Abstract
Physiological responses to abiotic stress in plants exhibit sexual differences. Females usually experience greater negative effects than males; however, little is known about the molecular mechanisms of sexual differences in abiotic stress responses. In the present study, transcriptional responses to salinity treatments were compared between male and female individuals of the poplar Populus yunnanensis. It was found that several functional groups of genes involved in important pathways were differentially expressed, including photosynthesis-related genes, which were mainly up-regulated in males but down-regulated in females. This gene expression pattern is consistent with physiological observations showing that salinity inhibited photosynthetic capacity more in females than in males. Furthermore, genes located in autosomes rather than in the female-specific region of the W chromosome are the major contributors to the sexual differences in the salinity tolerance of poplars. In conclusion, this study provided molecular evidence of sexual differences in the salinity tolerance of poplars. The identified sex-related genes in salinity tolerance and their functional groups will enhance our understanding of sexual differences in salinity stress at the transcription level.
Keywords: Next-generation sequencing, physiological responses, Populus, salinity tolerance, sexual differences, transcriptional profiling
Introduction
More than 800 Mha of land throughout the world are affected by salt (FAO, 2008). Among various salt types, sodium chloride is considered to be the most soluble and abundant salt. Although plants cannot escape from high salinity, they can trigger signals to alter their physiological characteristics and growth in order to survive (Zeller et al., 2009). Indeed, salinity stress affects virtually every aspect of plant physiology and metabolism. Therefore, an improvement of salinity tolerance in plants is an urgent issue and has been the target of several previous studies (Hasegawa et al., 2000; Zhu, 2000, 2001; Yokoi et al., 2002; Bartels and Sunkar, 2005; Munns, 2005; Munns and Tester, 2008). Many salt-responsive genes have been identified (Kawasaki et al., 2001; Seki et al., 2002; Rabbani et al., 2003; Dinneny et al., 2008) and several salinity-related signalling pathways have been predicted and confirmed (Knight and Knight, 2001; Zhu, 2002). Comparative transcriptional profiles of salt-sensitive (Arabidopsis thaliana) and salt-tolerant species (Thellungiella halophila) revealed a common set of genes in response to salt (Taji et al., 2004), and genes involved in photosynthesis, osmolyte production and transcription (Gong et al., 2005) exhibited species-specific differences, with only a few genes differentially regulated in the salt-tolerant T. halophila plants (Wong et al., 2006). Transcriptome studies also found that different functional categories of transcripts were differentially regulated at different time points (Kawasaki et al., 2001) and there is a cross-talk between signalling pathways in various stress conditions (Seki et al., 2002; Rabbani et al., 2003; Wong et al., 2006). These transcriptome studies have provided new knowledge of how to improve the salinity tolerance of plants.
Dioecious plants are an important component of terrestrial ecosystems, representing nearly 6% (14 620 of 240 000) of angiosperm species (Renner and Ricklefs, 1995). They play a crucial role in maintaining the stability of the structure and function in terrestrial ecosystems. Many dominant woody species, such as Populus, are dioecious. Sexual differences have been reported in several dioecious plants in response to salinity and other environmental stresses (Li et al., 2007; F. Chen et al., 2010; Zhang et al., 2011), and females usually show a lower tolerance capacity compared with males. For instance, P. yunnanensis males and females showed significant differences under salinity, drought, and, in particular, under a combination of both these stresses (L. Chen et al., 2010). Furthermore, male P. tremuloides trees were more responsive to elevated CO2 than females in terms of photosynthetic rates (Wang and Curtis, 2001). Sexual differences have also been observed in earlier studies on P. cathayana subjected to drought stress (Xu et al., 2008a , b ), UV-B radiation, and elevated CO2 (Zhao et al., 2009, 2011; Xu et al., 2010).
Because there is considerable variation in physiological responses between males and females, it is hypothesized that molecular responses may parallel salinity stress. However, molecular mechanisms underlying sex-related differences are largely unknown. Here the Solexa/Illumina’s digital gene expression (DGE) system (Blow, 2009; Morrissy et al., 2009) is used to investigate differential gene expression in responses to salinity stress between males and females of the poplar P. yunnanensis, which is naturally distributed at altitudes of 1300–2700 m in southwest China. The aims of the study are (i) to assess whether males are more tolerant to salinity stress than females; and (ii) to elucidate sex-related molecular mechanisms for salinity tolerance, and their possible relationship to sex chromosomes.
Materials and methods
Plant materials and experimental design
Two male and female trees of P. yunnanensis collected from a natural population in Meigu, Sichuan Province, China (28°18'N and 103°06'E), were used for a controlled intraspecific cross. From the F1 progeny, 10 male and 10 female individuals were vegetatively propagated to produce 50 male and 50 female cuttings, respectively. The cuttings were planted in 10 litre plastic pots filled with 8 kg of homogenized soil. After sprouting and growing for ∼2 months, healthy plants were replanted (one plant per pot). After another 2 months (July 2009), healthy plants with a similar crown size and equal height were chosen for the experiments. The experimental layout was completely randomized with two factors (sex and salt application). Therefore, there were finally four treatments: (i) males without salt application (control); (ii) females without salt application (control); (iii) males with salt application; and (iv) females with salt application. Nine plants of each sex were exposed to each treatment. Three replicates with three plants each were used to account for sampling errors. Salt-treated plants were firstly exposed to short-term low concentration salt pre-treatment (25 mM NaCl for 24 h) avoiding a large osmotic shock, and then to long-term salinity (50 mM NaCl for 7 d). Control plants were kept in well-watered conditions. After 8 d of salinity treatment, the fourth fully expanded and intact young leaves near the shoot apex of each plant were harvested and frozen immediately in liquid nitrogen, then stored at –80 °C for physiological monitoring and molecular analyses.
Measurements of gas exchange and chlorophyll fluorescence
Gas exchange and chlorophyll fluorescence traits were measured using the fourth fully expanded and intact leaves. The net photosynthesis rate (Pn), transpiration rate (E), and intercellular CO2 concentration (C i) were measured with a Li-Cor 6400, a portable photosynthesis measuring system (Li-Cor Inc., Lincoln, NE, USA). The gas exchange was measured in the morning (08:00–12:00 h) using the following conditions: leaf temperature, 25 °C; leaf-to-air vapour pressure deficit, 1.5±0.5 kPa; photosynthetic photon flux (PPF), 1400 μmol m−2 s−1; relative air humidity, 50%; and ambient CO2 concentration, 350±5 μmol mol−1. To measure chlorophyll fluorescence kinetic traits, the variable and maximum fluorescence (F v/F m) and qP (photochemical quenching coefficient) were determined using a PAM chlorophyll fluorometer (PAM 2100, Walz, Effeltrich, Germany) between 07:30 and 09:30 h (Kooten and Snel, 1990).
Determinations of chlorophyll, proline, and Cl− contents
Leaf discs (0.3 g) were treated with 80% chilled acetone (v/v) at 4 °C for ∼20 h until they changed to a white colour and were then quantified using a spectrometer (Unicam UV-330, Unicam, Cambridge, UK). Chlorophyll was extracted (Lichtenthaler, 1987) and the absorbances of chlorophyll a (Chl a) and Chl b were determined at 646 nm and 663 nm, respectively. Proline was measured by a spectrophotometric analysis at 515 nm using the ninhydrin reaction (Bates et al., 1973).
Cl− contents of leaves were analysed as described by Chen et al. (2001). Leaf samples were ground and passed through a 20 mesh screen after drying at 80 °C for 40 h. Dry powder (0.5 g) was extracted with 1 N HNO3. Abundant AgNO3 solution (0.025 N) was used to precipitate chloride from the aqueous extracts, and excess Ag+ was estimated by 0.02976 N NH4SCN titration. NH4Fe(SO4)2 was used as a colour indicator for determination of the isoionic point. The chloride concentration was calculated using the following formula: Cl− (mmol g−1 DW) = (NAgNO3V1–NNH4SCNV2)/DW, where DW (g) is the dry weight of plant tissue, V1 (ml) represents the total volume of AgNO3 solution in Cl− extracts, and V2 (ml) is the volume of NH4SCN solution used for excess Ag+ precipitation.
Physiological data analysis
Physiological data were analysed using the SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Two-way analyses of variance were performed to evaluate the interaction effect of sex and salinity conditions. Sexual differences were analysed using a model with salinity and sex as fixed effects. Significant individual differences among means of different treatments were determined by Tukey’s multiple range tests after conducting tests of homogeneity for variances. Differences were considered as statistically significant at the P < 0.05 level.
RNA isolation and DGE library preparation
Four DGE libraries representing four different treatments were prepared. Total RNA was extracted from three biological replicates using the E.Z.N.A.™ Plant RNA Kit (Omega, USA) according to the manufacturer’s instructions. Residual genomic DNA was removed with DNase I and the integrity of RNA was checked using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). RNA samples from nine individuals from each treatment were pooled with equal proportions. Double-stranded cDNA was synthesized using oligo(dT) beads. The cDNA was then digested with an anchoring restriction enzyme NlaIII, which recognizes and cuts 3' CATG. The digested cDNA samples were ligated to the Illumina-specific adaptor A, containing a recognition site for the enzyme MmeI. Following MmeI digestion, the Illumina-specific adaptor B containing a 2 bp degenerated 3' overhang was ligated. The adaptor-ligated cDNA tag library was then enriched by PCR primers annealing to the adaptor ends. The PCR program was as follows: 30 s at 98 °C, followed by 15 cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 15 s, and then 72 °C for 5 min. After gel purification, cDNA was used for cluster generation on a separated flow cell lane. Sequencing by synthesis was performed using the Illumina Genome Analyzer according to the manufacturer’s protocol. Image analysis, base calling, extraction of 17 bp tags, and tag counting were performed using the Illumina pipeline.
DGE data analysis
After data processing, raw sequences were transformed into clean tags. Sequences of 21 bp length (17 bp tag plus 4 bp NlaIII recognition site) were aligned against the poplar genome (Populus trichocarpa v1.1) (Tuskan et al., 2006) using the Short Oligonucleotide Alignment Program (SOAP) (Li et al., 2008). No more than a 1 bp mismatch was taken into account for the differences between species. Clean tags mapped to reference sequences from multiple genes were filtered. The remaining clean tags were designated as the distinct tags for further analysis. The number of distinct tags for each gene was calculated and then normalized to the TPM (number of transcripts per million clean tags) (‘t Hoen et al., 2008; Morrissy et al., 2009).
To identify differentially expressed genes (DEGs) between two samples, a rigorous algorithm was developed by BGI (Shenzhen, China), similar to ‘the significance of digital gene expression profiles’ (Audic and Claverie, 1997). A brief description of the algorithm is as follows: the number of distinct tags from gene A is denoted as x, and as every gene’s expression occupies only a small part of the library, the p(x) follows the Poisson distribution.
where λ is the real transcript of a gene.
