Abstract
Previous pharmacological results confirmed that haem oxygenase-1 (HO-1) is involved in protection of cells against ultraviolet (UV)-induced oxidative damage in soybean [Glycine max (L.) Merr.] seedlings, but there remains a lack of genetic evidence. In this study, the link between Arabidopsis thaliana HO-1 (HY1) and UV-C tolerance was investigated at the genetic and molecular levels. The maximum inducible expression of HY1 in wild-type Arabidopsis was observed following UV-C irradiation. UV-C sensitivity was not observed in ho2, ho3, and ho4 single and double mutants. However, the HY1 mutant exhibited UV-C hypersensitivity, consistent with the observed decreases in chlorophyll content, and carotenoid and flavonoid metabolism, as well as the down-regulation of antioxidant defences, thereby resulting in severe oxidative damage. The addition of the carbon monoxide donor carbon monoxide-releasing molecule-2 (CORM-2), in particular, and bilirubin (BR), two catalytic by-products of HY1, partially rescued the UV-C hypersensitivity, and other responses appeared in the hy1 mutant. Transcription factors involved in the synthesis of flavonoid or UV responses were induced by UV-C, but reduced in the hy1 mutant. Overall, the findings showed that mutation of HY1 triggered UV-C hypersensitivity, by impairing carotenoid and flavonoid synthesis and antioxidant defences.
Keywords: Antioxidant defences, Arabidopsis, carotenoid, flavonoid, hy1, UV-C hypersensitivity
Introduction
Although most harmful solar ultraviolet (UV)-C is effectively eliminated by the stratospheric ozone layer (Kazan and Manners, 2011), artificial UV-C irradiation is widely adopted for disinfection processes, including preventing cyanobacterial blooms, in many drinking water treatment plants (Bin Alam et al., 2001; USEPA, 2006; Sakai et al., 2009). UV-C stress, sharing the ELONGATED HYPOCOTYL5 (HY5)-mediated signalling pathway involved in UV-B signalling (Castells et al., 2010), is also an effective induction model for studying mechanisms of oxidative stress and programmed cell death (Renzing et al., 1996; Gao et al., 2008; Balestrazzi et al., 2009; Saxena et al., 2011). As it is highly energetic, UV-C radiation has deleterious effects on plant cells, including DNA damage, oxidation of cellular components, and impaired chloroplast function and thus reduced photosynthesis (Booij-James et al., 2000). Numerous studies have examined the different strategies adopted by plants to cope with the detrimental photooxidative processes caused by excessive UV radiation, including the regulation of flavonoid metabolism and the size of the xanthophyll cycle pool, as well as the manipulation of a variety of reactive oxygen species (ROS) scavengers (Landry et al., 1995; Kirchgeßner et al., 2003; Close et al., 2007). Nonetheless, the detailed molecular and biochemical mechanisms of the signalling pathways by which plants sense and relay UV-C signals remain to be elucidated.
Flavonoids are considered antioxidant molecules, which may serve as free radical scavengers by locating and neutralizing radicals before they can damage plant cells. It has been suggested that non-pigmented flavonoids play a central role in protecting against harmful UV light and excess visible light (Buer et al., 2010). The regulation of flavonoid biosynthesis in Arabidopsis thaliana has been well described by characterizing T-DNA insertion mutants (Qin et al., 2007; Buer et al., 2010; Castells et al., 2010; Supplementary Fig. S1 available at JXB online). The flavonoid biosynthetic pathway is a branch of the general phenylpropanoid pathway, the first committed step of which is catalysed by chalcone synthase (CHS). Subsequent reactions catalysed by chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3'-hydroxylase (F3'H), and flavonol synthase (FLS) lead to production of the basic flavonols, kaempferol and quercetin (Supplementary Fig. S1 available at JXB online). Genetic evidence has further confirmed that the Arabidopsis mutants transparent testa-4 (tt4; deficient in CHS) and transparent testa-5 (tt5; deficient in CHI) are unable to accumulate flavonoids, and are therefore more sensitive to UV-B and UV-C (Li et al., 1993; Filkowski et al., 2004). Both when growing normally and when exposed to high UV levels, fine regulation of flavonoid biosynthesis is accomplished by integrated modulation of multiple transcriptional factors, including WRKY, basic leucine zipper (bZIP), MADS-box, R2R3-MYB, and basic helix–loop–helix (bHLH) factors (Ramsay and Glover, 2005; Stracke et al., 2007, 2010). Under both visible light and UV-B, the bZIP transcriptional regulator HY5 is linked to CHS gene activation and accumulation of flavonoids (Oravecz et al., 2006). Recent studies have confirmed that the HY5-dependent regulation of the gene expression of the PRODUCTION OF FLAVONOL GLYCOSIDES (PFG) family of R2R3-MYB transcriptional factors, including PFG1/MYB12, PFG2/MYB11, and PEF3/MYB111, contributes to the establishment of UV-B tolerance (Stracke et al., 2010).
Other complementary and redundant mechanisms have also been shown to provide protection against photooxidative damage caused by UV irradiation, including carotenoid metabolism and a series of antioxidant defence enzymes responsible for ROS scavenging (Bartley and Scolnik, 1995). Using forward and reverse genetic approaches, the main pathway for carotenoid biosynthesis in Arabidopsis has been identified, and includes geranylgeranyl pyrophosphate synthase (GPS), phytoene synthase (PSY), phytoene desaturase (PDS), and zeta-carotene desaturase (ZDS). Enzymatic antioxidant systems also play a role in the efficient scavenging of ROS, including superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX). Further studies have shown that a Neurospora crassa mutant which is null for sod-1, encoding copper-zinc SOD, showed hypersensitivity to near-UV irradiation (Chary et al., 1994).
In animals, haem oxygenase (HO; EC 1.14.99.3) is a microsomal enzyme committed to haem degradation. Haem moieties from haemoproteins are cleaved by HO at the α-meso-carbon bridge, generating equimolar amounts of ferrous iron, carbon monoxide (CO), and biliverdin (BV). BV is subsequently reduced to bilirubin (BR) by cytosolic biliverdin reductase (Otterbein et al., 2003). To date, three isoforms (HO-1, HO-2, and HO-3) have been identified in animals. HO-1 has been confirmed as an inducible isoform by numerous stimuli, including haem as well as non-haem stimuli such as heavy metals, hormones, endotoxin, cytokines, ROS, heat shock, and UV-A/B/C irradiation (Applegate et al., 1991; Prawan et al., 2005; Loboda et al., 2008). Various HO-1 inducers have provided further support for the conclusion that HO-1, besides its role in haem degradation, may play an important role as a dynamic sensor of cellular oxidative stress and in maintaining cellular homeostasis (Bilban et al., 2008; Calabrese et al., 2010).
Several lines of evidence confirm the presence of HO in plants (Shekhawat and Verma, 2010). For example, in Arabidopsis, a small family of HOs with four members is classified into two subfamilies. The HO1 subfamily includes HY1, HO3, and HO4, while HO2 is the only member of the HO2 subfamily. It has been shown that HY1 is clearly the most highly expressed gene, followed by HO2, with both HO3 and HO4 expressed at low levels (Matsumoto et al., 2004). However, recent research has confirmed that HO2 is not a true HO (Gisk et al., 2010). Besides its participation in light signalling (Muramoto et al., 1999; Gisk et al., 2010; Shekhawat and Verma, 2010), plant HO and its catalytic product CO have been implicated in stomatal aperture regulation (Cao et al., 2007), root development (Xuan et al., 2008; Cao et al., 2011; Xu et al., 2011), and plant tolerance or acclimation to salinity (Xie et al., 2008, 2011a , b ), heavy metal exposure (Han et al., 2008), and UV-B irradiation (Yannarelli et al., 2006). For example, HO-1/CO mediates abscisic acid (ABA)-induced stomatal closure (Cao et al., 2007) and auxin-induced adventitious root development (Xuan et al., 2008). Importantly, the inducible response of HO-1 to the oxidative stress generated by UV-B exposure was first reported in soybean plants (Yannarelli et al., 2006). However, besides its antioxidant behaviour, the detailed roles of UV-induced HO-1 in plants and the underlying signalling mechanisms remain unclear.
In order to investigate the possible interconnection between HO signalling and UV-C response in plants, UV-C-induced phenotypic changes in the seedlings of hy1-100/ho2/ho3/ho4 single and/or double mutants and seedlings of a HY1 overexpression line were compared. It was found that only HY1 mutation (hy1-100) leads to UV-C hypersensitivity, consistent with observed decreases in flavonoid and carotenoid contents, and severe oxidative damage. Some contrasting responses, such as the enhancement of the contents of chlorophyll a, chlorophyll b, and carotenoid, were observed in the HY1 overexpression line, compared with the wild-type plants. Moreover, the data showed that the application of the CO donor carbon monoxide-releasing molecule-2 (CORM-2; Desmard et al., 2011; García-Arnandis et al., 2011) and BR, two catalytic by-products of HO, could reverse the above responses, including the UV-C hypersensitivity phenotype and the down-regulation of corresponding genes in the carotenoid and flavonoid biosynthesis pathways as well as antioxidant defences, in the hy1-100 mutant, with CORM-2 being especially effective . Transcriptional factors involved in the synthesis of flavonoid and UV responses, including HY5, HY5-homologue (HYH), MYB11, and MYB12, were induced by UV-C but reduced in the hy1-100 mutant.
Materials and methods
Plant materials, growth conditions, and chemicals
Arabidopsis thaliana hy1-100 (CS236, Col-0), hy1 (CS67, Ler), ho2 (SALK_025840, Col-0), ho3 (SALK_034321, Col-0), and ho4 (SALK_044934, Col-0) mutants were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/abrc), and Ler ecotype seeds were provided by C.Y. Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China). The homogenous HY1 overexpression line 35S:HY1-4 was used according to Xie et al. (2011b). Seeds were surface-sterilized and washed three times with sterile water for 20 min, then cultured in Petri dishes on solid Murashige and Skoog (MS) medium (pH 5.8). Plates containing seeds were kept at 4 °C for 2 d, and then transferred into a growth chamber with a 16/8 h (23/18 °C) day/night regime and 120 μmol m−2 s−1 irradiation.
All chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA) unless stated otherwise. The chemicals used for treatments were CORM-2 (0.1 μM), BR (100 μM), and FeSO4 (100 μM) (Cao et al., 2011; Desmard et al., 2011; García-Arnandis et al., 2011; Wu et al., 2011). The concentrations of CORM-2 and BR used in this study were determined in pilot experiments from which maximal induced responses were obtained.
Generation of the ho2/3, ho2/4, and ho3/4 mutants
Homozygous T-DNA insertion lines for ho2 (SALK_025840), ho3 (SALK_034321), and ho4 (SALK_044934) that prevented accumulation of the full-length corresponding mRNAs had been previously identified (Xie et al., 2011b). To generate the ho2/3, ho2/4, and ho3/4 double mutants, pollen of either ho2 or ho3 was transferred to the stigmas of emasculated flowers of ho3 and ho4 mutants. F1 plants were self-pollinated, and the resulting F2 individuals were genotyped using PCR primers specific for the presence of the double mutants (Supplementary Table S1 at JXB online).
UV-C tolerance assay and phenotype analysis
For UV-C tolerance analysis, 5-day-old seedlings were irradiated with 1.8 (0.5 W m−2 for 1 h), 3.6 (0.5 W m−2 for 2 h), or 5.4 (0.5 W m−2 for 3 h) kJ m−2 of UV-C (λ=254 nm) using a HL-2000 HybriLinker UV Crosslinker (UVP, USA), according to Castells et al. (2010), with some modifications. Alternatively, Arabidopsis seeds were cultured in MS medium with or without 0.1 μM CORM-2, 100 μM BR, and 100 μM Fe2+ pre-treatments for 5 d. Afterwards, seedlings were exposed (+) or not (–) to UV-C irradiation. After irradiation, plants were immediately returned to the growth chamber under 120 μmol m−2 s−1 light irradiation. The phenotypes, including the fresh weight and primary root growth rate (Castells et al., 2010; Xie et al., 2011b ), were then observed 5 days later.
Pigment analysis
To determine UV-absorbing pigments, seedling shoot or root tissues of each ecotype were ground in liquid nitrogen and extracted in methanol (1 ml per 0.1 g fresh weight). After centrifugation for 10 min at 14 000 g, the supernatant was diluted in methanol (1:10), and the absorbance was scanned between 220 nm and 450 nm using a DU 800 UV/VIS spectrophotometer (Beckman Coulter, USA). Flavonoids were extracted (Bieza and Lois, 2001) and quantified following Li et al. (1993) and Tossi et al. (2011). Chlorophyll a/b and carotenoids were extracted from leaves using 95% ethanol and quantified as described by Lichtenthaler (1987).
Histochemical staining and determination of the content of thiobarbituric acid-reactive substances (TBARS)
Histochemical detection of lipid peroxidation was performed with Schiff’s reagent as described by Han et al. (2008). Arabidopsis roots stained with Schiff’s reagent were washed thoroughly, then observed and photographed under a light microscope (model Stemi 2000-C; Carl Zeiss, Germany). Lipid peroxidation of seedlings was also estimated by measuring the amount of TBARS as previously described (Xie et al., 2008).
Real-time RT-PCR analysis
Total RNA was isolated using Trizol reagent (Invitrogen, Gaithersburg, MD, USA) according to the manufacturer’s instructions. Real-time quantitative reverse transcription-PCRs (RT-PCRs) were performed using a Mastercycler® ep realplex real-time PCR system (Eppendorf, Hamburg, Germany) with SYBR® Premix Ex Taq™ (TaKaRa Bio Inc., China) according to the manufacturer’s instructions. Using specific primers (Supplementary Table. S2 at JXB online), the expression levels of the genes are presented as values relative to the corresponding control samples at the indicated times or under the indicated conditions, after normalization to actin2/7 (accession number NM_121018) transcript levels.
Statistical analysis
Data are shown as the means ±SE (standard error) from at least three independent experimental replications. Both the analysis of variance (ANOVA) and multiple comparison were used in data analysis. In particular, the data of different genotype samples upon UV treatments were analysed using a general linear model for a completely randomized design with UV irradiation levels as one fixed factor. Differences in the phenotypic indicators among the wild-type, mutant, or overexpression lines were analysed by a nested ANOVA, with genotypes nested within UV irradiation treatments [model: phenotypic indicators = replication + treatment + sample (treatment)] or within recuperation times [model: phenotypic indicators = replication + time + sample (time)], followed by a multiple comparison. A threshold of P < 0.05 was set as the level of significance in tests. All the above procedures were programmed to run in a SAS version 9.1 environment (SAS Institute, Cary, NC, USA.
Results
UV-C hypersensitivity and HO transcripts
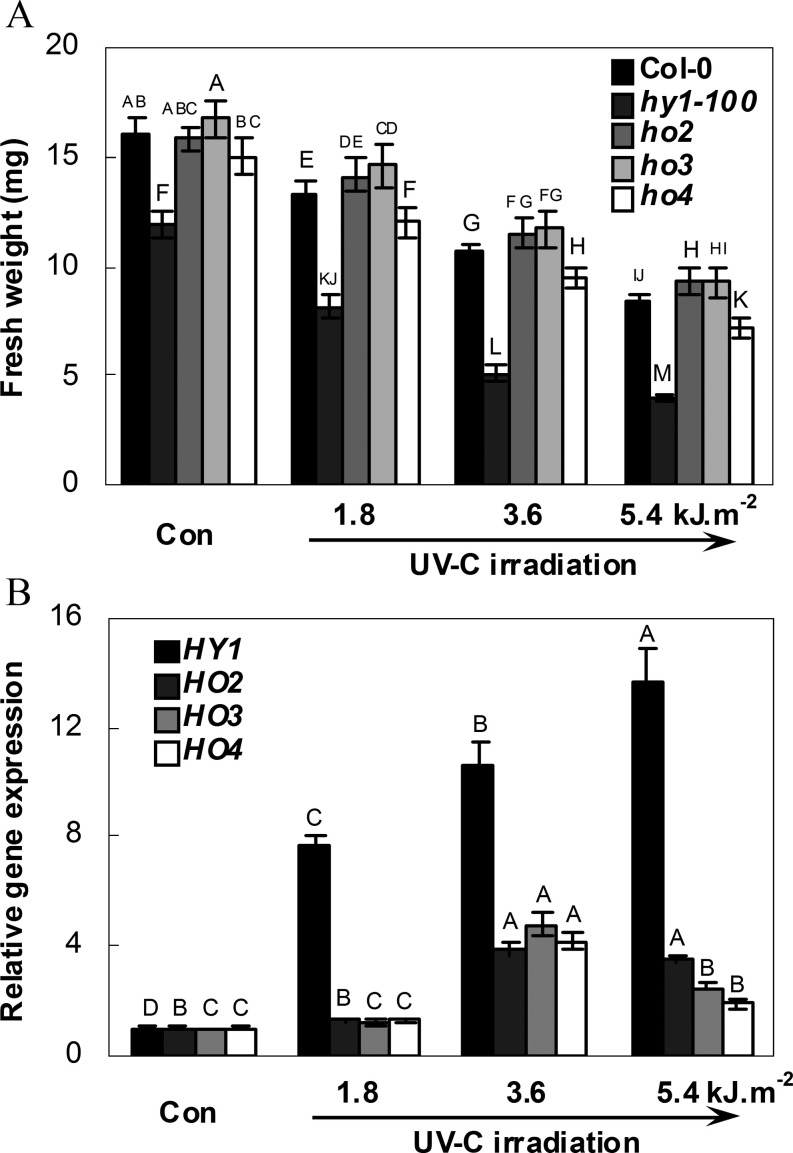
A previous study showed the involvement of Arabidopsis HY1 in salt acclimation (Xie et al., 2011b ). In this study, to assess the putative connection between UV-C hypersensitivity and HO, UV-C-induced phenotypic changes in hy1-100, ho2, ho3, and ho4 mutants were first analysed. The results shown in Fig. 1A confirmed that the growth of seedlings of the wild type (Col-0) is progressively inhibited with increasing levels of UV-C exposure (Castells et al., 2010). To identify the possible role of each HO in this process, the HY1, HO2, HO3, and HO4 mRNAs present in the wild type were investigated using quantitative RT-PCR (Fig. 1B). Compared with the UV-C-free condition, the maximal inducible expression of HY1 was observed in response to increasing doses of UV-C irradiation, while no inducible response was observed in HO2, HO3, and HO4 transcripts when exposed to 1.8 kJ m−2 UV-C. Although HO2, HO3, and HO4 were moderately induced by 3.6 kJ m−2 UV-C, levels of these transcripts then decreased at exposures of 5.4 kJ m−2 UV-C.
Fig. 1.
Seedling growth of the wild-type (Col-0), and hy1-100, ho2, ho3, and ho4 mutants (A) and expression analysis of four HO genes of wild-type roots (B) in response to increasing doses of UV-C irradiation. After exposure to corresponding doses of UV-C, seedlings of each ecotype were transferred to normal growth conditions for 5 d, after which the fresh weight per seedling was evaluated (A). After irradiation with different doses of UV-C, transcript levels of HY1 (At2g26670), HO2 (At2g26550), HO3 (At1g69720), and HO4 (At1g58300) of wild-type roots were then analysed using real-time PCR (B). The levels of HO transcripts are presented relative to the wild type in control conditions (Con, without UV-C irradiation). Data are means ±SE from three independent experiments. (A) Mean values were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within UV irradiation treatments [model: phenotypic indicator=replication+treatment+sample (treatment)]. Mean values with different letters denote a significant difference according to multiple comparison (P < 0.05). (B) Bars with different letters were significantly different within the different UV irradiation treatments at P < 0.05 according to multiple comparison.
