applied Genetics. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- Gao MJ, Parkin IAP, Lydiate DJ, Hannoufa A. An auxin-responsive SCARECROW-like transcriptional activator interacts with histone deacetylase. Plant Molecular Biology. 2004;55:417–431. doi: 10.1007/s11103-004-0892-9. [DOI] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. The Plant Cell. 2005;17:52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Saingery V, Chambrier P, Mayer U, Jurgens G, Berger F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiology. 2003;131:1661–1670. doi: 10.1104/pp.102.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman FD, Alonso AP, Schwender J, Shachar-Hill Y, Ohlrogge JB. Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiology. 2005;138:2269–2279. doi: 10.1104/pp.105.063628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef G, LaVallee BJ, Tenopir P, Tat M, Schweiger B, Kinney AJ, Van Gerpen JH, Clemente TE. A high-oleic-acid and low-palmitic-acid soybean: agronomic performance and evaluation as a feedstock for biodiesel. Plant Biotechnology Journal. 2009;7:411–421. doi: 10.1111/j.1467-7652.2009.00408.x. [DOI] [PubMed] [Google Scholar]
- Harwood JL. Recent advances in the biosynthesis of plant fatty acids. Biochimica et Biophysica Acta. 1996;1301:7–56. doi: 10.1016/0005-2760(95)00242-1. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana . The Plant Journal. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- Hua W, Li RJ, Zhan GM, Liu J, Li J, Wang XF, Liu GH, Wang HZ. Maternal control of seed oil content in Brassica napus: the role of silique wall photosynthesis. The Plant Journal. 2012;69:432–444. doi: 10.1111/j.1365-313X.2011.04802.x. [DOI] [PubMed] [Google Scholar]
- Inze D, De Veylder L. Cell cycle regulation in plant development. Annual Review of Genetics. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- Jain RK, Coffey M, Lai K, Kumar A, MacKenzie SL. Enhancement of seed oil content by expression of glycerol-3-phosphate acyltransferase genes. Biochemical Society Transactions. 2000;28:958–961. [PubMed] [Google Scholar]
- Jofuku KD, Omidyar PK, Gee Z, Okamuro JK. Control of seed mass and seed yield by the floral homeotic gene APETALA2 . Proceedings of the National Academy of Sciences, USA. 2005;102:3117–3122. doi: 10.1073/pnas.0409893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. The Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R. The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Molecular Biology. 2008;67:547–566. doi: 10.1007/s11103-008-9339-z. [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. The Plant Journal. 2003;36:94–104. doi: 10.1046/j.1365-313x.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Horiguchi G, Kim GT, Ohgishi M, Sakai T, Tsukaya H. The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by photoreceptors and sugar. Plant and Cell Physiology. 2005;46:213–223. doi: 10.1093/pcp/pci016. [DOI] [PubMed] [Google Scholar]
- Li RJ, Wang HZ, Mao H, Lu YT, Hua W. Identification of differentially expressed genes in seeds of two near-isogenic Brassica napus lines with different oil content. Planta. 2006;224:952–962. doi: 10.1007/s00425-006-0266-4. [DOI] [PubMed] [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiologia Plantarum. 2009;136:223–236. doi: 10.1111/j.1399-3054.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Hua W, Zhan GM, Wei F, Wang XF, Liu GH, Wang HZ. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of gene from Brassica napus . Plant Physiology and Biochemistry. 2010;48:9–15. doi: 10.1016/j.plaphy.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Lu C, Napier JA, Clemente TE, Cahoon EB. New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Current Opinion in Biotechnology. 2011;22:252–259. doi: 10.1016/j.copbio.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao HL, Sun SY, Yao JL, Wang CR, Yu SB, Xu CG, Li XH, Zhang QF. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proceedings of the National Academy of Sciences, USA. 2010;107:19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proceedings of the National Academy of Sciences, USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu JY, Tan HL, Zheng Q, et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiology. 2008;148:1042–1054. doi: 10.1104/pp.108.126342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. The Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ. Control of seed mass by APETALA2 . Proceedings of the National Academy of Sciences, USA. 2005;102:3123–3128. doi: 10.1073/pnas.0409858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi CH, Tanksley SD. Natural variation in an ABC transporter gene associated with seed size evolution in tomato species. PLoS Genetics. 2009;5:e1000347. doi: 10.1371/journal.pgen.1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta. 1989;975:384–394. [Google Scholar]