The total clean tag number of sample 1 is N1, and the total clean tag number of sample 2 is N2; gene A holds x tags in sample 1 and y tags in sample 2. The probability of gene A being expressed equally in two samples can be calculated as follows:
The P-value represents the significance of differential gene expression. The false discovery rate (FDR) determines the threshold of the P-value in multiple tests and analyses through manipulating the FDR value. If R differentially expressed genes were selected among which S genes really showed differential expression, then the other V genes are false positive. If the error ratio ‘Q=V/R’ must stay below a cut-off (e.g. 5%), then the FDR should be pre-set to a number not larger than 0.05 (Benjamini and Yekutieli, 2001). FDR < 0.001 and the absolute value of log-2 ratio ≥ 1 were the thresholds for the significance of a gene with differential expression. The full data set has been deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and is accessible through accession number GSE35223.
For the analysis of DEGs between males and females in response to salinity, singular enrichment analysis (SEA) and parametric analysis of gene set enrichment (PAGE, http://bioinfo.cau.edu.cn/agriGO/) were performed using plant GO slim with the P. trichocarpa gene models as background, followed by multiple testing with Bonferroni (corrected P-value ≤ 0.05) (Du et al., 2010). Functional annotation and gene ontology (GO) were also retrieved by querying with BLASTN against the Arabidopsis AGI transcripts database (http://arabidopsis.org/; TAIR10 release). Only BLASTN hits of e-value ≤ 1E-5 were considered further. The AGI number of each corresponding poplar DEG was categorized into functional groups and mapped using the MapMan (version 3.5.1; http://www.gabipd.de/projects/MapMan/) according to the standard protocol (Thimm et al., 2004; Usadel et al., 2005).
qRT-PCR
Ten transcripts from DGE libraries were validated by quantitative reverse transcription-PCR (qRT-PCR; Supplementary Table S1 available at JXB online ) using the same RNA samples as in the DGE library construction. Total RNA was treated with DNase I and used for cDNA synthesis with ReverTra Ace Moloney murine leukaemia virus reverse transcriptase (Toyobo, Osaka, Japan) and oligo(dT)18 primer. Synthesized cDNAs were diluted to a final volume of 100 μl, and 1 μl was used as a template for qRT-PCRs. qRT-PCR was performed in triplicate on the Opticon Monitor 3 DNA engine (Bio-Rad). The detailed cycling conditions were described in the manufacturer’s instructions of SYBR® Premix Ex Taq™ II (TaKaRa, Dalian, China). Sequences of the gene-specific primers are presented in Supplementary Table S1. The gene encoding ubiquitin-conjugating enzyme (NCBI accession number: XM_002307243.1) with a similar intensity value across all DGE libraries was used as the reference gene for normalization. Gene expression was evaluated using the comparative cycle threshold method, and data were expressed as the mean ±SE (Livak and Schmittgen, 2001).
Results
Sexual differences in physiological traits
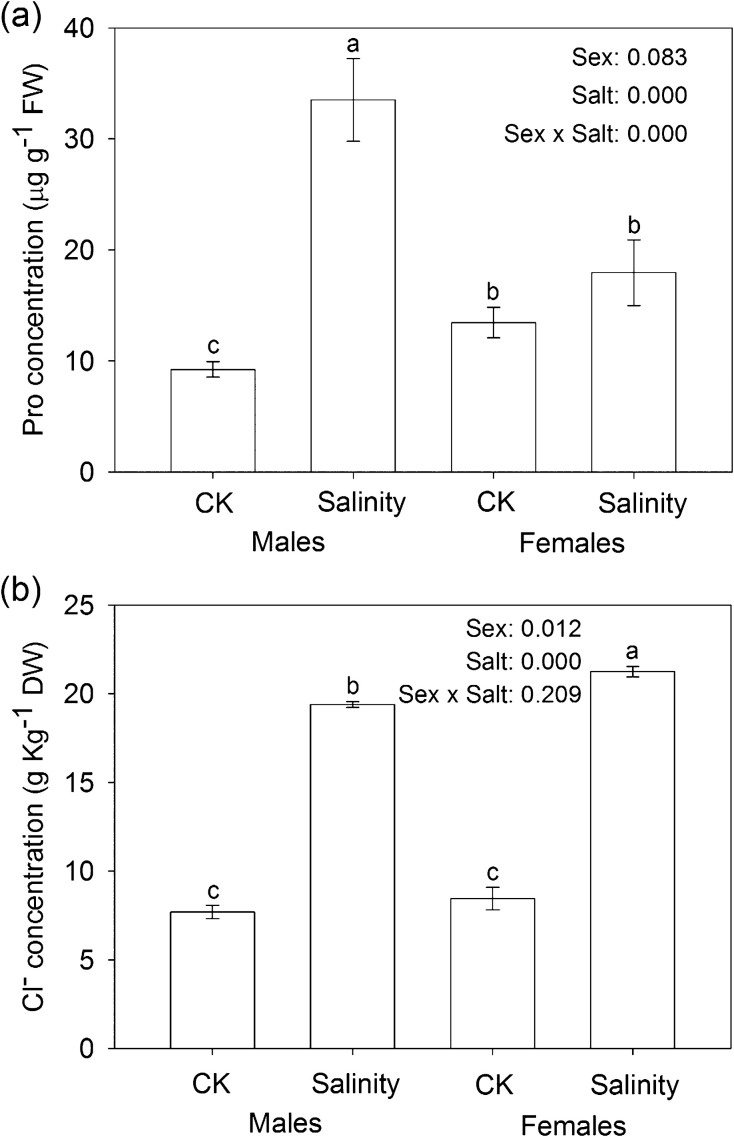
Under salinity stress, the photosynthetic capacity, Pn, E, Ci, total chlorophyll content (TC), F v/F m, and qP significantly decreased; while the proline and Cl− contents, and the Chl a/b ratio increased in both males and females (Table 1). Significant sexual differences in these physiological traits were also observed, among which Pn, E, Ci, F v/F m, and qP decreased more in females than in males, whereas the Chl a/b ratio increased less in males than in females. Furthermore, salinity stress significantly increased the proline content in males but it had less effect on females (Fig. 1a); while it significantly increased the Cl− content in females compared with that in males (Fig. 1b). In addition, the interaction between sex and salinity stress was significant in Pn, E, Ci, F v/F m, qP, Chl a/b ratio, and proline content (Fig. 1a and Table 1).
Table 1.
Sexual differences in photosynthetic responses to salinity stress in P. yunnanensis
| Pn (μmol m−2s−1 ) | E (mmol m−2s−1) | C i (μmol mol−1 ) | F v/F m | qP | TC (mg g−1 ) | Chl a/b | |
| Control | |||||||
| Male | 16.17±0.38 a | 4.22±0.13 a | 256±2.65 a | 0.82±0.009 a | 0.88±0.02 a | 1.23±0.01 a | 2.10±0.05 c |
| Female | 14.97±0.46 a | 4.22±0.15 a | 267±9.17 a | 0.82±0.004 a | 0.88±0.013 a | 1.22±0.01 a | 1.93±0.02 b |
| 50 mM NaCl | |||||||
| Male | 12.33±0.09 b | 3.49±0.11 b | 236±5.69 ab | 0.78±0.004 b | 0.81±0.002 b | 1.01±0.04 b | 2.58±0.09 c |
| Female | 9.44±0.09 c | 2.89±0.10 c | 206±8.66 c | 0.69±0.008 c | 0.69±0.018 c | 0.93±0.15 c | 5.05±0.40 a |
| P (sex) | 0.000 *** | 0.046 * | 0.214 NS | 0.000*** | 0.001** | 0.065 NS | 0.000 *** |
| P (salt) | 0.000 *** | 0.000 *** | 0.000 *** | 0.000*** | 0.000*** | 0.000 *** | 0.000 *** |
| P (sex×salt) | 0.025 * | 0.042 * | 0.020 * | 0.000*** | 0.002** | 0.174 ns | 0.000 *** |
Different letters represent statistical significance between treatments (means ±SE, n=3) at P < 0.05 according to Tukey’s multiple range tests. Significant values of the factorial analysis (ANOVA) for the effects of sex, salt, and sex×salt interaction are denoted as follows: NS, non-significant; *P < 0.05; **P < 0.01; ***P < 0.001
Pn, net photosynthesis rate; E, transpiration rate; C i, intercellular CO2 concentration; F v/F m, variable and maximum fluorescence; qP, photochemical quenching coefficient; TC, total chlorophyll content; Chl a/b, the chlorophyll a/b ratio.
Fig. 1.
Salt-induced accumulation of (a) proline and (b) Cl− (mean ±SE, n=3) in males and females. Different letters above the bars represent statistically significant differences between treatments at P < 0.05 according to Tukey’s multiple range tests. Significance values of the factorial analysis (ANOVA) are denoted as follows: sex, sex effect; salt, salt effect; sex×salt, sex×salt interaction effects.
Transcriptional profiles
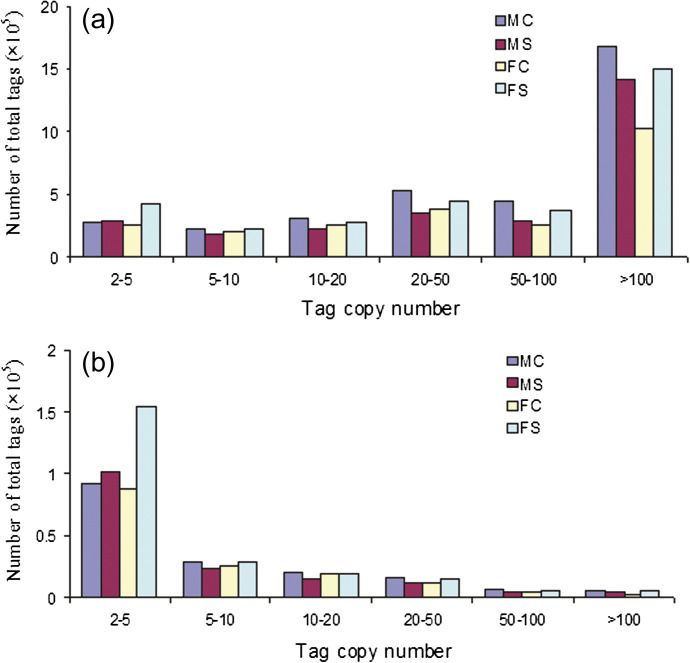
Four DGE libraries were sequenced using the Solexa technology. After data processing according to DGE system requirements, the total number of clean tags per library ranged from ∼2.3 million to 3.4 million, and distinct tags representing unique nucleotide sequences varied from ∼0.15 million to 0.22 million. Figure 2 shows the distribution profiles of the clean and distinct tags. Tags with an abundance of >100 copies in quantity and <5 copies in variety were dominant. A total of 4224 and 5980 DEGs were detected in males and females, respectively. These DEGs were involved in metabolism, regulation, and other aspects. A large proportion (∼77%) of the DEGs were down-regulated in response to salinity stress. To confirm the DGE library data, 10 transcripts were selected for qRT-PCR analysis. The expression tendency of these transcripts was highly consistent when these two methods were compared (Supplementary Fig. S1 at JXB online).
Fig. 2.