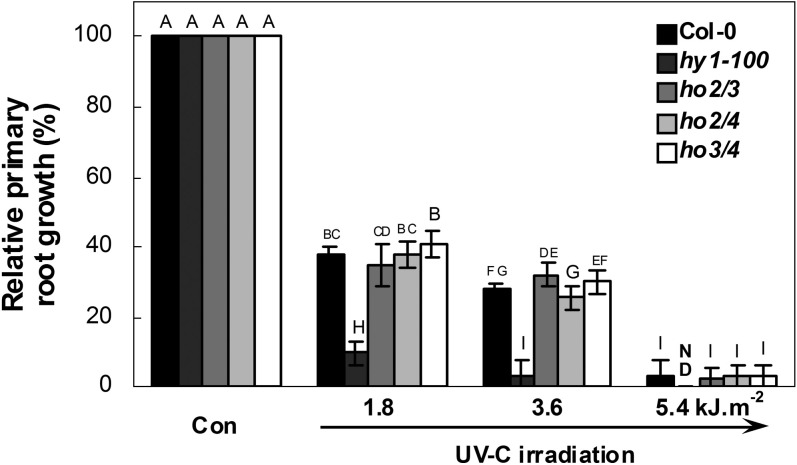
Further observation showed that the fresh weight of the hy1-100 mutant was 25.78±1.8% less than that of the wild type (Col-0) under normal growth conditions (Con, Fig. 1A). The relative decreases in fresh weight in wild-type and hy1-100 mutant plants upon UV-C exposure were then compared. The results further showed that fresh weight inhibition (mean ±SE) of hy1-100 mutant plants was 30.7±4.8, 57.0±3.0 and 66.5±1.3% at exposures of 1.8, 3.6, and 5.4 kJ m−2, respectively, in comparison with 16.7±3.5, 33.3±2.3, and 47.1±1.6% in wild-type plants (original data are shown in Fig. 1A), suggesting that the hy1-100 mutant was hypersensitive to UV-C. In comparison, no such severe inhibition appeared in the ho4 mutant. However, similar and/or slightly increased fresh weight was observed in ho2 and ho3 mutants, in comparison with the wild-type plants. Further redundancy tests showed that in contrast to the responses of hy1-100, the double mutants ho2/3, ho2/4, and ho3/4 did not show UV-C hypersensitivity, as revealed by changes in relative primary root growth, compared with the wild type (Fig. 2). These data clearly indicate that loss of HY1 increases sensitivity to UV-C irradiation.
Fig. 2.
Redundancy analysis of HO2, HO3, and HO4. After exposure to increasing doses of UV-C and transfer to normal growth conditions for 5 d, the primary root growth rate of each ecotype was measured, taking the growth rate of each ecotype grown in MS medium (Con) as 100%. Data are means ±SE from three independent experiments. Mean values were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within UV irradiation treatments [model: phenotypic indicator=replication+treatment+sample (treatment)]. Mean values with different letters denote a significant difference according to multiple comparison (P < 0.05). ND, not detected.
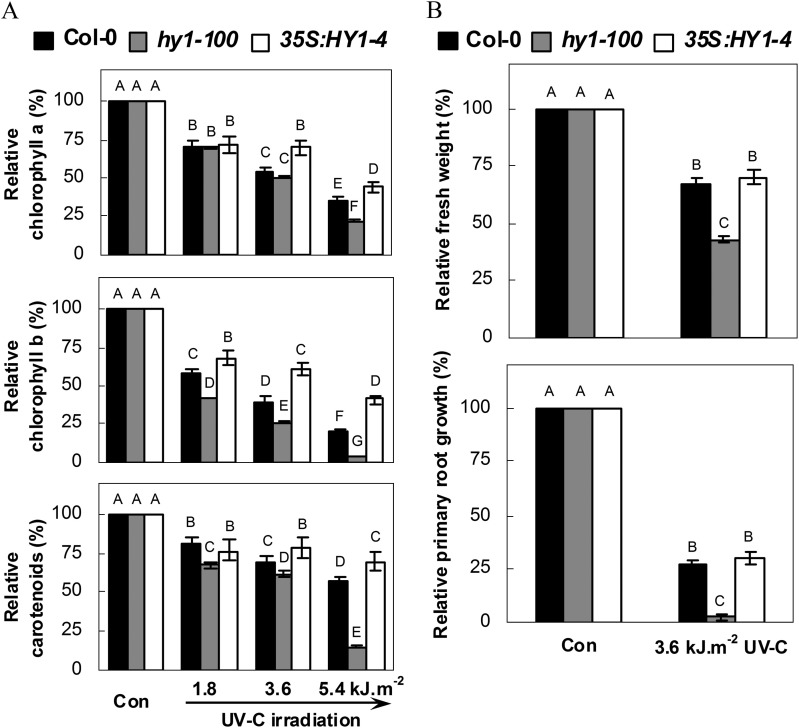
This conclusion was further confirmed by measuring the chlorophyll a, chlorophyll b, and carotenoid contents after UV-C exposure. The transgenic overexpressing plant line 35S:HY1-4 (Xie et al., 2011b ) was used as a positive control. In the experiments, in the wild-type UV-C irradiation resulted in dose-dependent losses of chlorophyll a and b, and reduced carotenoid contents were observed (Fig. 3A). The hy1-100 mutant exhibited a similar UV-C-hypersensitive tendency, consistent with the observed inhibition of seedling growth (Fig. 1) and changes in relative primary root growth (Fig. 2). Similarly, the Arabidopsis hy1 mutant (Ler background) also displayed a UV-C-hypersensitive phenotype (Supplementary Fig. S2 at JXB online). Contrasting responses, such as the alleviation of chlorophyll a, chlorophyll b, and carotenoid loss, were observed in the HY1 overexpression line. However, no significant differences were observed between the wild-type plants and the overexpressing plant line 35S:HY1-4 regarding changes in the inhibition of seedling growth or relative primary root growth (Fig. 3B). Taken together, these results further confirm that hy1 mediates Arabidopsis UV-C hypersensitivity. Therefore, a moderate dose of UV-C irradiation (3.6 kJ m−2) was used to investigate the role of hy1 in UV-C hypersensitivity throughout this study.
Fig. 3.
Effects of UV-C irradiation on relative chlorophyll a, chlorophyll b, and carotenoid contents (A) and seedling growth (B) of wild-type, hy1-100 mutant, and HY1-overexpressing line 35S:HY1-4 seedlings. After exposure to increasing doses of UV-C, seedlings of each ecotype were transferred to normal growth conditions for 5 d, and measured (n=15). Corresponding seedlings without UV-C irradiation were regarded as a control (Con, 100%). Data are means ±SE from three independent experiments. Mean values were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within UV irradiation treatments [model: phenotypic indicator=replication+treatment+sample (treatment)]. Mean values with different letters denote a significant difference according to multiple comparison (P < 0.05).
Changes in carotenoid biosynthetic genes
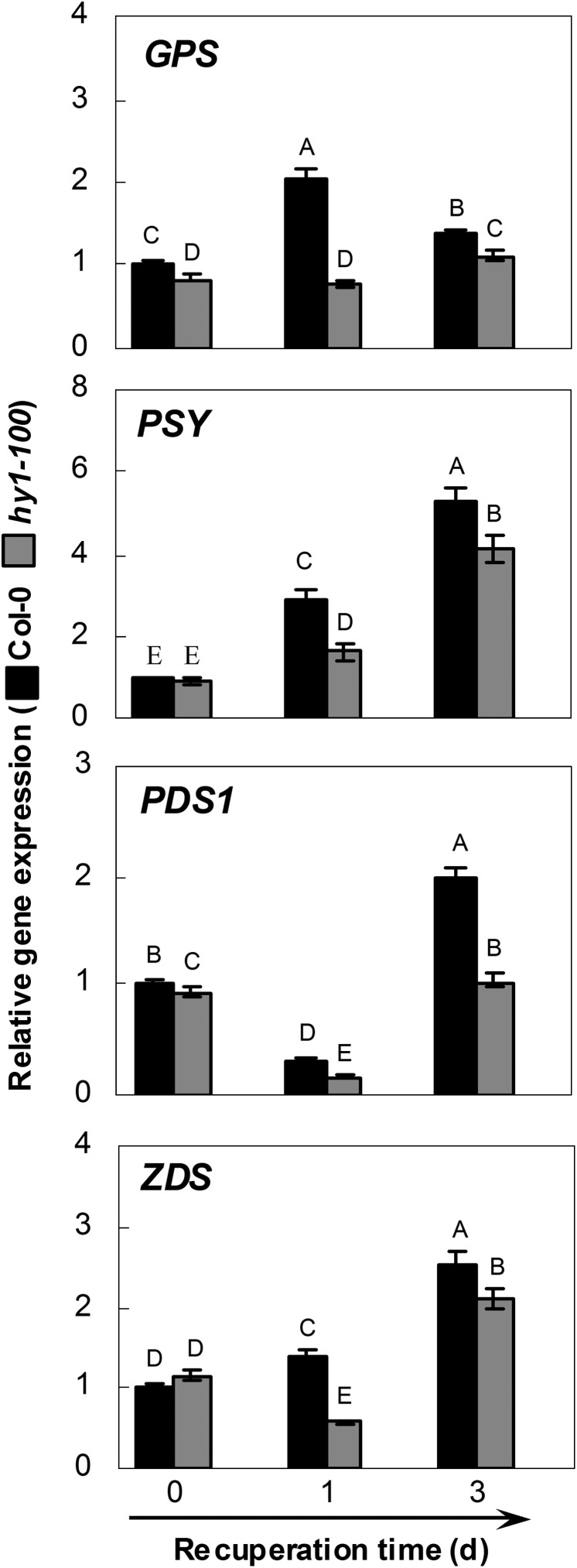
Results from microarray data showed four representative carotenoid biosynthetic genes, namely ZDS, GPS, PSY (in particular), and PDS1, which were induced in wild-type seedling roots during the 24 h period of UV-B irradiation (Supplementary Fig. S3 at JXB online). These inducible genes were then compared in order to study further the mechanisms of hy1-mediated UV-C hypersensitivity. A similar inducible phenomenon was observed in the wild type after 3 d recuperation following treatment with 3.6 kJ m−2 UV-C (Fig. 4). However, the expression of the above four genes was down-regulated in the hy1-100 mutant during the recovery test compared with the wild type, consistent with the changes in carotenoid contents (Fig. 3A).
Fig. 4.