- Rogalski M, Carrer H. Engineering plastid fatty acid biosynthesis to improve food quality and biofuel production in higher plants. Plant Biotechnology Journal. 2011;9:554–564. doi: 10.1111/j.1467-7652.2011.00621.x. [DOI] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genetics. 2006;2:e210. doi: 10.1371/journal.pgen.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiology. 2004;136:2700–2709. doi: 10.1104/pp.104.047977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. The Plant Journal. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature. 2004;432:779–782. doi: 10.1038/nature03145. [DOI] [PubMed] [Google Scholar]
- Schwender J, Shachar-Hill Y, Ohlrogge JB. Mitochondrial metabolism in developing embryos of Brassica napus . Journal of Biological Chemistry. 2006;281:34040–34047. doi: 10.1074/jbc.M606266200. [DOI] [PubMed] [Google Scholar]
- Shen B, Sinkevicius KW, Selinger DA, Tarczynski MC. The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Physiology and Biochemistry. 2006;60:377–387. doi: 10.1007/s11103-005-4110-1. [DOI] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nature Genetics. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genetics. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Takano-Kai N, Jiang H, Kubo T, et al. Evolutionary history of GS3, a gene conferring grain size in rice. Genetics. 2009;182:1323–1334. doi: 10.1534/genetics.109.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HL, Yang XH, Zhang FX, et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiology. 2010;156:1577–1588. doi: 10.1104/pp.111.175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng W, Han Y, Du Y, Sun D, Zhang Z, Qiu L, Sun G, Li W. QTL analysis of seed weight during the development of soybean (Glycine max L. Merr.) . Heredity. 2009;102:372–380. doi: 10.1038/hdy.2008.108. [DOI] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB. Metabolic engineering of fatty acid biosynthesis in plants. Metabolic Engineering. 2002;4:12–21. doi: 10.1006/mben.2001.0204. [DOI] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) . Development. 1996;122:1589–1600. doi: 10.1242/dev.122.5.1589. [DOI] [PubMed] [Google Scholar]
- Van der Knaap E, Kim JH, Kende H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiology. 2000;122:695–704. doi: 10.1104/pp.122.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker T, Kinney AJ. Variations in the biosynthesis of seed-storage lipids. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:335–361. doi: 10.1146/annurev.arplant.52.1.335. [DOI] [PubMed] [Google Scholar]
- Wang ET, Wang JJ, Zhu XD, et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nature Genetics. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang L, et al. The genome of the mesopolyploid crop species Brassica rapa . Nature Genetics. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. The Plant Journal. 2008;56:432–444. doi: 10.1111/j.1365-313X.2008.03616.x. [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. The Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H, Yang L, Liu Z, Ding J, Pan A, Zhang D. Novel reference gene, high-mobility-group protein I/Y, used in qualitative and real-time quantitative polymerase chain reaction detection of transgenic rapeseed cultivars. Journal of AOAC International. 2005;88:577–584. [PubMed] [Google Scholar]
- Weng JF, Gu SH, Wan XY, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Research. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- Weselake RJ, Taylor DC, Rahman MH, Shah S, Laroche A, McVetty PB, Harwood JL. Increasing the flow of carbon into seed oil. Biotechnology Advances. 2009;27:866–878. doi: 10.1016/j.biotechadv.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Wynn AN, Rueschhoff EE, Franks RG. Transcriptomic characterization of a synergistic genetic interaction during carpel margin meristem development in Arabidopsis thaliana . PLoS One. 2011;6:e26231. doi: 10.1371/journal.pone.0026231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Yao QH, Xu ZH, Xue HW. Development of an efficient method for the isolation of factors involved in gene transcription during rice embryo development. The Plant Journal. 2004;38:348–357. doi: 10.1111/j.1365-313X.2004.02037.x. [DOI] [PubMed] [Google Scholar]
- Zhang DF, Li B, Jia GQ, Zhang TF, Dai JR, Li JS, Wang SC. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.) Plant Science. 2008;175:809–817. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.