Distribution of clean tag copy numbers in males and females. (a) Distribution of all clean tags. (b) Distribution of distinct clean tags, which represent unique nucleotide sequences. MC, males with 0 mM NaCl; MS, males with 50 mM NaCl; FC, females with 0 mM NaCl; FS, females with 50 mM NaCl. (This figure is available in colour at JXB online.)
Functional categories of DEGs
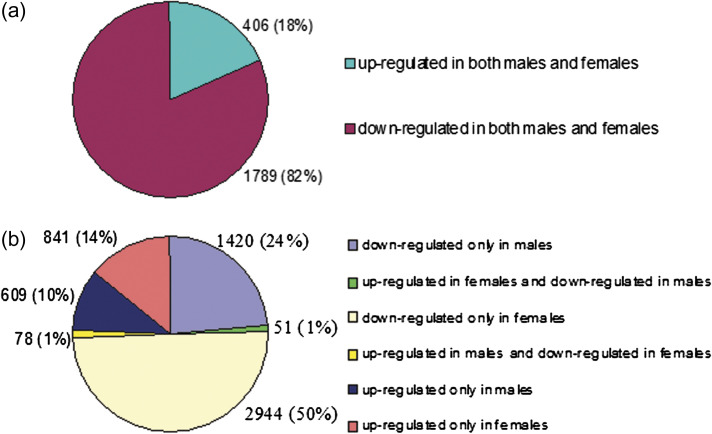
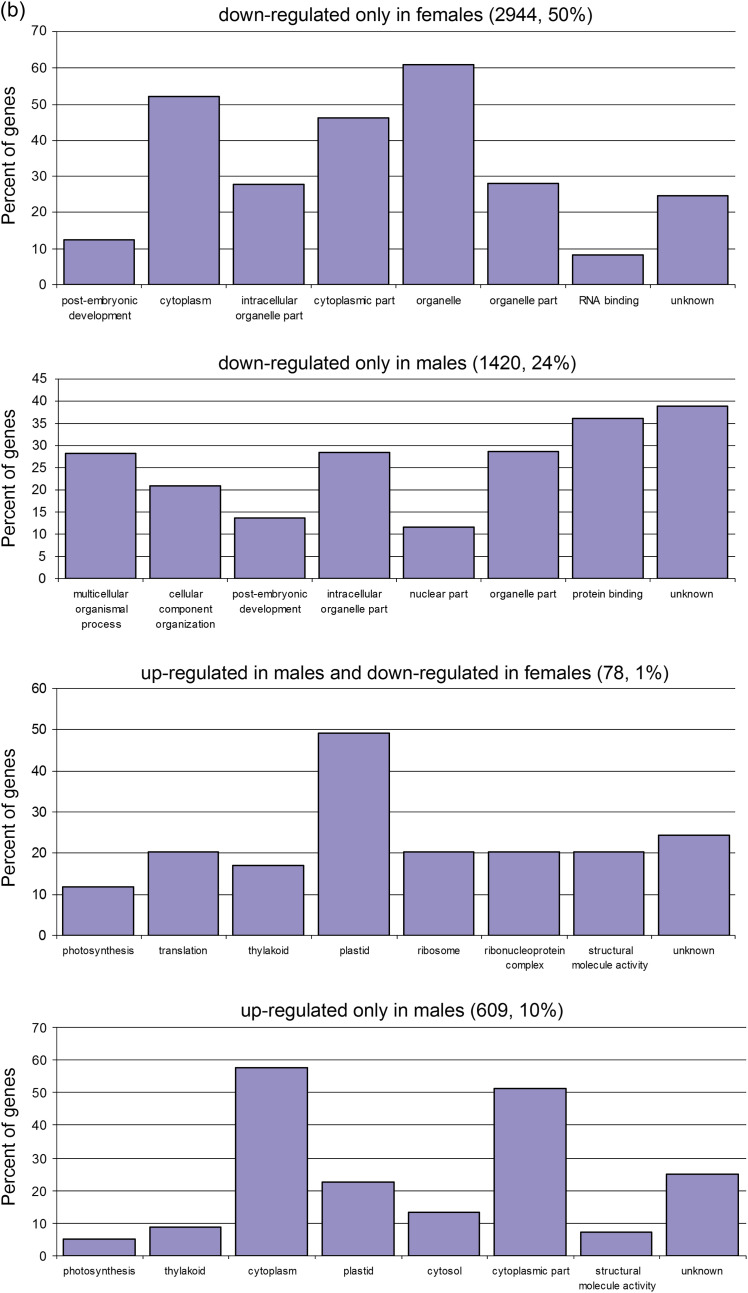
Two strategies were used to identify functional gene groups differentially expressed in the two sexes under salinity. First, an overview of the main results was obtained by PAGE, and significant GO terms were listed (Tables 2 and 3). It was found that the most enriched term in the biological process was ‘photosynthesis’ (GO: 0015979), which was up-regulated under salinity stress in males. In contrast, ‘cellular macromolecule metabolic process’ (GO: 0044260), ‘developmental process’ (GO: 0032502), and ‘anatomical structure development’ (GO: 0048856) were down-regulated in females. In the molecular function, ‘DNA binding’ (GO: 0003677), ‘nuclease activity’ (GO: 0004518), and ‘nucleic acid binding’ (GO: 0003676) were detected more frequently in males; while ‘motor activity’ (GO: 0003774), ‘chromatin binding’ (GO: 0003682), ‘pyrophosphatase activity’ (GO: 0016462), etc. were relatively abundant in females. The most enriched terms of the cellular component in males and females were ‘nuclear lumen’ (GO: 0031981) and ‘intracellular part’ (GO: 0044424), respectively. The second strategy was based on the gene expression levels in males and females. The DEGs were classified into two groups: the salinity-induced co-regulated DEGs in both males and females (Figs 3a, 4a) and the sexually different DEGs (Figs 3b, 4b). A total of 1420 and 609 DEGs were found to be down- and up-regulated, respectively, only in males, while 2944 and 841 DEGs were down- and up-regulated, respectively, only in females. Interestingly, 78 DEGs were up-regulated in males but down-regulated in females (UMDF), while 51 DEGs were regulated in the opposite way (UFDM).
Table 2.
Overview of significant GO terms analysed by the parametric analysis of gene set enrichment (PAGE) in males
| GO term | Onto | No. | Description | Z-score | Mean | FDR |
| GO: 0015979 | P | 63 | Photosynthesis | 5.6 | 0.74 | 1.6e-06 |
| GO: 0006350 | P | 361 | Transcription | –3.6 | –2.8 | 0.023 |
| GO: 0010467 | P | 703 | Gene expression | –3.7 | –2.6 | 0.018 |
| GO: 0043170 | P | 1381 | Macromolecule metabolic process | –3.8 | –2.5 | 0.0094 |
| GO: 0040029 | P | 42 | Regulation of gene expression, epigenetic | –3.9 | –4.5 | 0.0088 |
| GO: 0006259 | P | 171 | DNA metabolic process | –4.6 | –3.5 | 0.00038 |
| GO: 0006807 | P | 908 | Nitrogen compound metabolic process | –4.7 | –2.7 | 0.00016 |
| GO: 0016043 | P | 600 | Cellular component organization | –4.9 | –2.9 | 8.8e-05 |
| GO: 0006139 | P | 779 | Nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | –5.6 | –2.9 | 1.2e-06 |
| GO: 0003677 | F | 361 | DNA binding | –3.5 | –2.8 | 0.014 |
| GO: 0004518 | F | 46 | Nuclease activity | –3.7 | –4.2 | 0.0076 |
| GO: 0003676 | F | 723 | Nucleic acid binding | –3.8 | –2.6 | 0.0038 |
| GO: 0031981 | C | 199 | Nuclear lumen | –3.5 | –3.1 | 0.02 |
| GO: 0005856 | C | 152 | Cytoskeleton | –3.6 | –3.2 | 0.018 |
| GO: 0044428 | C | 311 | Nuclear part | –5 | –3.2 | 3.2e-05 |
| GO: 0005634 | C | 806 | Nucleus | –5 | –2.8 | 2.3e-05 |
P, biological process; F, molecular function; C, cellular component. Mean, mean log-2 expression ratio, >0 represents up-regulation, <0 represents down-regulation.
Table 3.
Overview of significant GO terms analysed by the parametric analysis of gene set enrichment (PAGE) in females
| GO term | Onto | No. | Description | Z-score | Mean | FDR |
| GO: 0044260 | P | 1727 | Cellular macromolecule metabolic process | –3.4 | –2.7 | 0.049 |
| GO: 0032502 | P | 1147 | Developmental process | –3.5 | –2.8 | 0.043 |
| GO: 0048856 | P | 887 | Anatomical structure development | –3.5 | –2.9 | 0.037 |
| GO: 0048869 | P | 368 | Cellular developmental process | –3.5 | –3.2 | 0.032 |
| GO: 0043170 | P | 1960 | Macromolecule metabolic process | –3.6 | –2.7 | 0.027 |
| GO: 0030154 | P | 299 | Cell differentiation | –3.6 | –3.3 | 0.025 |
| GO: 0000003 | P | 609 | Reproduction | –3.8 | –3 | 0.011 |
| GO: 0009653 | P | 412 | Anatomical structure morphogenesis | –3.9 | –3.2 | 0.0063 |
| GO: 0009790 | P | 416 | Embryonic development | –4.1 | –3.2 | 0.0038 |
| GO: 0006807 | P | 1290 | Nitrogen compound metabolic process | –4.2 | –2.9 | 0.0024 |
| GO: 0006139 | P | 1094 | Nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | –4.5 | –2.9 | 0.00054 |
| GO: 0016043 | P | 792 | Cellular component organization | –4.9 | –3.1 | 8.3e-05 |
| GO: 0007049 | P | 309 | Cell cycle | –5.3 | –3.7 | 9.3e-06 |
| GO: 0006259 | P | 230 | DNA metabolic process | –5.7 | –4 | 9e-07 |
| GO: 0003774 | F | 53 | Motor activity | –3.4 | –4.5 | 0.019 |
| GO: 0003682 | F | 55 | Chromatin binding | –3.5 | –4.5 | 0.015 |
| GO: 0016462 | F | 357 | Pyrophosphatase activity | –4.6 | –3.4 | 0.00013 |
| GO: 0016817 | F | 364 | Hydrolase activity, acting on acid anhydrides | –4.7 | –3.4 | 9.8e-05 |
| GO: 0016818 | F | 361 | Hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides | –4.7 | –3.4 | 9.4e-05 |
| GO: 0017111 | F | 349 | Nucleoside-triphosphatase activity | –5 | –3.5 | 2.1e-05 |
| GO: 0000166 | F | 993 | Nucleotide binding | –5.2 | –3.1 | 4.9e-06 |
| GO: 0044424 | C | 2977 | Intracellular part | –3.5 | –2.6 | 0.024 |
| GO: 0005622 | C | 3057 | Intracellular | –3.7 | –2.6 | 0.013 |
| GO: 0032991 | C | 856 | Macromolecular complex | –3.8 | –2.9 | 0.0079 |
| GO: 0005856 | C | 199 | Cytoskeleton | –3.9 | –3.6 | 0.0043 |
| GO: 0005730 | C | 113 | Nucleolus | –4 | –4 | 0.0036 |
| GO: 0043227 | C | 2464 | Membrane-bound organelle | –4 | –2.7 | 0.0027 |
| GO: 0043231 | C | 2461 | Intracellular membrane-bound organelle | –4 | –2.7 | 0.0027 |
| GO: 0005634 | C | 1088 | Nucleus | –4.1 | –2.9 | 0.0025 |
| GO: 0005654 | C | 162 | Nucleoplasm | –4.1 | –3.8 | 0.0019 |
| GO: 0043229 | C | 2652 | Intracellular organelle | –4.2 | –2.7 | 0.0014 |
| GO: 0043226 | C | 2654 | Organelle | –4.2 | –2.7 | 0.0014 |
| GO: 0044446 | C | 1223 | Intracellular organelle part | –4.4 | –2.9 | 0.00051 |
| GO: 0044422 | C | 1227 | Organelle part | –4.4 | –2.9 | 0.00048 |
| GO: 0031974 | C | 343 | Membrane-enclosed lumen | –4.8 | –3.5 | 6.9e-05 |
| GO: 0070013 | C | 340 | Intracellular organelle lumen | –4.9 | –3.5 | 5e-05 |
| GO: 0043233 | C | 340 | Organelle lumen | –4.9 | –3.5 | 5e-05 |
| GO: 0031981 | C | 262 | Nuclear lumen | –5.7 | –3.9 | 7.4e-07 |
| GO: 0043232 | C | 571 | Intracellular non-membrane-bound organelle | –6.1 | –3.5 | 4.1e-08 |
| GO: 0043228 | C | 571 | Non-membrane-bound organelle | –6.1 | –3.5 | 4.1e-08 |
| GO: 0044428 | C | 406 | Nuclear part | –7 | –3.9 | 1.8e-10 |
P, biological process; F, molecular function; C, cellular component. Mean, mean log-2 expression ratio, >0 represents up-regulation, <0 represents down-regulation.