Effects of UV-C irradiation on the gene expression in wild-type and hy1-100 mutant seedling roots. The transcript levels of representative carotenoid biosynthetic genes Geranylgeranyl Pyrophosphate Synthase (GPS; At2g34630), Phytoene synthase (PSY; At5g17230), Phytoene Desaturase1 (PDS1; At4g14210), and Zeta-carotene desaturase (ZDS; At3g04870) analysed by real-time PCR. After exposure to 3.6 kJ m−2 of UV-C, seedlings of each ecotype were allowed to recuperate under normal growth conditions, and samples were collected at the indicated times. Expression levels of each gene are presented relative to wild-type samples at 0 d. Mean values were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within different treatment times [model: phenotypic indicator=replication+time+sample (time)]. Mean values with different letters denote a significant difference according to multiple comparison (P < 0.05).
Changes in flavonoid metabolism
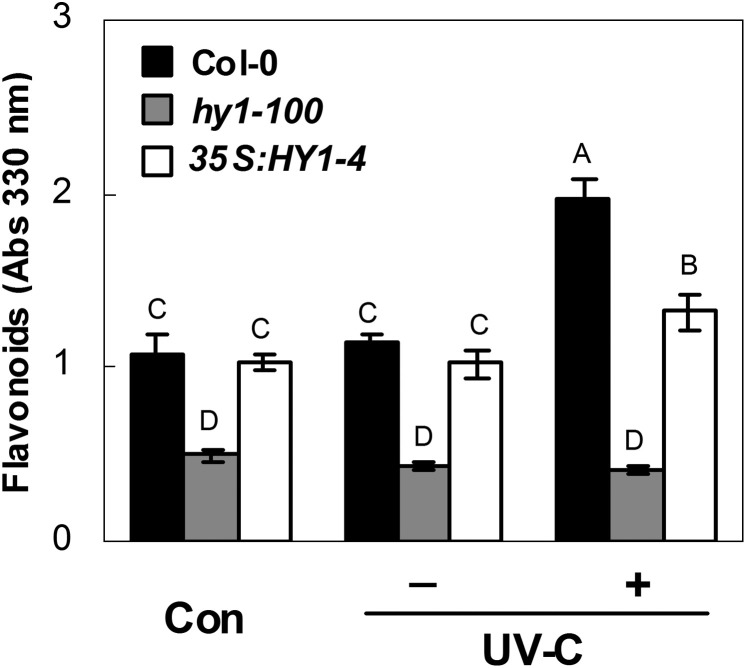
It is well known that flavonoids can act as sunscreen pigments to reduce the penetration of UV irradiation (Castells et al., 2010). Production of flavonoids from phenylalanine and acetyl-CoA is stimulated by increased UV-C (Filkowski et al., 2004). In the next experiment, it was shown that under normal growth conditions, hy1-100 extracts from shoot and root tissues (the latter in particular) had significantly lower levels of UV-absorbing compounds in the 220–450 nm range, the UV-A, UV-B, and part of the UV-C wavelength range (Supplementary Fig. S4A at JXB online). In particular, the absorbance measurement at 330 nm showed a 53.9% decrease in flavonoid contents in the root tissue of hy1-100 compared with the wild type (Con; Fig. 5). This finding also perfectly matches the changes in the four representative flavonoid biosynthetic genes, namely CHS (in particular), CHI, F3H, and FLS (Supplementary Fig. S4B at JXB online). When irradiated, the decrease of flavonoid in the hy1-100 mutant, with a reduction to 21.4% compared with the wild type, was greater compared with the HY1 overexpression line, with 67.9% compared with the wild type.
Fig. 5.
Effects of 3.6 kJ m−2 UV-C irradiation on the flavonoid contents of wild-type, hy1-100 mutant, and HY1-overexpressing line 35S:HY1-4 seedling roots. Five-day-old seedlings were exposed (+) or not (–) to UV-C irradiation. Afterwards, the flavonoid contents were determined. Samples before UV-C irradiation constituted the control (Con). Data are means ±SE from three independent experiments. Mean values were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within UV irradiation treatments [model: phenotypic indicator=replication+treatment+sample (treatment)]. Mean values with different letters denote a significant difference according to multiple comparison (P < 0.05).
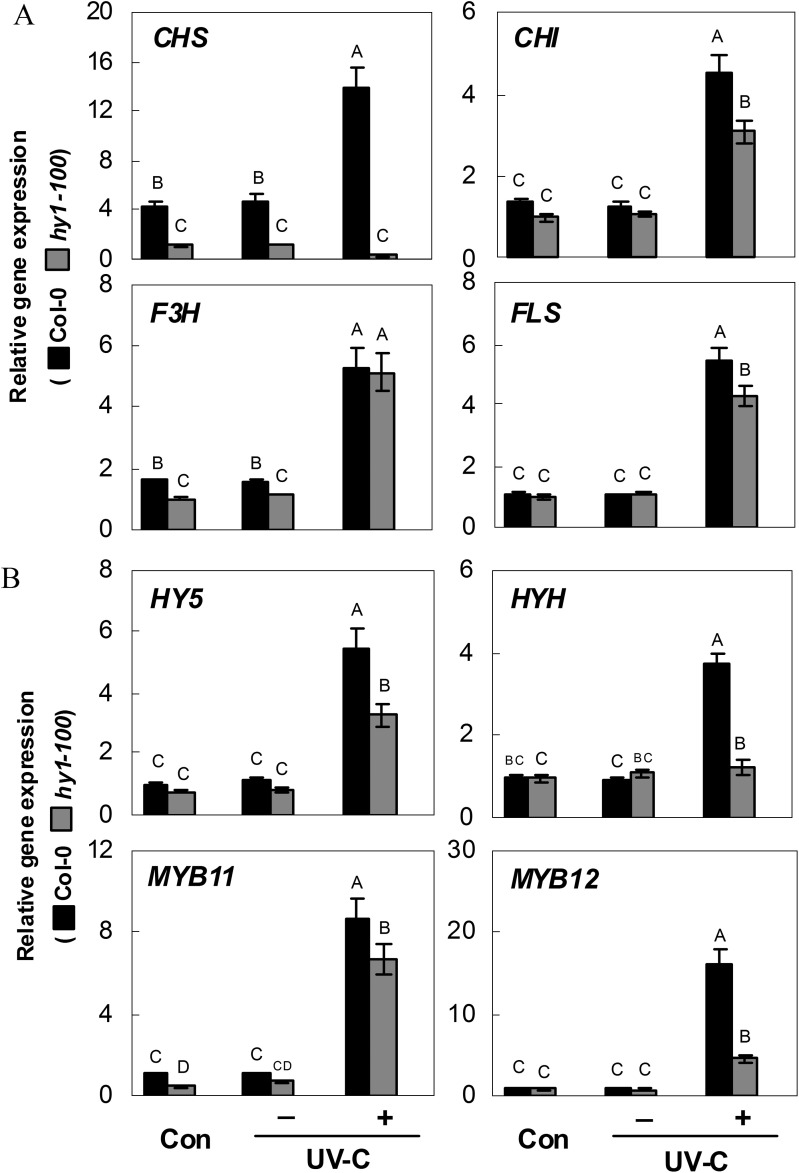
To determine whether the modulation of CHS, and other representative genes of the flavonoid biosynthetic pathway, in hy1-100 mutant seedling roots could explain these changes in flavonoid contents, quantitative RT-PCR analysis of CHS, CHI, F3H, and FLS mRNAs was performed on the same root extracts. This analysis showed that, compared with the wild type, expression of CHS is significantly down-regulated in hy1-100 roots exposed to UV-C irradiation, while F3H is unaffected, and CHI and FLS are moderately decreased (Fig. 6A). Similarly, further analysis revealed that expression of HY5, HYH, and MYB11/12, the genes encoding four transcription factors responsible for the regulation of flavonoid biosynthesis and UV responses, was reduced to 57.3±6.7, 32.5±4.8, 76.3±8.8. and 28.5±2.5%, respectively, compared with the wild type (Fig. 6B). Therefore, the down-regulated expression of early functional and related transcription factor genes in the flavonoid synthetic pathway may result in decreased levels of both kaempferol and quercetin flavonoids, which serve as precursors for the downstream flavonol compounds (Supplementary Fig. S1 at JXB online).
Fig. 6.
Effects of 3.6 kJ m−2 UV-C irradiation on the expression of genes encoding representative enzymes in the flavonoid biosynthetic pathway (A; Chalcone Synthase, CHS, At5g13930; Chalcone Isomerase, CHI, At3g55120; Flavanone 3-Hydroxylase, F3H, At3g51240; and Flavonol Synthase, FLS, At5g08640) and related transcription factors (B; Elongated Hypocotyl5, HY5, At5g11260; HY5 homologue, HYH, At3g17609; MYB11, At3g62610; and MYB12, At2g47460) in wild-type and hy1-100 mutant seedling roots. Five-day-old seedlings were exposed (+) or not (–) to UV-C irradiation. Samples before UV-C irradiation constituted the control (Con). Expression levels of each gene are presented relative to the hy1-100 mutant under normal growth conditions. Mean values were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within UV irradiation treatments [model: phenotypic indicator=replication+treatment+sample (treatment)]. Mean values with different letters denote a significant difference according to multiple comparison (P < 0.05).
Changes in hy1-mediated oxidative damage and ROS-scavenging enzyme genes
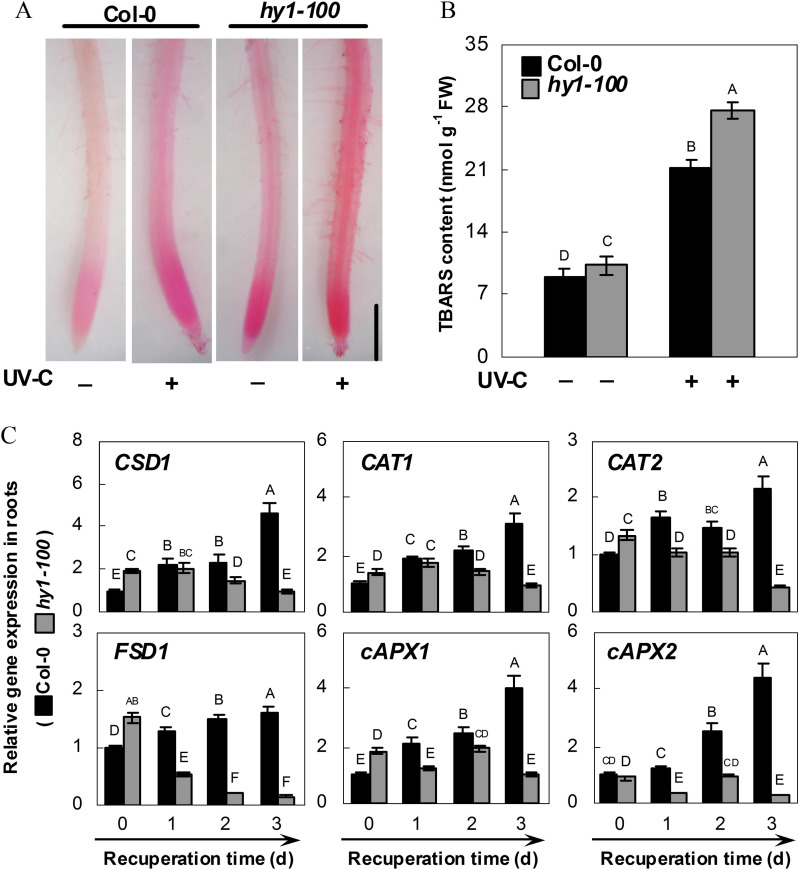
To determine whether wild-type and hy1-100 mutant plants differ in terms of oxidative stress induced by UV-C irradiation, assessment of lipid peroxidation in root tissues was performed using histochemical staining with Schiff’s reagent. As shown in Fig. 7A, lipid peroxidation staining in the wild-type or hy1-100 plants upon UV-C irradiation was more extensive than that under control conditions. The changes in the amount of TBARS showed that lipid peroxidation in the hy1-100 mutant increases to a greater extent than that in the wild type in response to UV-C irradiation (Fig. 7B). These results indicate that hy1-100 plants are less effective in counteracting UV-C-induced lipid peroxidation than wild-type plants.
Fig. 7.
Effect of 3.6 kJ m−2 UV-C irradiation on lipid peroxidation (A), TBARS content (B), and the expression of antioxidant defence genes (C) in the wild-type and the hy1-100 mutant. Five-day-old seedlings were exposed (+) or not (–) to UV-C irradiation. Seedlings of each ecotype were allowed to recuperate under normal growth conditions for 2 d. Afterwards, the TBARS content was determined (B). After another 1 d, corresponding roots were stained with Schiff’s reagent, and immediately photographed under a light microscope. Bar=0.5 mm (A). Transcript levels of Copper/zinc Superoxide Dismutase (CSD1; At1g08830), Catalase1 (CAT1; At1g20630), Catalase2 (CAT2; At4g35090); Fe Superoxide Dismutase1 (FSD1; At4g25100), cytosolic Ascorbate Peroxidase1 (cAPX1; At1g07890), and cytosolic Ascorbate Peroxidase2 (cAPX2; At3g09640) of root tissues of each ecotype were analysed by real-time PCR at the indicated times (C). Expression levels of each gene are presented relative to wild-type samples at 0 d. Mean values were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within UV irradiation treatments [model: phenotypic indicator=replication+treatment+sample (treatment)] (B) or within UV irradiation treatment times [model: phenotypic indicator=replication+time+sample (time)] (C). Mean values with different letters denote a significant difference according to multiple comparison (P < 0.05).
Because the data indicated a possible link between hy1-100 and UV-C-triggered lipid peroxidation, the responses of representative antioxidant enzyme genes to UV-C were tested. Time-course analysis showed that compared with the control samples (T0), significant increases in the transcripts of CSD1, CAT1, CAT2, FSD1, cAPX1, and cAPX2 were observed in wild-type seedling root (Fig. 7C) and shoot (Supplementary Fig. S5 at JXB online) tissues, after 3 d of recuperation. In contrast, compared with the wild type, significant down-regulation was seen in hy1-100 mutants, all of which was consistent with the histochemical staining and TBARS content results obtained previously (Fig. 7A, B).
Mutation of HY1-induced responses was rescued by CO and BR
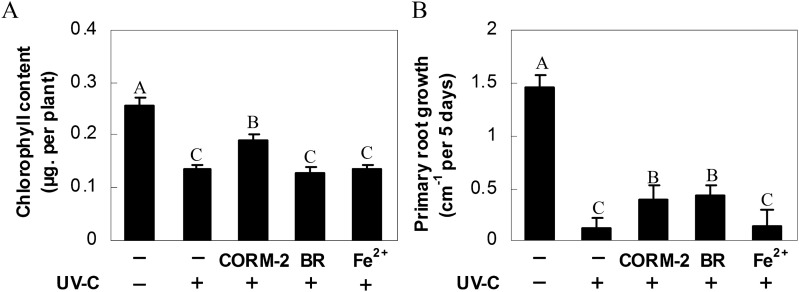
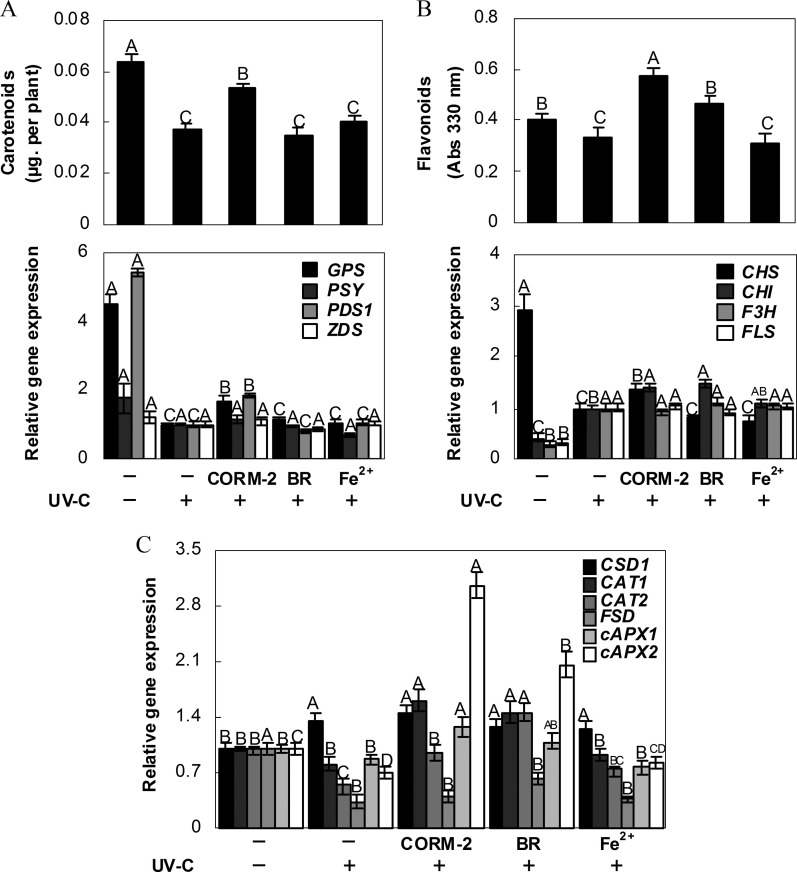
If the loss of HY1 function increases sensitivity to UV-C irradiation, feeding hy1-100 mutant plants with HO by-products might, at least partially, rescue UV-C-induced degradation of chlorophyll as well as the inhibition of primary root growth. As expected, exposure to the CO donor, CORM-2, significantly alleviated the decrease in chlorophyll (Fig. 8A), carotenoid, and flavonoid contents (Fig. 9A, B), and the inhibition of primary root growth (Fig. 8B), although no obvious response was found to the addition of Fe2+. Meanwhile, treatment with BR only alleviated the inhibition of primary root growth and the decrease in flavonoid content. After the addition of CORM-2, there were comparative increases in the levels of GPS and PDS1, and CHS and CHI mRNA in hy1-100 seedlings. Meanwhile, only CHI transcript was induced in BR-treated samples (Fig. 9B). In contrast, no significant changes were observed in the carotenoid and flavonoid synthetic genes in Fe2+-treated mutant plants subjected to UV-C stress, compared with those stressed but not treated with Fe2+. These changes in synthetic gene expression were consistent with the partial restoration of carotenoid content and synthesis of flavonoids following UV-C irradiation.
Fig. 8.
Effects of carbon monoxide-releasing molecule-2 (CORM-2), bilirubin (BR), and FeSO4 (Fe2+) pre-treatments on hy1-100 seedling growth under 3.6 kJ m−2 UV-C irradiation. Arabidopsis seeds were cultured in MS medium with or without 0.1 μM CORM-2, 100 μM BR, and 100 μM Fe2+ for 5 d. Afterwards, seedlings were exposed (+) or not (–) to UV-C irradiation. Chlorophyll content (A) and primary root growth rate (B) were then determined after another 5 d growth under normal conditions. Data are means ±SE from three independent experiments. Bars with different letters are significantly different compared with the control sample without UV-C irradiation at the P < 0.05 level according to multiple comparison.
Fig. 9.
Effects of CORM-2, BR, and Fe2+ pre-treatments on carotenoid and flavonoid metabolism as well as antioxidant enzyme genes in hy1-100 seedlings exposed to 3.6 kJ m−2 UV-C irradiation. Arabidopsis seeds were cultured in MS medium with or without 0.1 μM CORM-2, 100 M BR, and 100 μM FeSO4 for 5 d. Afterwards, seedlings were exposed (+) or not (–) to UV-C irradiation. Carotenoid (A) and flavonoid (B) contents and the expression of corresponding biosynthesis genes as well as the representative antioxidant enzyme genes (C) were determined after another 5 d growth under normal conditions. Data are means ±SE from four independent experiments. Bars with different letters are significantly different within the different treatments at the P < 0.05 level according to multiple comparison.
Changes in the antioxidant enzyme genes exhibited similar patterns (Fig. 9C). For example, in comparison with the stress alone samples, CAT1, CAT2, cAPX1, and cAPX2 transcripts were induced significantly in CORM-2-treated mutant roots. BR also brought about increases in CAT1, CAT2, and cAPX2 mRNA levels. However, no obvious difference was observed when plants were treated with Fe2+ together with UV-C stress.