Fig. 3.
Classifications of (a) co-regulated and (b) sexually different DEGs up- or down-regulated in males and females under salinity stress. (This figure is available in colour at JXB online.)
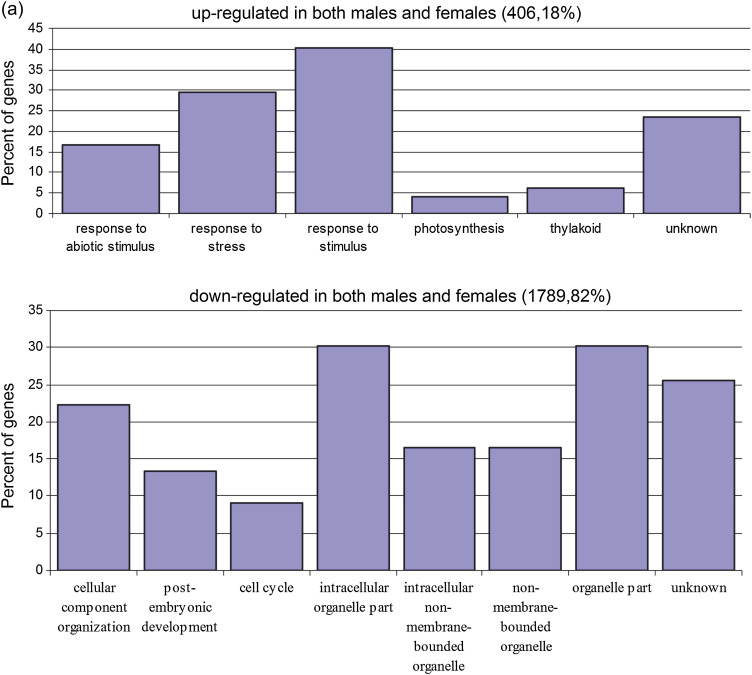
Fig. 4.
SEA analysis implying significant GO terms of (a) co-regulated and (b) sexually different DEGs up- or down-regulated in males and females under salinity stress, excluding categories ‘up-regulated only in females’ (only one: vacuole) and ‘up-regulated in females and down-regulated in males’ (none). (This figure is available in colour at JXB online.)
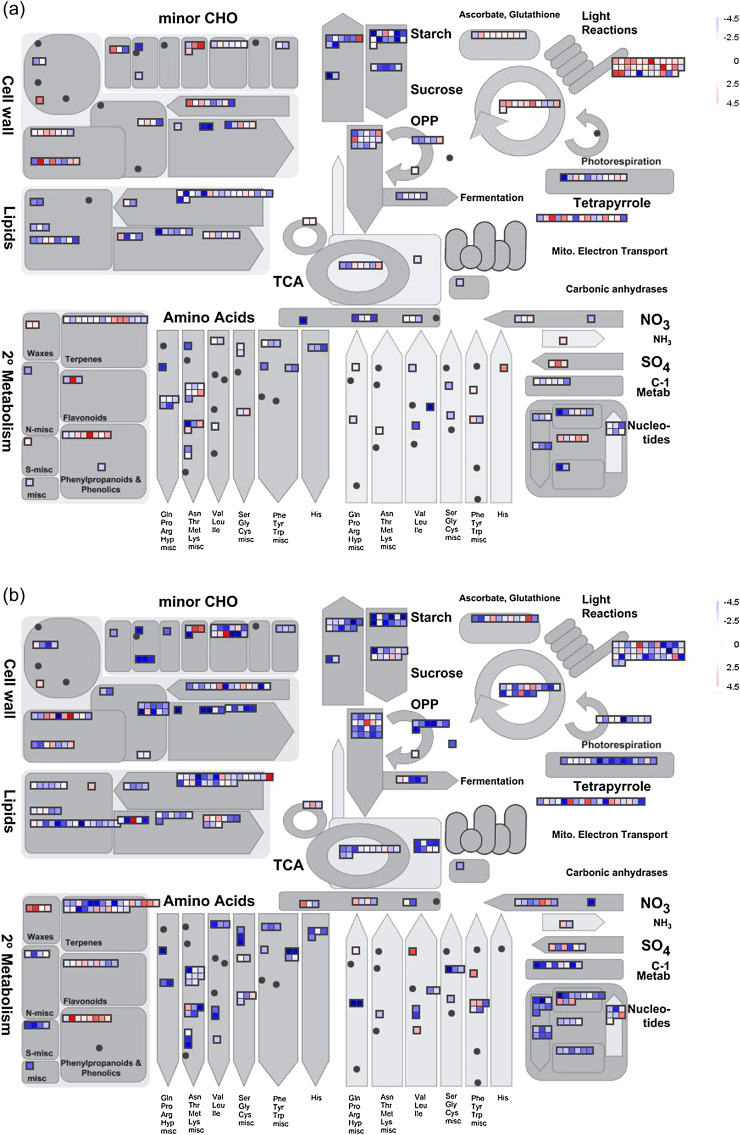
To test whether the DEGs above are directly related to sexual differences in response to salinity stress, they were compared with the Arabidopsis genome, and the best matches and AGI numbers were obtained. Further MapMan analysis indicated that these DEGs were widely involved in plant metabolic and regulatory pathways (Fig. 5; Supplementary Fig. S2 at JXB online), including DEGs related to metabolism, such as photosynthesis, major and minor carbohydrates, cell wall, lipids, and amino acids. DEGs related to plant regulation mostly concerned transcription factors, protein modification and degradation, and hormones. These results suggested that salinity stress has a great influence on poplars, which changed the transcript abundance of many genes to tackle salinity. These results also indicated that a set of common functions and pathways are induced in the salinity responses, regardless of the sex. Nevertheless, distinct differences between males and females were shown in some pathways, such as photosynthesis and transcription (see the later sections), highlighting sexual differences in salinity responses.
Fig. 5.
Overview of the DEGs assigned to ‘metabolism’ by MapMan. (a) Change of transcript levels in males under salinity stress. (b) Change of transcript levels in females under salinity stress. Data were processed according to the standard protocol of MapMan. The scale indicates DEGs significantly up- (red) or down-regulated (blue) in response to salinity stress.
Sexual differences of co-regulated DEGs under salinity stress
Most co-regulated DEGs in both males and females were down-regulated under salinity stress. Due to different gene expression levels, these DEGs were divided into three groups. The first group consisted of 34 genes with similar expression patterns in both males and females, such as SPL7, TOR, GI, and DEX1. These genes were mostly involved in development. Genes in the second group represented a cluster with greater expression in males, including eight ribosomal protein genes (RPS3C, RPS4D, RPS17C, RPS18C, RPS30B, RPL15A, RPL31B, and MEE49) and genes with function in amino acid metabolism (such as MAT3, MTO1, and CBL), minor carbohydrates, signalling, redox, and DNA repair. The third group of genes up-regulated in females had functions in starch degradation, tetrapyrrole synthesis, post-transcriptional modification of proteins, light signalling, DNA synthesis, protein targeting, and transport.
Genes up-regulated only in males or in females
Under salinity stress, 609 genes were up-regulated only in males (Supplementary Table S2 at JXB online), such as genes encoding ketoacyl ACP synthase, enoyl ACP reductase, long chain fatty acid-CoA ligase, and acyl-CoA-binding protein, which are involved in fatty acid (FA) synthesis and elongation. Two genes, GGT1 and ALAAT2, encoding aminotransferases involved in central amino acid metabolism and two genes encoding galactosyltransferase family proteins were also detected only in males. More importantly, five genes encoding photosystem II (PSII) polypeptide subunits were exclusively up-regulated in males, providing possible reasons why males maintained relatively higher F v/F m than females under salinity (Table 1).
There were 841 genes up-regulated only in females (Supplementary Table S3 at JXB online), including genes related to lipid metabolism (nine genes), stress (four genes), miscellaneous enzyme families (22 genes), and other proteins (66 genes). This result suggested that females increase gene expression of regulatory pathways during salinity. The functional categories FA synthesis and FA elongation were also detected in females, but the identified genes in females were different from those in males. In females, genes involved in FA synthesis and FA elongation were KCS11 and MCD, which encode β-ketoacyl-CoA synthase and malonyl-CoA decarboxylase, respectively. The salinity stress-altered gene expression pattern involved miscellaneous enzyme families, including genes encoding a universal stress protein family protein, pollen Ole e 1 allergen and extensin family protein, and germin-like protein 5. Interestingly, DEGs identified only in females included genes related to four functional groups of proteins: protein synthesis (12 genes, e.g. RPS9C, RPS13A, RPS19C, RPS20A, RPS24A, and RPS29C), targeting (four genes, e.g. NTF2 and ALB3), post-translational modification (18 genes, e.g. CKA2, CIPK25, and PP2C), and degradation (32 genes, e.g. EGY2, PUX3, and PAD1). This result suggested that genes encoding enzymes and other proteins could play key roles in the salinity tolerance of female poplars, and their different expression patterns may make a significant contribution to the different physiological responses in males and females under salinity stress.