Discussion
HY1, an indispensable endogenous modulator of plant UV-C tolerance
UV irradiation causes non-specific damage responses in living organisms, including decreased biomass accumulation, acceleration of chlorophyll degradation, and overproduction of ROS (Gao and Zhang, 2008; Gao et al., 2008; He et al., 2008; Castells et al., 2010; Tao et al., 2010). Previous pharmacological tests have shown that HO-1 is involved in soybean defence against the oxidative stress generated by UV-B radiation, and that ROS are involved in the mechanisms inducing HO-1 (Yannarelli et al., 2006; Santa-Cruz et al., 2010). In this study, several consequences of UV-C hypersensitivity were determined (Bashandy et al., 2009; Castells et al., 2010), including inhibition of the increase in fresh weight (Fig. 1) and relative primary root growth (Fig. 2), and the degradation of chlorophyll a/b (Fig. 3). It was discovered that HY1 plays a central role, acting as an indispensable endogenous modulator of plant UV-C tolerance. This conclusion was supported by genetic and pharmacological evidence. For example, compared with ho2/ho3/ho4 mutants, the maximal UV-C-hypersensitive phenotype was observed in hy1-100 plants (Col-0 background; Fig. 1). The Arabidopsis hy1 mutant from the Ler background also displayed a similar hypersensitive phenotype (Supplementary Fig. S2 at JXB online). The maximal inducible expression of HY1 in the wild type increased in a dose-dependent manner with increasing doses of UV-C, compared with HO2/HO3/HO4 transcripts. A functional redundancy analysis of ho2/3, ho2/4, and ho3/4 double mutants further confirmed that UV-C-induced maximal hypersensitivity was hy1 specific (Fig. 2). As expected (Cao et al., 2011; Wu et al., 2011), the addition of the CO donors CORM-2 (in particular) and BR, two catalytic by-products of HY1, could partially rescue the hypersensitive phenotype of the hy1 mutant (Fig. 8). Together, these results indicate that the Arabidopsis HY1 mutation is required for UV-C hypersensitivity.
A very recent study has provided genetic and pharmacological evidence that hy1-100 showed maximal sensitivity to salinity and no acclimation response, whereas plants overexpressing HY1 (35S:HY1-3/4) showed tolerance characteristics (Xie et al., 2011b ). However, it should be noted that, except for the alleviation of chlorophyll a, chlorophyll b, and carotenoid degradation upon exposure to UV-C (Fig. 3A), no visible hyposensitivity phenotype was observed in the HY1-overexpressing line 35S:HY1-4 (Fig. 3B). To account for this discrepancy, it is suggested that additional factors may be required for HY1 to function in promoting plant tolerance against UV-C irradiation, and that its signalling mechanism might be different from that for salt tolerance/acclimation. Similar phenomena regarding the lack of a phenotypic change in a gain-of-function mutant compared with the wild type were also reported in a regulatory network involving YAB3, WOX3, and KNOX genes, required for leaf development in rice (Dai et al., 2007). Additionally, the possibility of the fact that the protective action of HO-1 may be restricted to a narrow threshold could not be easily ruled out (Suttner and Dennery, 1999; Bauer et al., 2008).
It was previously reported that Arabidopsis HY1 was required for phytochrome chromophore biosynthesis (Muramoto et al., 1999; Davis et al., 2001), and that phytochrome A and light-stable phytochromes appeared to be directly involved in the regulation of specific genes in the carotenoid biosynthetic pathway (von Lintig et al., 1997). Additionally, the yellow-green-2 mutant of tomato and the hy1 mutant of Arabidopsis, both of which are defective in HO-1, display the reduced chlorophyll phenotype as a result of reduced 5-aminolaevulinic acid (ALA) formation, one of the two pivotal control points of tetrapyrrole biosynthesis (Terry and Kendrick, 1999; Davis et al., 2001; Cornah et al., 2003). These results might partially explain why the alleviation or aggravation of chlorophyll a, chlorophyll b, or carotenoid degradation on exposure to UV-C was observed in the HY1-overexpressing line 35S:HY1-4 or hy1-100 mutant seedlings, respectively (Fig. 3A).
Decreases in carotenoid and flavonoid biosynthesis and antioxidant defences: three major mechanisms responsible for hy1-induced UV-C hypersensitivity
The degree of UV-C-induced damage to plants is strongly correlated with the efficiency of their UV-stimulated photooxidative protection and repair mechanisms (Bieza and Lois, 2001; Frohnmeyer and Staiger, 2003; Castells et al., 2010). Plants have evolved survival mechanisms to eliminate the overproduction of ROS induced by UV irradiation, including mechanisms to modulate the metabolism of carotenoids and flavonoids, and to activate antioxidant defences (Li et al., 1993; Gao and Zhang, 2008; Liu et al., 2011; Tossi et al., 2011). In plants, carotenoids are the essential group of pigments responsible for quenching the ROS (singlet oxygen, etc.) induced by exposure to UV radiation (Bartley and Scolnik, 1995). The common carotenoid biosynthetic pathway has been described, and carotenoids serve as precursors in the biosynthesis of vitamin A and ABA (Qin et al., 2007). In this study, it was shown that the decreases in carotenoid content in wild-type, hy1-100 mutant, and HY1 overexpression line plants were approximately positively correlated with UV-C dose levels (Fig. 3A). For example, the relative carotenoid content of the hy1-100 mutant decreased greatly (86.7%) after irradiation with 5.4 kJ m−2 UV-C, compared with ∼42.3% in wild-type plants. In contrast, in comparison with corresponding wild-type plants, relative carotenoid levels rose notably in HY1 overexpression line plants at the 3.6 kJ m−2 and 5.4 kJ m−2 UV-C exposures tested. In addition, four carotenoid biosynthesis genes (Fig. 4) differentially decreased in the hy1-100 mutant seedling roots 1 d after recuperation. These results, combined with the severe chlorophyll degradation in hy1-100 exposed to UV-C (Fig. 3A), are consistent with those reported by Moliné et al. (2009), who found that enhanced carotenoid biosynthesis in yeast might be an early response to combat UV exposure, and also with a previous study where nuclear-encoded photosynthetic genes were repressed in a bleached carotenoid-deficient mutant (Mayfield and Taylor, 1984). Functional complementation has also confirmed that disruption of Arabidopsis PDS3 results in albino and dwarf phenotypes by impairing chlorophyll, carotenoid, and gibberellin biosynthesis (Qin et al., 2007). In contrast, pre-treatments with CORM-2, a CO donor, significantly increased GPS and PDS1 transcripts (Fig. 9A) as well as chlorophyll content (Fig. 8A), and weakened the UV-C hypersensitivity as shown by the inhibition of primary root growth in the hy1-100 mutant (Fig. 8B). No significant responses were observed when Fe2+ was added. The above results clearly suggest that the UV-C hypersensitivity of hy1-100 might be mainly due to the lack of CO production, one of HY1’s enzymatic products. Additionally, the possible antioxidant role of BR in the attenuation of UV-C hypersensitivity (Figs 8, 9) cannot be easily ruled out (Noriega et al., 2004; Santa-Cruz et al., 2010).
Flavonoids are considered ubiquitous in plants and affect many facets of their physiology, including UV absorbance, modulation of ROS levels, defence, and hormone transport (Buer et al., 2010). Genetic studies indicate that flavonoid-deficient mutants are susceptible to UV exposure. The cytoprotective functions of flavonoids are unique and are active in protecting against long-term light stress, but do not protect against rapid light intensity fluctuations (Li et al., 1993; Bartley and Scolnik, 1995; Havaux and Kloppstech, 2001). These previous studies suggest that reductions in flavonoid biosynthesis might lead to UV hypersensitivity. In the second experiment in the present study, mutation of hy1-induced UV-C hypersensitivity was accompanied by reduced flavonoid contents (Fig. 5) as well a reduction in their biosynthesis (Fig. 6). For example, the expression level of CHS, which catalyses the first committed step of flavonoid biosynthesis, was obviously inhibited in the hy1-100 mutant regardless of UV-C exposure. Similarly, four well-known CHS-related transcription factor genes (Stracke et al., 2007), HY5, HYH, MYB11, and MYB12 (in particular), displayed significantly reduced tendencies in the hy1-100 mutant upon UV-C treatment, compared with the corresponding wild-type plants (Fig. 6B). In contrast, some of the responses noted above could be blocked by pre-treatment with CORM-2 (in particular) and BR (Fig. 9B). These results are consistent with the partial reversal of hypersensitivity in the hy1-100 mutant (Fig. 9B), suggesting that the mutation of Arabidopsis HY1-induced UV-C hypersensitivity might be due to the down-regulation of flavonoid biosynthesis. A significant, but much weaker reduction in flavonoid content was observed in the HY1 overexpression line, compared with the wild type (Fig. 5). These results also suggest that besides its fundamental role in acting as an antioxidant enzyme, as previously reported (Noriega et al., 2004), HY1 itself and/or its catalytic products might be involved in flavonoid metabolism (Fig. 9B).
Previous studies reported that the up-regulation of soybean HO-1 served as a cell protection mechanism against UV-induced oxidative damage (Yannarelli et al., 2006). UV irradiation is known to induce ROS generation, resulting in oxidative stress, and UV tolerance is partially correlated with an efficient antioxidant system (Gao and Zhang, 2008; Gao et al., 2008). Previous work also illustrated that the Arabidopsis ascorbic acid-deficient mutant vitamin c-1 (vtc1) was more sensitive to supplementary UV-B treatment than wild-type plants, and ascorbic acid was considered an important antioxidant for UV-B radiation (Gao and Zhang, 2008). Subsequently, it was observed that hy1-100 mutants suffered severe oxidative stress upon UV-C irradiation, compared with the wild type, and this was confirmed by histochemical staining for the detection of lipid peroxidation in root apexes (Fig. 7A) and TBARS content determination (Fig. 7B). This result also suggests that mutation of Arabidopsis HY1 makes it more vulnerable to UV-C-induced oxidative stress than wild-type plants. Meanwhile, this phenotype was ascribed to decreasing expression levels of transcripts encoding antioxidant enzymes, including CSD1, CAT1/2, FSD1, and cAPX1/2 (Fig. 7C; Supplementary Fig. S5 at JXB online), which were further blocked differentially when CORM-2 (in particular) or BR was added, separately (Fig. 9C). This finding further confirms that HO-1 could confer beneficial cytoprotection against oxidative damage in both animals and plants (Yannarelli et al., 2006; Paine et al., 2010; Santa-Cruz et al., 2010; Kim et al., 2011).
In summary, hy1-100, an Arabidopsis mutant with a defect in HY1, displayed a hypersensitivity to UV-C irradiation compared with wild-type plants, as indicated by: (i) a greater inhibition of seedling growth and primary root growth; (ii) marked degradation of chlorophyll; (iii) lower biosynthesis of carotenoids and flavonoids; (iv) a greater accumulation of lipid peroxidation; and (v) the down-regulation of the antioxidant system. However, the addition of the CO donors CORM-2 (in particular) and BR, two catalytic by-products of HY1, could partially rescue the above responses in the hy1 mutant. These genetic, molecular, and biochemical results provide the first indication, to our knowledge, that the mutation of HY1-triggered UV-C hypersensitivity is associated with decreases in carotenoid and flavonoid synthesis, as well as antioxidant defences (Fig. 10). A recently published paper has reported a causal relationship between the UV-C tolerance of tobacco plants and Ca2+/H+ exchanger-mediated fast cytosolic Ca2+ extrusion (within 30 min; Shabala et al., 2011). Meanwhile, ROS production is also known to be critically dependent on cytosolic Ca2+ levels (Lecourieux et al., 2002). Although the identification of the interrelationship between the above signalling components and HY1 might be beyond the scope of this work, in view of the fact that no visible hyposensitivity phenotype was observed in the HY1-overexpressing line (Fig. 3B), further genetic and molecular investigations will be needed to explore the possible endogenous factors (Ca2+ or ROS signalling, etc.) required for HY1 function in promoting plant tolerance to UV-C irradiation.
Fig. 10.
Schematic representation of the proposed hy1-mediated UV-C hypersensitivity signalling pathway. Mutation of HY1 triggered decreases in carotenoid and flavonoid synthesis, as well as antioxidant defence, further leading to UV-C hypersensitivity. The dashed line and the question mark denote the possible involvement of other undescribed signalling components, such as Ca2+ and ROS signals.
Supplementary data
Supplementary data are available at JXB online.
Figure S1 . Scheme of the flavonoid biosynthetic pathway in Arabidopsis.
Figure S2 . Primary root growth of wild-type (Ler) and hy1 mutant seedlings in response to 3.6 kJ m−2 UV-C irradiation.
Figure S3 . Representation of carotenoid biosynthetic genes in Arabidopsis seedling root tissues upon UV-B irradiation.
Figure S4 . The hy1-100 mutant underproduces UV-absorbing compounds.
Figure S5 . Effects of 3.6 kJ m−2 UV-C on expression of antioxidant defence genes.
Table S1 . The sequences of PCR primers for genotyping.
Table S2 . The sequences of PCR primers for real-time RT-PCR.
Acknowledgments
This work was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 project, and the Fundamental Research Funds for the Central Universities (grant nos KYZ200905 and KYJ200912). We sincerely thank Dr Tianqing Zheng (Institute of Crop Sciences/National Key Facility for Crop Gene Resources and Genetic Improvement, Chinese Academy of Agricultural Sciences) and Professor Rongzhan Guan (State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University) for their kind help rendered in statistical analysis.
References
- Applegate LA, Luscher P, Tyrrell RM. Induction of heme oxygenase: a general response to oxidant stress in cultured mammalian cells. Cancer Research. 1991;51:974–978. [PubMed] [Google Scholar]
- Balestrazzi A, Locato V, Bottone MG, Gara LD, Biggiogera M, Pellicciari C, Botti S, Gesù D, Donà M, Carbonera D. Response to UV-C radiation in topo I-deficient carrot cells with low ascorbate levels. Journal of Experimental Botany. 2009;61:575–585. doi: 10.1093/jxb/erp323. [DOI] [PubMed] [Google Scholar]
- Bartley GE, Scolnik DA. Plant carotenoids: pigments for photoprotection, visual attraction, and human health. The Plant Cell. 1995;7:1027–1038. doi: 10.1105/tpc.7.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T, Taconnat L, Renou JP, Meyer Y, Reichheld JP. Accumulation of flavonoids in an ntra ntrb mutant leads to tolerance to UV-C. Molecular Plant. 2009;2:249–258. doi: 10.1093/mp/ssn065. [DOI] [PubMed] [Google Scholar]
- Bauer M, Huse K, Settmacher U, Claus RA. The heme oxygenase–carbon monoxide system: regulation and role in stress response and organ failure. Intensive Care Medicine. 2008;34:640–648. doi: 10.1007/s00134-008-1010-2. [DOI] [PubMed] [Google Scholar]
- Bieza K, Lois R. An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiology. 2001;126:1105–1115. doi: 10.1104/pp.126.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilban M, Haschemi A, Wegiel B, Chin BY, Wagner O, Otterbein LE. Heme oxygenase and carbon monoxide initiate homeostatic signaling. Journal of Molecular Medicine. 2008;86:267–279. doi: 10.1007/s00109-007-0276-0. [DOI] [PubMed] [Google Scholar]
- Bin Alam MDZ, Otaki M, Furumai H, Ohgaki S. Direct and indirect inactivation of Microcystis aeruginosa by UV-radiation. Water Research. 2001;35:1008–1014. doi: 10.1016/s0043-1354(00)00357-2. [DOI] [PubMed] [Google Scholar]
- Booij-James IS, Dube SK, Jansen MA, Edelman M, Mattoo AK. Ultraviolet-B radiation impacts light-mediated turnover of the photosystem II reaction center heterodimer in Arabidopsis mutants altered in phenolic metabolism. Plant Physiology. 2000;124:1275–1284. doi: 10.1104/pp.124.3.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA. Flavonoids: new roles for old molecules. Journal of Integrative Plant Biology. 2010;52:98–111. doi: 10.1111/j.1744-7909.2010.00905.x. [DOI] [PubMed] [Google Scholar]
- Calabrese V, Cornelius C, Dinkova-Kostova AT, Calabrese EJ, Mattson MP. Cellular stress responses, the hormesis paradigm, and vitagenes: novel targets for therapeutic intervention in neurodegenerative disorders. Antioxidants and Redox Signaling. 2010;13:1763–1811. doi: 10.1089/ars.2009.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao ZY, Geng BB, Xu S, Xuan W, Nie L, Shen WB, Liang YC, Guan RZ. BnHO1, a haem oxygenase-1 gene from Brassica napus, is required for salinity and osmotic stress-induced lateral root formation. Journal of Experimental Botany. 2011;62:4675–4689. doi: 10.1093/jxb/err190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao ZY, Huang BK, Wang QY, Xuan W, Ling TF, Zhang B, Chen X, Shen WB. Involvement of carbon monoxide produced by heme oxygenase in ABA-induced stomatal closure in Vicia faba and its proposed signal transduction pathway. Chinese Science Bulletin. 2007;52:2365–2373. [Google Scholar]
- Castells E, Molinier J, Drevensek S, Genschik P, Barneche F, Bowler C. Det1-1-induced UV-C hyposensitivity through UVR3 and PHR1 photolyase gene over-expression. The Plant Journal. 2010;63:392–404. doi: 10.1111/j.1365-313X.2010.04249.x. [DOI] [PubMed] [Google Scholar]
- Chary P, Dillon D, Schroeder AL, Natvig DO. Superoxide dismutase (sod-1) null mutants of Neurospora crassa: oxidative stress sensitivity, spontaneous mutation rate and response to mutagens. Genetics. 1994;137:723–730. doi: 10.1093/genetics/137.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close DC, MuArthur C, Hagerman AE, Davies NW, Beadle CL. Phenolic acclimation to ultraviolet-A irradiation in Eucalyptus nitens seedlings raised across a nutrient environment gradient. Photosynthetica. 2007;45:36–42. [Google Scholar]
- Cornah JE, Terry MJ, Smith AG. Green or red: what stops the traffic in the tetrapyrrole pathway. Trends in Plant Science. 2003;8:224–230. doi: 10.1016/S1360-1385(03)00064-5. [DOI] [PubMed] [Google Scholar]
- Dai M, Hu Y, Zhao Y, Liu H, Zhou D. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiology. 2007;144:380–390. doi: 10.1104/pp.107.095737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD. The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiology. 2001;126:656–669. doi: 10.1104/pp.126.2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmard M, Foresti R, Daqoussat M, et al. Differential antibacterial activity against Pseudomonas aeruginosa by carbon monoxide-releasing molecules. Antioxidants and Redox Signaling. 2012;16:153–163. doi: 10.1089/ars.2011.3959. [DOI] [PubMed] [Google Scholar]
- Filkowski J, Kovalchuk O, Kovalchuk I. Genome stability of vtc1, tt4, and tt5 Arabidopsis thaliana mutants impaired in protection against oxidative stress. The Plant Journal. 2004;38:60–69. doi: 10.1111/j.1365-313X.2004.02020.x. [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H, Staiger D. Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiology. 2003;133:1420–1428. doi: 10.1104/pp.103.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao CJ, Xing D, Li LL, Zhang LR. Implication of reactive oxygen species and mitochondrial dysfunction in the early stages of plant programmed cell death induced by ultraviolet-C overexposure. Planta. 2008;227:755–767. doi: 10.1007/s00425-007-0654-4. [DOI] [PubMed] [Google Scholar]
- Gao Q, Zhang LX. Ultraviolet-B-induced oxidative stress and antioxidant defense system responses in ascorbate-deficient vtc1 mutants of Arabidopsis thaliana . Journal of Plant Physiology. 2008;165:138–148. doi: 10.1016/j.jplph.2007.04.002. [DOI] [PubMed] [Google Scholar]
- García-Arnandis I, Guillén MI, Gomar F, Casteión MA, Alcaraz MJ. Control of cell migration and inflammatory mediators production by CORM-2 in osteoarthritic synoviocytes. PLoS One. 2011;6:e24591. doi: 10.1371/journal.pone.0024591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisk B, Yasui Y, Kohchi T, Frankenberg-Dinkel N. Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochemical Journal. 2010;425:425–434. doi: 10.1042/BJ20090775. [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang J, Chen XY, Gao ZZ, Xuan W, Xu S, Ding X, Shen WB. Carbon monoxide alleviates cadmium-induced oxidative damage by modulating glutathione metabolism in the roots of Medicago sativa . New Phytologist. 2008;177:155–166. doi: 10.1111/j.1469-8137.2007.02251.x. [DOI] [PubMed] [Google Scholar]