Genes down-regulated only in males or in females
There were 1420 genes down-regulated only in males (Supplementary Table S4 at JXB online), including genes related to cell functions, such as hormone metabolism, transcription, and DNA synthesis. In hormone metabolism, the genes found mainly had functions in the synthesis and degradation of auxin, abscisic acid, brassinosteroid, and jasmonate, such as ARG1, ABA3, XF1, and OPR2. Two signal transduction genes, and genes encoding squalene monooxygenase and ethylene-responsive family protein, were also detected. This result suggested that male poplars changed their phytohormone levels to cope with salinity stress, and the genes detected only in males may further expand the sexually different adaption. Three genes involved in lipid degradation (IBR3, MFP2, and FAR4), and 15 genes related to chromatin structure involving DNA synthesis, such as DPB2, SLD5, CHR4, RHL2, NRP1, EMB2411, and MAA3, were also down-regulated. This observation suggested that lipids and chromatin of male poplars could develop reorganization in response to salinity.
In contrast, 2944 genes were down-regulated only in females, including five major pathways: minor carbohydrate (16 genes), lipid (40 genes), hormone (24 genes), protein (39 genes), and cell (85 genes). In the pathway ‘cell’, 85 genes fall into four groups: organization (32 genes), division (23 genes), cycle (15 genes), and vesicle transport (15 genes). This result suggested that genes with a role in the cell were significantly depressed in female poplars when exposed to salinity stress. In addition, seven genes involved in metal handling, such as FRO4, MTO3, and MT3, and two genes, GAUT8 and GAUT1, encoding polygalacturonate 4-α-galacturonosyltransferase and galacturonic acid transferase involved in pectin synthesis, were also down-regulated in females under salinity stress. Other genes which were down-regulated only in females are listed in Supplementary Table S5 at JXB online.
Genes with complex regulation patterns in males and females
A small number of genes exhibited opposite expression patterns in males and females under salinity stress. In the category UMDF, DEGs were mainly related to cell wall formation, photosynthesis, and protein pathways (Table 4). Eight DEGs in photosynthesis (Fig. 4) were involved in light reactions (Chl a/b-binding protein 2, CAB2; light-harvesting complex II Chl a/b-binding protein 2, LHCB2.2; chlorophyll-binding protein D1, PSBA; photosystem II PsbY protein, PSBY; gamma subunits of chloroplast ATP synthase, ATPC1; photosystem I subunit PsaN, PSAN; and leaf ferredoxin, FED A), the Calvin cycle (ribulose bisphosphate carboxylase small chain 1A, RBCS1A), and photorespiration (RBCS1A).
Table 4.
Functional categories and gene expression patterns of 59 DEGs up-regulated in males but down-regulated in females under salinity stress
Populus gene model ID 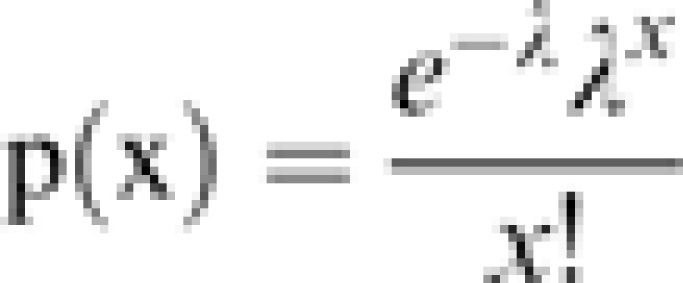
|
AGI | AGI annotation and function description | Log-2 ratio | |
| MS/MC | FS/FC | |||
| Cell wall | ||||
| estExt_Genewise1_v1.C_LG_XIV0850 | AT2G45470 | AGP8; fasciclin-like arabinogalactanprotein 8 | 1.09 | –1.71 |
| eugene3.00140737 | AT3G62830 | UXS2; UDP-glucuronate decarboxylase activity, catalytic activity, dTDP-glucose 4,6-dehydratase activity | 1.66 | –1.3 |
| gw1.III.2269.1 | AT5G47500 | MNJ7.9; pectin lyase-like superfamily protein, pectinesterase activity | 1.39 | –2.27 |
| Lipids | ||||
| fgenesh4_pm.C_scaffold_44000016 | AT3G15730 | PLD alpha 1; phospholipase D activity | 1.04 | –1.15 |
| estExt_fgenesh4_pm.C_LG_VI0352 | AT3G11170 | FAD7; omega-3 fatty acid desaturase activity | 1.3 | –1.56 |
| Redox | ||||
| estExt_fgenesh4_pg.C_LG_IX1399 | AT1G07890 | Cytosolic ascorbate peroxidase APX1; L-ascorbate peroxidase activity | 1.26 | –1.4 |
| TCA | ||||
| grail3.0010030801 | AT3G17240 | mtLPD2; dihydrolipoyl dehydrogenase activity | 1.68 | –1.12 |
| Photosynthesis | ||||
| estExt_fgenesh4_kg.C_LG_IV0017 | AT1G67090 | RBCS1A; ribulose-bisphosphate carboxylase activity | 2.3 | –1.3 |
| eugene3.00110470 | AT1G29920 | CAB2; chlorophyll binding | 2.55 | –1.37 |
| estExt_fgenesh4_pm.C_LG_II0962 | AT2G05070 | LHCB2.2; chlorophyll binding | 1.31 | –2.47 |
| gw1.XIII.2252.1 | ATCG00020 | PSBA; chlorophyll binding | 1.21 | –2.83 |
| estExt_Genewise1_v1.C_LG_X5024 | AT1G67740 | PSBY; manganese ion binding | 1.68 | –1.63 |
| estExt_Genewise1_v1.C_LG_V2453 | AT5G64040 | PSAN; calmodulin binding | 1.9 | –1.35 |
| eugene3.00040033 | AT4G04640 | ATPC1; enzyme regulator activity | 3.28 | –1.45 |
| estExt_fgenesh4_kg.C_1630003 | AT1G60950 | FED A; electron carrier activity | 1.17 | –1.65 |
| Amino acid | ||||
| eugene3.00090483 | AT5G11880 | DAPD; diaminopimelate decarboxylase activity | 1.83 | –3.09 |
| eugene3.00050687 | AT1G22410 | DAHP; 3-deoxy-7-phosphoheptulonate synthase activity | 1.17 | –2.41 |
| C1-metabolism | ||||
| grail3.0066005802 | AT3G03780 | ATMS2; 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity, methionine synthase activity | 1.18 | –1.54 |
| grail3.0050014702 | AT1G02500 | SAM1; methionine adenosyltransferase activity | 2.38 | –1.65 |
| eugene3.00012227 | AT4G13930 | SHM4; catalytic activity, glycine hydroxymethyltransferase activity | 1.08 | –1.51 |
| Nucleotides | ||||
| fgenesh4_pg.C_LG_XII000913 | AT5G63310 | NDPK2; ATP binding, protein binding, nucleoside diphosphate kinase activity | 1.54 | –1.85 |
| Protein | ||||
| eugene3.00410149 | AT1G35340 | T9I1.11; ATP-dependent peptidase activity | 2.29 | –1.73 |
| grail3.0028002001 | AT5G60360 | AALP; cysteine-type peptidase activity | 2.02 | –1.19 |
| fgenesh4_pm.C_LG_II000602 | AT1G74970 | RPS9; nuclear encoded component of the chloroplast ribosome | 1.87 | –3.16 |
| gw1.XI.877.1 | AT1G32990 | PRPL11; mutant has decreased effective quantum yield of photosystem II | 1.25 | –2.38 |
| gw1.XVI.542.1 | AT3G54210 | F24B22.170; structural constituent of ribosome | 1.11 | –1.61 |
| eugene3.00060477 | AT1G58380 | XW6; structural constituent of ribosome | 1.25 | –3.37 |
| estExt_fgenesh4_pm.C_LG_IV0210 | AT3G02560 | RPS7B; structural constituent of ribosome | 1.97 | –1.91 |
| estExt_fgenesh4_pg.C_LG_V0222 | AT3G49910 | RPL26A; structural constituent of ribosome | 1.58 | –1.63 |
| eugene3.00061117 | AT1G70600 | F24J13.17; structural constituent of ribosome | 1.29 | –1.57 |
| estExt_fgenesh4_pg.C_410046 | AT4G22380 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein; RNA binding | 1.23 | –2.09 |
| estExt_Genewise1Plus.C_LG_VII0502 | AT4G17300 | NS1; asparagine-tRNA ligase activity | 1.12 | –2.03 |
| grail3.0002059601 | AT1G04940 | TIC20; Tic20 is believed to function as a component of the protein-conducting channel at the inner envelope membrane | 2.95 | –1.9 |
| estExt_fgenesh4_pg.C_LG_IX1267 | AT2G28800 | ALB3; P–P-bond-hydrolysis-driven protein transmembrane transporter activity | 2.09 | –2.6 |
| Hormones | ||||
| e_gw1.XIX.2349.1 | AT4G19170 | NCED4; similar to nine-cis-epoxycarotenoid dioxygenase | 3.78 | –2.34 |
| Signalling | ||||
| estExt_Genewise1_v1.C_LG_XV2501 | AT5G61790 | CNX1; calcium ion binding, unfolded protein binding | 1.38 | –1.59 |
| Stress | ||||
| gw1.XIV.3121.1 | AT1G05850 | POM1; chitinase activity | 1.49 | –1.2 |
| estExt_fgenesh4_pg.C_1500058 | AT5G49910 | HSC70-7; protein binding | 1.98 | –1.7 |
| Development | ||||
| gw1.VI.1538.1 | AT1G67440 | EMB1688; GTPase activity | 2.66 | –2.03 |
| Not assigned | ||||
| gw1.XVIII.2260.1 | AT2G26900 | F12C20.6; bile acid:sodium symporter activity, transporter activity | 1.05 | –1.49 |
| grail3.0061011301 | AT2G31400 | GUN1; DNA binding | 1.31 | –3.12 |
| eugene3.00570020 | AT4G35760 | F4B14.2; NAD(P)H dehydrogenase (quinone) activity | 1.2 | –1.04 |
| e_gw1.VI.585.1 | AT2G23990 | ENODL11; electron carrier activity | 1.85 | –4.67 |
| eugene3.00012979 | AT1G79090 | YUP8H12R.29; molecular function unknown | 3.97 | –4.21 |
| eugene3.00870040 | AT1G65230 | T23K8.14; molecular function unknown | 1.2 | –1.03 |
| estExt_Genewise1_v1.C_LG_XVI3139 | AT2G36885 | Unknown | 2.34 | –1.72 |
| estExt_fgenesh4_pg.C_LG_II2402 | AT3G07090 | T1B9.26; PPPDE putative thiol peptidase family protein | 1.68 | –1.19 |
| estExt_fgenesh4_pg.C_LG_IX0791 | AT3G49720 | T16K5.70; molecular function unknown | 1.03 | –2.87 |
| estExt_fgenesh4_pm.C_LG_II0597 | AT1G47740 | T2E6.19; PPPDE putative thiol peptidase family protein | 1.65 | –1.76 |
| estExt_Genewise1Plus.C_LG_III1018 | AT1G32080 | F3C3.12; membrane protein, putative | 1.31 | –1.39 |
| fgenesh4_pg.C_scaffold_137000002 | AT5G55930 | OPT1; oligopeptide transporter activity | 2.57 | –2.28 |
| fgenesh4_pg.C_LG_XIV000399 | AT4G01030 | F3I3.50; pentatricopeptide (PPR) repeat-containing protein | 1.6 | –3.11 |
| eugene3.00012101 | AT3G01780 | TPLATE; a cytokinesis protein targeted to the cell plate, binding | 1.67 | –1.81 |
| eugene3.00170125 | AT5G65260 | MQN23.21; RNA binding, nucleic acid binding, nucleotide binding | 1.19 | –1.67 |
| estExt_Genewise1_v1.C_LG_IX2088 | AT3G48690 | CXE12; carboxylesterase activity | 1.55 | –3.46 |
| estExt_fgenesh4_pg.C_LG_II0348 | AT1G06200 | F9P14.6; peptidase S24/S26A/S26B/S26C family protein | 1.41 | –1.4 |
| estExt_fgenesh4_pg.C_LG_VII0502 | AT2G22170 | T26C19.17; lipase/lipooxygenase, PLAT/LH2 family protein | 1.21 | –1.14 |
| estExt_fgenesh4_pg.C_290237 | AT4G17730 | SYP23; SNAP receptor activity | 1.78 | –1.4 |
| e_gw1.I.2717.1 | AT3G01660 | F4P13.20; methyltransferase activity | 1.03 | –1.05 |
MC, males with 0 mM NaCl; MS, males with 50 mM NaCl; FC, females with 0 mM NaCl; FS, females with 50 mM NaCl; NS, not statistically significant.
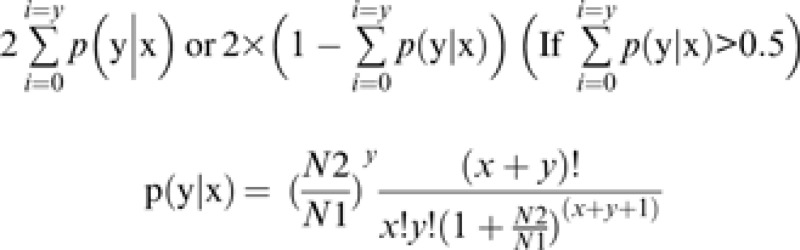
Genes belonging to the UFDM category were regulated in the opposite way to those in UMDF. RNA and protein were two important pathways in this category (Table 5). Three DEGs involved in the pathway of RNA processing and transcription regulation were down-regulated in males but up-regulated in females (Table 5). Genes in this category included HNH endonuclease domain-containing protein, MXM12.5; TRAF-type zinc finger-related protein, F21M12.31; and A20/AN1-like zinc finger family protein, SAP5.
Table 5.
Functional categories and gene expression patterns of 28 DEGs down-regulated in males but up-regulated in females under salinity stress
| Populus gene model ID | AGI | AGI annotation and function description | Log-2 ratio | |
| MS/MC | FS/FC | |||
| Cell wall | ||||
| fgenesh4_pg.C_LG_XVI000539 | AT3G52370 | FLA15; fasciclin-like arabinogalactanprotein 15 precursor | –2.37 | 1.5 |
| Lipids | ||||
| fgenesh4_pm.C_LG_VIII000723 | AT3G23510 | MEE5.5; cyclopropane-fatty-acyl-phospholipid synthase activity | –2.69 | 1.01 |
| Major CHO metabolism | ||||
| estExt_fgenesh4_pg.C_LG_II1784 | AT2G47470 | UNE5; protein disulphide isomerase activity | –1.3 | 2.63 |
| fgenesh4_pm.C_LG_VI000400 | AT4G26140 | BGAL12; beta-galactosidase activity, catalytic activity, hydrolase activity, hydrolyzing O-glycosyl compounds | –1.57 | 1.24 |
| fgenesh4_pg.C_LG_IX001086 | AT1G50460 | HKL1; hexokinase activity | –2.94 | 2.01 |
| RNA | ||||
| eugene3.00120596 | AT5G07810 | MXM12.5; endonuclease activity, helicase activity | –2.4 | 1.26 |
| grail3.0003096203 | AT1G09920 | F21M12.31; TRAF-type zinc finger-related | –2 | 1.12 |
| eugene3.00011778 | AT3G12630 | A20/AN1-like zinc finger family protein; DNA binding, zinc ion binding | –2.35 | 1.34 |
| Protein | ||||
| estExt_fgenesh4_pm.C_LG_XIII0008 | AT1G11910 | APA1; aspartic proteinase, endopeptidase activity | –1.3 | 2.29 |
| estExt_Genewise1_v1.C_570194 | AT3G51350 | F26O13.3; aspartic-type endopeptidase activity | –1.77 | 2.11 |
| estExt_Genewise1Plus.C_44690001 | AT2G35780 | SCPL26; serine-type carboxypeptidase activity | –2.22 | 2.98 |
| estExt_fgenesh4_pm.C_LG_XVIII0083 | AT5G25760 | PEX4; ubiquitin-protein ligase activity | –1.53 | 1.58 |
| estExt_Genewise1_v1.C_LG_I8663 | AT5G41700 | UBC8; ubiquitin-protein ligase activity | –1.57 | 1.5 |
| estExt_fgenesh4_pm.C_280017 | AT5G20570 | RBX1; protein binding | –1.92 | 0.88 |
| Signalling | ||||
| grail3.0033033502 | AT4G11880 | AGL14; DNA binding, sequence-specific DNA binding transcription factor activity | –1.49 | 1.69 |
| estExt_Genewise1_v1.C_LG_V0143 | AT1G43860 | F28H19.11; sequence-specific DNA binding transcription factor activity | –1.22 | 1.77 |
| eugene3.00131252 | AT4G18700 | CIPK12; kinase, protein kinase, protein serine/threonine kinase activity | –1.47 | 1.03 |
| eugene3.00400111 | AT1G02130 | ARA5; GTP binding | –1.59 | 1.25 |
| grail3.0001009101 | AT4G35860 | ATGB2; GTP binding | –1.26 | 1.41 |
| Stress | ||||
| estExt_Genewise1_v1.C_LG_II2154 | AT4G33300 | ADR1-L1; ATP binding | –1.99 | 1.68 |
| Large enzyme families | ||||
| eugene3.00141430 | AT5G43940 | HOT5; S-(hydroxymethyl) glutathione dehydrogenase activity, S-nitrosoglutathione reductase activity | –1.64 | 2.56 |
| Not assigned | ||||
| gw1.I.9523.1 | AT1G15520 | PDR12; ATPase activity, coupled to transmembrane movement of substances | –1.46 | 1.68 |
| eugene3.01470017 | AT4G31480 | F3L17.50; binding, clathrin binding, structural molecule activity | –1.18 | 1.08 |
| estExt_fgenesh4_pm.C_LG_I1158 | AT5G55940 | EMB2731; molecular function unknown | –3.85 | 3.27 |
| gw1.VII.1749.1 | AT4G39970 | T5J17.140; catalytic activity, hydrolase activity | –2.3 | 1.25 |
| estExt_fgenesh4_pm.C_LG_VI0047 | AT4G16480 | INT4; carbohydrate transmembrane transporter activity, myo-inositol:hydrogen symporter activity, sugar:hydrogen symporter activity | –1.01 | 1.08 |
| estExt_fgenesh4_pg.C_LG_VIII0748 | AT2G02990 | RNS1; endoribonuclease activity, ribonuclease activity | –1.4 | 3.75 |
| e_gw1.XVIII.760.1 | AT5G11700 | T22P22.90; similar to glycine-rich protein, putative | –2.55 | 1.72 |
MC, males with 0 mM NaCl; MS, males with 50 mM NaCl; FC, females with 0 mM NaCl; FS, females with 50 mM NaCl; NS, not statistically significant.
Genes located in the female-specific region of the W chromosome
In order to investigate further the molecular mechanisms underlying salinity-induced gene expression and different sexual responses, genes located in the female-specific region of the W chromosome (FSW) (Yin et al., 2008) were analysed. Surprisingly, none of the identified differentially expressed genes in females was located in the FSW. Thus, it appears that the genes located in autosomes rather than in the FSW make a major contribution to sexual differences in the salinity tolerance of poplars.
Discussion
Sexual differences in responses to salinity can occur in osmotic and ionic phases and involve accumulation of Cl− and proline
Previous studies have clearly demonstrated that salt tolerance of plants includes two main phases, the osmotic stress and the salt-specific effect (Munns, 2005). In P. yunnanensis, Cl− rather than Na+ content exhibited a large increment in the two sexes, but more significantly in females than in males (Fig. 1b). This phenomenon was consistent with an earlier study (L. Chen et al., 2010) suggesting that Cl− is the more toxic ion, as also observed in grapevine and citrus (Storey and Walker, 1999). Na+ can be withheld effectively in woody roots and stems, while Cl− continues to accumulate in leaves as the major toxic ion. Accordingly, increased transcription was observed of a gene encoding a chloride channel (CLC) family protein (CLC-c) with osmotic adjustment function under high salinity (Diédhiou and Golldack, 2006; Li et al., 2006), but only in females. This observation suggests that under the same salt conditions, specific signals triggered to cope with Cl− influx accumulated more in females than in males. In this way, males are more efficient in Cl− management than females.
Interestingly, salinity significantly increased the proline content but only in males (Fig. 1a), and the interaction effect between sex and salinity stress was significant (P=0.000). Proline is a proteinogenic amino acid with an exceptional conformational rigidity. It accumulates in many plant species in response to environmental stress, and it plays a crucial role in osmotic protection (Yamada et al., 2005; Schat et al., 2006; Szabados and Savouré, 2010). However, little is known about the detailed links between proline increment and Cl− accumulation. Previous physiological studies revealed an interactive relationship between proline increment and Cl− accumulation during the osmotic phase (F. Chen et al., 2010; L. Chen et al., 2010). The significantly lower Cl− content in males is presumably partially due to the higher accumulation of proline in males than in females.
Transcriptional profiling analysis revealed a massive influence of salinity on proline metabolism, including induction of several key enzymes, such as pyrroline-5-carboxylate synthetase (P5CS) (Székely et al., 2007). In plants, proline is synthesized mainly from glutamate, which is reduced to glutamate-semialdehyde (GSA) by P5CS, and then converted to pyrroline-5-carboxylate (P5C) (Hu et al., 1992; Savouré et al., 1995). P5C reductase (P5CR) further reduces the P5C intermediate to proline (Szoke et al., 1992; Verbruggen et al., 1993). Alternatively, plants can synthesize proline from ornithine, which is firstly transaminated by ornithine-δ-aminotransferase (OAT) to produce GSA and P5C (Roosens et al., 1998). During osmotic stress, proline biosynthesis is controlled by P5CS1 (Székely et al., 2007). In the present study, the transcript of P5CS1 was found more frequently in females while OAT was found more frequently in males, suggesting that males and females adopted distinct strategies that trigger different signals to synthesize more osmoprotectant proline to cope with the predicament. However, no obvious transcriptional changes were found in other key genes, such as P5CS2 (Székely et al., 2007), P5CR (Szoke et al., 1992), PDH1, PDH2 (Kiyosue et al., 1996), and P5CDH (Deuschle et al., 2001). Because males accumulate more proline than females, the OAT pathway could be important in the salinity tolerance of poplars.