- Havaux M, Kloppstech K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npg and tt mutants. Planta. 2001;213:953–966. doi: 10.1007/s004250100572. [DOI] [PubMed] [Google Scholar]
- He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. Journal of Biological Chemistry. 2008;283:774–783. doi: 10.1074/jbc.M704185200. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. The interplay between light and jasmonate signaling during defence and development. Journal of Experimental Botany. 2011;62:4087–4100. doi: 10.1093/jxb/err142. [DOI] [PubMed] [Google Scholar]
- Kim YM, Pae HO, Park JE, Lee YC, Woo JM, Kim NH, Choi YK, Lee BS, Kim SR, Chung HT. Heme oxygenase in the regulation of vascular biology: from molecular mechanisms to therapeutic opportunities. Antioxidants and Redox Signaling. 2011;14:137–167. doi: 10.1089/ars.2010.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgeßner HD, Reichert K, Hauff K, Steinbrecher R, Schnitzler JP, Pfündel EE. Light and temperature, but not UV radiation, affect chlorophylls and carotenoids in Norway spruce needles (Picea abies (L.) Karst.) Plant, Cell and Environment. 2003;26:1169–1179. [Google Scholar]
- Landry LG, Chapple CC, Last RL. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiology. 1995;109:1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux D, Mazars C, Pauly N, Ranjeva R, Pugin A. Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. The Plant Cell. 2002;14:2627–2641. doi: 10.1105/tpc.005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. The Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods in Enzymology. 1987;148:350–382. [Google Scholar]
- Liu C, Cai L, Han X, Ying T. Temporary effect of postharvest UV-C irradiation on gene expression profile in tomato fruit. Gene. 2011;486:56–64. doi: 10.1016/j.gene.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Loboda A, Jazwa A, Grochot-Przeczek A, Rutkowski AJ, Cisowski J, Agarwal A, Jozkowicz A, Dulak J. Heme oxygenase-1 and the vascular bed: from molecular mechanisms to therapeutic opportunities. Antioxidants and Redox Signaling. 2008;10:1767–1812. doi: 10.1089/ars.2008.2043. [DOI] [PubMed] [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiology. 2004;135:2379–2391. doi: 10.1104/pp.104.042408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Taylor WC. Carotenoid-deficient maize seedlings fail to accumulate light-harvesting chlorophyll a/b binding protein (LHCP) mRNA. European Journal of Biochemistry. 1984;144:79–84. doi: 10.1111/j.1432-1033.1984.tb08433.x. [DOI] [PubMed] [Google Scholar]
- Moliné M, Libkind D, Diéguez Mdel C, van Broock M. Photoprotective role of carotenoids in yeast: response to UV-B of pigmented and naturally-occurring albino strains. Journal of Photochemistry and Photobiology. B, Biology. 2009;95:156–161. doi: 10.1016/j.jphotobiol.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of mutation in a plastid heme oxygenase. The Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega GO, Balestrasse KB, Batlle A, Tomaro ML. Heme oxygenase exerts a protective role against oxidative stress in soybean leaves. Biochemical and Biophysical Research Communications. 2004;323:1003–1008. doi: 10.1016/j.bbrc.2004.08.199. [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R. CONSTITUTIVELY PHOTOMORPHOGENICI is required for the UV-B response in Arabidopsis . The Plant Cell. 2006;18:1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterbein LE, Soares MP, Yamashita K, Bach FH. Heme oxygenase-1: unleashing the protective properties of heme. Trends in Immunology. 2003;24:449–455. doi: 10.1016/s1471-4906(03)00181-9. [DOI] [PubMed] [Google Scholar]
- Paine A, Eiz-Vesper B, Blasczyk R, Immenschuh S. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochemical Pharmacology. 2010;80:1895–1903. doi: 10.1016/j.bcp.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Prawan A, Kundu JK, Surh YJ. Molecular basis of heme oxygenase-1 induction: implications for chemoprevention and chemoprotection. Antioxidants and Redox Signaling. 2005;7:1688–1703. doi: 10.1089/ars.2005.7.1688. [DOI] [PubMed] [Google Scholar]
- Qin G, Gu HY, Ma LG, Peng YB, Deng XW, Chen ZL, Qu LJ. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Research. 2007;17:471–482. doi: 10.1038/cr.2007.40. [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Renzing J, Hansen S, Lane DP. Oxidative stress is involved in the UV activation of p53. Journal of Cell Science. 1996;109:1105–1112. doi: 10.1242/jcs.109.5.1105. [DOI] [PubMed] [Google Scholar]
- Sakai H, Katayama H, Oguma K, Ohgaki S. Kinetics of Microcystis aeruginosa growth and intracellular microcystins release after UV irradiation. Environmental Science and Technology. 2009;43:896–901. doi: 10.1021/es802246x. [DOI] [PubMed] [Google Scholar]
- Santa-Cruz DM, Pacienza NA, Polizio AH, Balestrasse KB, Tomaro ML, Yannarelli GG. Nitric oxide synthase-like dependent NO production enhances heme oxygenase up-regulation in ultraviolet-B-irradiated soybean plants. Phytochemistry. 2010;71:1700–1707. doi: 10.1016/j.phytochem.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Saxena SC, Joshi PK, Grimm B, Arora S. Alleviation of ultraviolet-C-induced oxidative damage through overexpression of cytosolic ascobate peroxidase. Biologia. 2011;66:1052–1059. [Google Scholar]
- Shabala S, Baekgaard L, Shabala L, Fuglsang A, Babourina O, Palmgren MG, Cuin TA, Rengel Z, Nemchinov LG. Plasma membrane Ca2+ transporters mediate virus-induced acquired resistance to oxidative stress. Plant, Cell and Environment. 2011;34:406–417. doi: 10.1111/j.1365-3040.2010.02251.x. [DOI] [PubMed] [Google Scholar]
- Shekhawat GS, Verma K. Haem oxygenase (HO): an overlooked enzyme of plant metabolism and defence. Journal of Experimental Botany. 2010;61:2255–2270. doi: 10.1093/jxb/erq074. [DOI] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant, Cell and Environment. 2010;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttner DM, Dennery PA. Reversal of HO-1 related cytoprotection with increased expression is due to reactive iron. FASEB Journal. 1999;13:1800–1809. doi: 10.1096/fasebj.13.13.1800. [DOI] [PubMed] [Google Scholar]
- Tao Y, Zhang XH, Au DW, Mao X, Yuan K. The effects of sub-lethal UV-C irradiation on growth and cell integrity of cyanobacteria and green algae. Chemosphere. 2010;78:541–547. doi: 10.1016/j.chemosphere.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore deficient aurea and yellow-green-2 mutants of tomato. Plant Physiology. 1999;119:143–152. doi: 10.1104/pp.119.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi V, Amenta M, Lamattina L, Cassia R. Nitric oxide enhances plant ultraviolet-B protection up-regulating gene expression of the phenylpropanoid biosynthetic pathway. Plant, Cell and Environment. 2011;34:909–921. doi: 10.1111/j.1365-3040.2011.02289.x. [DOI] [PubMed] [Google Scholar]
- USEPA. Ultraviolet disinfection guidance manual for the final long term 2 enhanced surface water treatment rule. Appendix C, 2006:1–12.. http://www.epa.gov/safewater/disinfection/It2/compliance.html. [Google Scholar]
- von Lintig J, Welsch R, Bonk M, Giuliano G, Batschauer A, Kleinig H. Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings. The Plant Journal. 1997;12:625–634. doi: 10.1046/j.1365-313x.1997.00625.x. [DOI] [PubMed] [Google Scholar]
- Wu MZ, Huang JJ, Xu S, Ling TF, Xie YJ, Shen WB. Haem oxygenase delays programmed cell death in wheat aleurone layers by modulation of hydrogen peroxide metabolism. Journal of Experimental Botany. 2011;62:235–248. doi: 10.1093/jxb/erq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YJ, Cui WT, Yuan XX, Shen WB, Yang Q. Heme oxygenase-1 is associated with wheat salinity acclimation by modulating reactive oxygen species homeostasis. Journal of Integrative Plant Biology. 2011a;53:653–670. doi: 10.1111/j.1744-7909.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Xie YJ, Ling TF, Han Y, et al. Carbon monoxide enhances salt tolerance by nitric oxide-mediated maintenance of ion homeostasis and up-regulation of antioxidant defence in wheat seedling roots. Plant, Cell and Environment. 2008;31:1864–1881. doi: 10.1111/j.1365-3040.2008.01888.x. [DOI] [PubMed] [Google Scholar]
- Xie YJ, Xu S, Han B, Wu MZ, Yuan XX, Han Y, Gu Q, Xu DK, Yang Q, Shen WB. Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. The Plant Journal. 2011b;66:280–292. doi: 10.1111/j.1365-313X.2011.04488.x. [DOI] [PubMed] [Google Scholar]
- Xu S, Zhang B, Cao ZY, Ling TF, Shen WB. Heme oxygenase is involved in cobalt chloride-induced lateral root development in tomato. Biometals. 2011;24:181–191. doi: 10.1007/s10534-010-9386-1. [DOI] [PubMed] [Google Scholar]
- Xuan W, Zhu FY, Xu S, Huang BK, Ling TF, Qi JY, Ye MB, Shen WB. The heme oxygenase/carbon monoxide system is involved in the auxin-induced cucumber adventitious rooting process. Plant Physiology. 2008;148:881–893. doi: 10.1104/pp.108.125567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannarelli GG, Noriega GO, Batlle A, Tomaro ML. Heme oxygenase up-regulation in ultraviolet-B irradiated soybean plants involves reactive oxygen species. Planta. 2006;224:1154–1162. doi: 10.1007/s00425-006-0297-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.