Sexual differences in photosynthetic capacity under salinity stress
Photosynthesis is closely related to the salinity tolerance of poplars (Wang et al., 2007; F. Chen et al., 2010; L. Chen et al., 2010). The present results showed that salinity significantly inhibits gas exchange in both males and females because of lowered P n, E, and C i (Table 1). Similar observations have been reported for P. cathayana (F. Chen et al., 2010) and P. popularis (Wang et al., 2007). The mean values of P n and E decreased by 37% and 31.5% in females, but by only 23.7% and 17.3% in males, respectively (Table 1). This observation suggested that the inhibitory effects of salinity were more significant in females than in males, and the result is consistent with an earlier study (L. Chen et al., 2010). Chlorophyll fluorescence, as an indicator of the photochemical efficiency of PSII, can provide insights into both the ability and extent to which plants tolerate environmental stresses (Maxwell and Johnson, 2000; Xu et al., 2008a ). It was observed that F v/F m and qP decreased more in females than in males, indicating less photodamage to PSII reaction centres in males. Chlorophyll degradation is an important symptom of leaf senescence. In P. yunnanensis, the total chlorophyll content significantly decreased in both males and females, but the Chl a/b ratio significantly increased only in females. This result implies that salinity aggravates the senescence of leaves particularly in females, which will further affect photosynthesis and increase sexual differences in salinity responses.
Salinity significantly changed gene expression related to photosynthesis, a similar result to that detected in physiological responses. A total of 56 and 66 genes were detected to have altered transcription levels in response to salinity stress in males and females, respectively, among which 78.5% in males but only 19.7% in females were up-regulated (Supplementary Tables S6, S7 at JXB online). It was concluded that genes with high up-regulation may enable males to maintain higher photosynthetic capacity than females under salinity stress. In addition to co-regulated genes, sexually differently expressed genes were also mainly up-regulated in males (25 up- and one down-regulated) but down-regulated in females (34 down- and two up-regulated). This result suggests that expression of genes involved in photosynthesis differed between males and females, and males can up-regulate the gene expression level in order to cope with salinity stress. Furthermore, sex-related genes up-regulated only in males had functions in light-harvesting complex I (LHC-I), LHC-II, PSI, and PSII polypeptide subunits, ATP synthase, and so on, while sex-related DEGs detected only in females included genes with additional functions (cytochrome b 6/f, ferredoxin reductase, plastocyanin, etc).
Of eight DEGs up-regulated in males but down-regulated in females in the category ‘photosynthesis’ (Table 4), seven DEGs had a role in light reactions (Supplementary Fig. S3 at JXB online). These genes may directly contribute to sexual differences in photosynthetic capacity. Salinity down-regulated PSBA in females, which encodes D1 protein involved in repairing the damaged PSII. A previous study on Synechocystis (Allakhverdiev et al., 2002) showed up-regulation of PSBA in males, suggesting that PSBA can repair the damaged PSII. In contrast, down-regulation of PSBA to low levels aggravates the decline of PSII in females. PSBY, the single nuclear gene, is imported into chloroplasts, where it is processed into two integral membrane proteins with identical topology (PSBY-1 and PSBY-2). The protein appears to bind manganese, but its role is not well understood. Meetam et al. (1999) reported that the PSBY protein is not essential for oxygenic photosynthesis. However, sexual differences in PSBY expression patterns do exist between males and females.
CAB2 and LHCB2.2 are included in the category of photosynthesis. CAB2 encodes LHC proteins that constitute the antenna system of the photosynthetic apparatus. Both of them are closely related to chlorophyll binding, which suggests that transcription of these two genes to high levels in males may help to enhance their photosynthetic capacity in response to salinity stress. In the redox chain, ATPC1 encoding the gamma subunit of chloroplast ATP synthase was significantly up-regulated in males but down-regulated in females. The absence of ATPC1 decreased ATP synthase activity, which restricted overall rates of leaf photosynthesis (Wu et al., 2007). Thus, different gene expression patterns of ATPC1 contribute to salinity-induced sexual differences in photosynthesis. PSAN, encoding the only subunit of PSI and located entirely in the thylakoid lumen, may be involved in the interaction between plastocyanin and the PSI complex (Haldrup et al., 1999), which together with FED A is related to photosynthetic electron transport.
Sexual differences in amino acid metabolism and transcription under salinity stress
Metabolism of amino acids plays a key role in the salinity tolerance of plants (Sanchez et al., 2008). In the present study, 59 and 81 salinity-responsive DEGs related to amino acid metabolism were identified in males and females, respectively (Supplementary Table S6 at JXB online). These DEGs are mainly involved in the alanine–glycine pathway, alanine–glutamate pathway, GABA (γ-aminobutyric acid) metabolism, N metabolism, homoserinethreonine synthesis, and sulphate assimilation. Nearly half of the identified DEGs (23 in males and 45 in females) showed obvious sexual differences in gene expression patterns. For instance, genes encoding glutamate decarboxylase (GAD1 and GAD5) and glutamate dehydrogenase (GDH1 and GDH2) involved in GABA metabolism were detected only in females. Three genes that function in homoserine dehydrogenase, kinase, and dehydrogenase were down-regulated only in females. Interestingly, two genes encoding DAHP (Dyer et al., 1990) and DAPD (Less and Galili, 2008), respectively, were up-regulated in males but down-regulated in females, suggesting different regulation strategies in amino acid metabolism between males and females under salinity stress. DAHP is the first enzyme in a series of metabolic steps, such as the shikimate pathway, responsible for biosynthesis of some amino acid (Dyer et al., 1990). Different expression patterns of DAHP may produce diverse amino acid components and contents, leading to sexual differences under salinity stress.
Previous studies have shown that a significant alternation of transcription-related gene expression was closely linked to plant stress tolerance (Luo et al., 2009; Brinker et al., 2010; Cohen et al., 2010). The present results provided new insights into sexual differences in transcription related to the salinity tolerance of plants. During salinity stress, 324 and 416 DEGs related to transcription factors were differentially expressed in males and females, respectively, most of which were down-regulated, such as MYB domain, NAC domain, Aux/IAA, basic helix–loop–helix, and the C2H2 zinc finger. This result suggested that salinity suppressed the normal transcription of genes. In addition, the transcript abundances of several genes were altered only in males during salinity stress, including genes involved in chromatin assembly or disassembly (CMT2), brassinosteroid signalling transduction (BEH3), DNA-binding bromodomain (F4I1.24), asymmetric cell division (HSFB4), circadian rhythm (ELF3), regulation of transcription (LUH), and DNA-mediated transformation (ASF1B). More transcription factors were identified in females than in males, such as genes encoding WRKY domain, zinc finger (GATA type), and ABI3/VP1-related B3-domain-containing transcription factors. This observation implied that the influence of salinity stress on transcription was more severe or more extensive in the female transcriptome when compared with that of males.
Genes located in the female-specific region of the W chromosome are not the main cause of sexual differences in salinity responses
Sex chromosomes of plants, when present, are distinctive not only because of their gender-determining role (Charlesworth, 1985) but also due to genomic features (Ming and Moore, 2007). In the present study, numerous DEGs related to sexual differences in physiological responses were detected. The presence of ZW sex chromosomes in poplar (Yin et al., 2008) provides an opportunity to ascertain whether the DEGs identified in this study are associated with sex chromosomes. Recombination suppression in male- and female-specific chromosomal regions is a hallmark of sex chromosomes (Ming et al., 2011). The female-specific region of the poplar W chromosome (FSW) is a 706 kb region with no counterpart on the Z chromosome. Surprisingly, based on the chromosomal location of the identified DEGs, salinity-induced DEGs of females were mainly located in autosomes rather than within the FSW. This discovery suggested that molecular responses of P. yunnanensis under salinity stress mainly rely on genes located in autosomes, and gene regulation related to the FSW makes less of a contribution to sexual differences in the plant’s salinity tolerance. Does this conclusion simply prove and supplement the established conclusion that the principal function of the sex chromosome is to reinforce dioecy? Dioecy in plants means that unisexual flowers are produced on two types of individuals, staminate flowers on males and pistillate flowers on females (Ming et al., 2011). Genes controlling staminate or pistillate flower development are randomly distributed in the genome and scattered on every chromosome (Wellmer et al., 2004). It is proposed here that more attention should be paid to genes located in autosomes rather than only on sex chromosomes when relating sexual differences and sex chromosomes.
In conclusion, the present study showed that P. yunnanensis males and females exhibit distinct sexual differences in response to salinity stress. A series of changes in physiological and biochemical traits were identified between the two sexes, clearly showing that salinity stress restrained normal metabolism and regulation, particularly in females. Transcriptional profiling analysis revealed many genes in response to salinity stress, most of which were down-regulated. Some genes with different expression patterns in males and females may cause the differences visible in growth responses, including genes involved in photosynthesis, amino acid metabolism, and transcription. Functional genes located in autosomes rather than in the FSW could play a key role in poplar salinity tolerance. Identified DEGs with different expression patterns between males and females are excellent targets for further functional studies in order to understand more specific molecular mechanisms of sexual differences in plant salt tolerance.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. qRT-PCR validation of DGE library data
Figure S2. Overview of the DEGs assigned to ‘regulation’ by MapMan.
Figure S3. Overview of the DEGs assigned to ‘photosynthesis’ by MapMan.
Table S1. Primer sequences of candidate genes used for validation in qRT-PCR.
Table S2. Genes up-regulated only in males under salinity stress.
Table S3. Genes up-regulated only in females under salinity stress.
Table S4. Genes down-regulated only in males under salinity stress.
Table S5. Genes down-regulated only in females under salinity stress.
Table S6. Comparisons of three major pathways between males and females.
Table S7. Differentially regulated genes involved in photosynthesis under salinity stress.
Acknowledgments
The research was supported by the National Key Basic Research Program of China (No. 2012CB416901) and the Key Program of the National Natural Science Foundation of China (No. 30930075).
References
- Allakhverdiev SI, Nishiyama Y, Miyairi S, Yamamoto H, Inagaki N, Kanesaki Y, Murata N. Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis . Plant Physiology. 2002;130:1443–1453. doi: 10.1104/pp.011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie J. The significance of digital gene expression profiles. Genome Research. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences. 2005;24:23–58. [Google Scholar]
- Bates L, Waldren R, Teare I. Rapid determination of free proline for water-stress studies. Plant and Soil. 1973;39:205–207. [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- Blow N. Transcriptomics: the digital generation. Nature. 2009;458:239–242. doi: 10.1038/458239a. [DOI] [PubMed] [Google Scholar]
- Brinker M, Brosche M, Vinocur B, Abo-Ogiala A, Fayyaz P, Janz D, Ottow EA, Cullmann AD, Saborowski J, Kangasjarvi J. Linking the salt transcriptome with physiological responses of a salt-resistant Populus species as a strategy to identify genes important for stress acclimation. Plant Physiology. 2010;154:1697–1709. doi: 10.1104/pp.110.164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Distribution of dioecy and self-incompatibility in angiosperms. In: Greenwood PJ, Harvey PH, Slatkin M, editors. Evolution: essays in honour of John Maynard Smith. Cambridge: Cambridge University Press; 1985. pp. 237–268. [Google Scholar]
- Chen F, Chen L, Zhao H, Korpelainen H, Li C. Sex-specific responses and tolerances of Populus cathayana to salinity. Physiologia Plantarum. 2010;140:163–173. doi: 10.1111/j.1399-3054.2010.01393.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang S, Zhao H, Korpelainen H, Li C. Sex-related adaptive responses to interaction of drought and salinity in. Populus yunnanensis . Plant, Cell and Environment. 2010;33:1767–1778. doi: 10.1111/j.1365-3040.2010.02182.x. [DOI] [PubMed] [Google Scholar]
- Chen S, Li J, Wang S, Hüttermann A, Altman A. Salt, nutrient uptake and transport, and ABA of Populus euphratica; a hybrid in response to increasing soil NaCl. Trees-Structure and Function. 2001;15:186–194. [Google Scholar]
- Cohen D, Bogeat-Triboulot MB, Tisserant E, et al. Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics. 2010;11:630. doi: 10.1186/1471-2164-11-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Hellmann H, Däschner K, Binder S, Frommer WB. A nuclear gene encoding mitochondrial Δ1-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. The Plant Journal. 2001;27:345–356. doi: 10.1046/j.1365-313x.2001.01101.x. [DOI] [PubMed] [Google Scholar]
- Diédhiou C, Golldack D. Salt-dependent regulation of chloride channel transcripts in rice. Plant Science. 2006;170:793–800. [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer WE, Weaver LM, Zhao J, Kuhn DN, Weller SC, Herrmann KM. A cDNA encoding 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Solanum tuberosum L. Journal of Biological Chemistry. 1990;265:1608–1614. [PubMed] [Google Scholar]
- FAO. FAO Land and Plant Nutrition Management Service. 2008 http://www.fao.org/ag/agl/agll/spush. [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana . The Plant Journal. 2005;44:826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Haldrup A, Naver H, Scheller HV. The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. The Plant Journal. 1999;17:689–698. doi: 10.1046/j.1365-313x.1999.00419.x. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Hu C, Delauney AJ, Verma D. A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proceedings of the National Academy of Sciences, USA. 1992;89:9354–9358. doi: 10.1073/pnas.89.19.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ. Gene expression profiles during the initial phase of salt stress in rice. The Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. The Plant Cell. 1996;8:1323–1335. doi: 10.1105/tpc.8.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Knight MR. Abiotic stress signalling pathways: specificity and cross-talk. Trends in Plant Science. 2001;6:262–267. doi: 10.1016/s1360-1385(01)01946-x. [DOI] [PubMed] [Google Scholar]
- Kooten O, Snel J. The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynthesis Research. 1990;25:147–150. doi: 10.1007/BF00033156. [DOI] [PubMed] [Google Scholar]
- Less H, Galili G. Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiology. 2008;147:316–330. doi: 10.1104/pp.108.115733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Xu G, Zang RG, Korpelainen H, Berninger F. Sex-related differences in leaf morphological and physiological responses in Hippophae rhamnoides along an altitudinal gradient. Tree Physiology. 2007;27:399–406. doi: 10.1093/treephys/27.3.399. [DOI] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Li WY, Wong FL, Tsai SN, Phang TH, Shao G, Lam HM. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant, Cell and Environment. 2006;29:1122–1137. doi: 10.1111/j.1365-3040.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods in Enzymology. 1987;148:350–382. [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo ZB, Janz D, Jiang X, Gobel C, Wildhagen H, Tan Y, Rennenberg H, Feussner I, Polle A. Upgrading root physiology for stress tolerance by ectomycorrhizas: insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiology. 2009;151:1902–1917. doi: 10.1104/pp.109.143735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence—a practical guide. Journal of Experimental Botany. 2000;51:659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- Meetam M, Keren N, Ohad I, Pakrasi HB. The PsbY protein is not essential for oxygenic photosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiology. 1999;121:1267–1272. doi: 10.1104/pp.121.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Bendahmane A, Renner S. Sex chromosomes in land plants. Annual Review of Plant Biology. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. [DOI] [PubMed] [Google Scholar]
- Ming R, Moore PH. Genomics of sex chromosomes. Current Opinion in Plant Biology. 2007;10:123–130. doi: 10.1016/j.pbi.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A. Next-generation tag sequencing for cancer gene expression profiling. Genome Research. 2009;19:1825–1835. doi: 10.1101/gr.094482.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. Genes and salt tolerance: bringing them together. New Phytologist. 2005;167:645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annual Review of Plant Biology. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Rabbani M, Maruyama K, Abe H, Khan M, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiology. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS, Ricklefs RE. Dioecy and its correlates in the flowering plants. American Journal of Botany. 1995;82:596–606. [Google Scholar]
- Roosens NHCJ, Thu TT, Iskandar HM, Jacobs M. Isolation of the ornithine-δ-aminotransferase cDNA and effect of salt stress on its expression in Arabidopsis thaliana. Plant Physiology. 1998;117:263–271. doi: 10.1104/pp.117.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez DH, Siahpoosh MR, Roessner U, Udvardi M, Kopka J. Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiologia Plantarum. 2008;132:209–219. doi: 10.1111/j.1399-3054.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- Savouré A, Jaoua S, Hua XJ, Ardiles W, Van Montagu M, Verbruggen N. Isolation, characterization, and chromosomal location of a gene encoding the [Delta] 1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana. FEBS Letters. 1995;372:13–19. doi: 10.1016/0014-5793(95)00935-3. [DOI] [PubMed] [Google Scholar]
- Schat H, Sharma S, Vooijs R. Heavy metal-induced accumulation of free proline in a metal-tolerant and a nontolerant ecotype of Silene vulgaris . Physiologia Plantarum. 2006;101:477–482. [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Storey R, Walker RR. Citrus and salinity. Scientia Horticulturae. 1999;78:39–81. [Google Scholar]
- Székely G, Abrahám E, Cséplo A, et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. The Plant Journal. 2007;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- Szabados L, Savouré A. Proline: a multifunctional amino acid. Trends in Plant Science. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Szoke A, Miao G-H, Hong Z, Verma DPS. Subcellular location of Δ1-pyrroline-5-carboxylate reductase in root/nodule and leaf of soybean. Plant Physiology. 1992;99:1642–1649. doi: 10.1104/pp.99.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiology. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- ’t Hoen P, Ariyurek Y, Thygesen H, Vreugdenhil E, Vossen R, de Menezes R, Boer J, van Ommen G, den Dunnen J. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Research. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiology. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Villarroel R, Van Montagu M. Osmoregulation of a pyrroline-5-carboxylate reductase gene in Arabidopsis thaliana. Plant Physiology. 1993;103:771–781. doi: 10.1104/pp.103.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RG, Chen SL, Deng L, Fritz E, Huttermann A, Polle A. Leaf photosynthesis, fluorescence response to salinity and the relevance to chloroplast salt compartmentation and anti-oxidative stress in two poplars. Trees-Structure and Function. 2007;21:581–591. [Google Scholar]
- Wang XZ, Curtis PS. Gender-specific responses of Populus tremuloides to atmospheric CO2 enrichment. New Phytologist. 2001;150:675–684. [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. The Plant Cell. 2004;16:1314–1326. doi: 10.1105/tpc.021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C, Li Y, Labbe A, Guevara D, Nuin P, Whitty B, Diaz C, Golding G, Gray G, Weretilnyk E. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiology. 2006;140:1437–1450. doi: 10.1104/pp.105.070508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Ortiz-Flores G, Ortiz-Lopez A, Ort DR. A point mutation in atpC1 raises the redox potential of the Arabidopsis chloroplast ATP synthase γ-subunit regulatory disulfide above the range of thioredoxin modulation. Journal of Biological Chemistry. 2007;282:36782–36789. doi: 10.1074/jbc.M707007200. [DOI] [PubMed] [Google Scholar]
- Xu X, Peng GQ, Wu CC, Korpelainen H, Li CY. Drought inhibits photosynthetic capacity more in females than in males of. Populus cathayana . Tree Physiology. 2008a;28:1751–1759. doi: 10.1093/treephys/28.11.1751. [DOI] [PubMed] [Google Scholar]
- Xu X, Yang F, Xiao XW, Zhang S, Korpelainen H, Li CY. Sex-specific responses of Populus cathayana to drought and elevated temperatures. Plant, Cell and Environment. 2008b;31:850–860. doi: 10.1111/j.1365-3040.2008.01799.x. [DOI] [PubMed] [Google Scholar]
- Xu X, Zhao H, Zhang X, H nninen H, Korpelainen H, Li C. Different growth sensitivity to enhanced UV-B radiation between male and female. Populus cathayana. Tree Physiology. 2010;30:1489–1498. doi: 10.1093/treephys/tpq094. [DOI] [PubMed] [Google Scholar]
- Yamada M, Morishita H, Urano K, Shiozaki N, Yamaguchi-Shinozaki K, Shinozaki K, Yoshiba Y. Effects of free proline accumulation in petunias under drought stress. Journal of Experimental Botany. 2005;56:1975–1981. doi: 10.1093/jxb/eri195. [DOI] [PubMed] [Google Scholar]
- Yin T, DiFazio SP, Gunter LE, Zhang X, Sewell MM, Woolbright SA, Allan GJ, Kelleher CT, Douglas CJ, Wang M. Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Research. 2008;18:422–430. doi: 10.1101/gr.7076308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S, A.Bressan R, Hasegawa PM. Salt stress tolerance of plants. JIRCAS Working Report. 2002:25–33. [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D, Laubinger S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. The Plant Journal. 2009;58:1068–1082. doi: 10.1111/j.1365-313X.2009.03835.x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Jiang H, Peng S, Korpelainen H, Li C. Sex-related differences in morphological, physiological, and ultrastructural responses of Populus cathayana to chilling. Journal of Experimental Botany. 2011;62:675–686. doi: 10.1093/jxb/erq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Li Y, Duan B, Korpelainen H, Li C. Sex-related adaptive responses of Populus cathayana to photoperiod transitions. Plant, Cell and Environment. 2009;32:1401–1411. doi: 10.1111/j.1365-3040.2009.02007.x. [DOI] [PubMed] [Google Scholar]
- Zhao H, Xu X, Zhang Y, Korpelainen H, Li C. Nitrogen deposition limits photosynthetic response to elevated CO2 differentially in a dioecious species. Oecologia. 2011;165:41–54. doi: 10.1007/s00442-010-1763-5. [DOI] [PubMed] [Google Scholar]
- Zhu J-K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiology. 2000;124:941–948. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K. Plant salt tolerance. Trends in Plant Science. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- Zhu J-K. Salt and drought signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.