Abstract
Corticotropin-releasing factor receptor type 2 (CRFR2) is highly expressed in skeletal muscle (SM) tissue where it is suggested to inhibit interactions between insulin signaling pathway components affecting whole-body glucose homeostasis. However, little is known about factors regulating SM CRFR2 expression. Here, we demonstrate the exclusive expression of CRFR2, and not CRFR1, in mature SM tissue using RT-PCR and ribonuclease protection assays and report a differential expression of CRF receptors during C2C12 myogenic differentiation. Whereas C2C12 myoblasts exclusively express CRFR1, the C2C12 myotubes solely express CRFR2. Using cAMP luciferase assays and calcium mobilization measurements, we further demonstrate the functionality of these differentially expressed receptors. Using luciferase reporter assays we show a differential activation of CRFR promoters during myogenic differentiation. Transfections with different fragments of the 5′-flanking region of the mCRFR2β gene fused to a luciferase reporter gene show a promoter-dependent expression of the reporter gene and reveal the importance of the myocyte enhancer factor 2 consensus sequence located at the 3′-proximal region of CRFR2β promoter. Furthermore, we demonstrate that CRFR2 gene transcription in the mature mouse is stimulated by both high-fat diet and chronic variable stress conditions. Performing a whole-genome expression microarray analysis of SM tissues obtained from CRFR2-null mice or wild-type littermates revealed a robust reduction in retinol-binding protein 4 expression levels, an adipokine whose serum levels are elevated in insulin-resistant states. In correlation with the SM CRFR2β levels, the SM retinol-binding protein 4 levels were also elevated in mice subjected to high-fat diet and chronic variable stress conditions. The current findings further position the SM CRFR2 pathways as a relevant physiological system that may affect the known reciprocal relationship between psychological and physiological challenges and the metabolic syndrome.
CRF receptors are differentially expressed during myogenic differentiation, and mature skeletal muscle CRFR2 expression is up-regulated by either high-fat diet or chronic variable stress conditions.
Abdominal obesity and insulin resistance have each been proposed as the primary factors underlying metabolic syndrome (1,2). Skeletal muscle (SM) comprises the largest insulin-sensitive tissue in humans, and thus, insulin resistance in this organ impacts whole-body glucose homeostasis (3). Insulin resistance in SM was proposed to promote atherogenic dyslipidemia by decreasing muscle glycogen synthesis and elevating hepatic de novo lipid synthesis and very-low-density lipoprotein production (2).
The corticotropin-releasing factor (CRF)/urocortin (Ucn) family of peptides and receptors is involved in the maintenance and adaptive responses necessary for energy homeostasis (4,5,6,7,8,9,10,11). The CRF/Ucn family of neuropeptides signals through the activation of two G protein-coupled receptors, CRF receptor type 1 (CRFR1) (12,13,14) and CRF receptor type 2, CRFR2 (15,16,17,18). Mouse CRFR2 has three apparent splice variants, which results in two putative receptor proteins of 411 and 431 amino acids (CRFR2α and CRFR2β, respectively) and in a 422-amino acid insertion-variant (iv) with dominant-negative activity. In rodents, CRFR2α is predominantly expressed in the brain (19). The CRFR2β splice variant is expressed primarily in the SM, the heart, the brain choroid plexus, the gastrointestinal tract, and the skin (17,20,21) whereas ivCRFR2β is exclusively expressed in the heart (22).
In SM tissue, CRFR2β was suggested to be involved in different cellular processes. SM CRFR2β activation was suggested to impede glucose metabolism. CRFR2-null mice have enhanced glucose tolerance, increased insulin sensitivity and are protected from high-fat diet-induced insulin resistance (6). Ucn2, which is highly expressed in SM tissue (23) and most likely serves as the endogenous ligand for SM CRFR2β, inhibits the interactions between insulin-signaling pathway components and insulin-induced glucose uptake in cultured SM cells, and in C2C12 myotubes (8). The Ucn2-null mice exhibit increased insulin sensitivity and are protected from fat-induced insulin resistance (8). In addition, CRFR2β activation was demonstrated to increase SM mass (24), reduce SM mass loss in atrophying SM due to denervation or casting, and to increase nonatrophying SM mass (25).
Given the importance of CRFR2 in regulating the central stress response and its beneficial effect on cardiovascular function (26), the regulation of its hypothalamic and heart expression has been extensively studied (Refs. 27,28,29,30,31 and Refs. 22 and 32,33,34,35, respectively). However, little is known regarding factors regulating SM CRFR2β expression. Here, we demonstrate the differential expression of CRFR1 and CRFR2 mRNA during C2C12 myogenic differentiation. The functional signaling of those receptors was determined, and promoter analysis studies demonstrated the importance of muscle-specific transcription factors putative binding sites. Additionally, we show the in vivo regulation of SM CRFR2β mRNA by chronic physiological or psychological stressors and its association with insulin-resistant states.
Results
Differential expression of CRFR1 and CRFR2 during myogenic differentiation
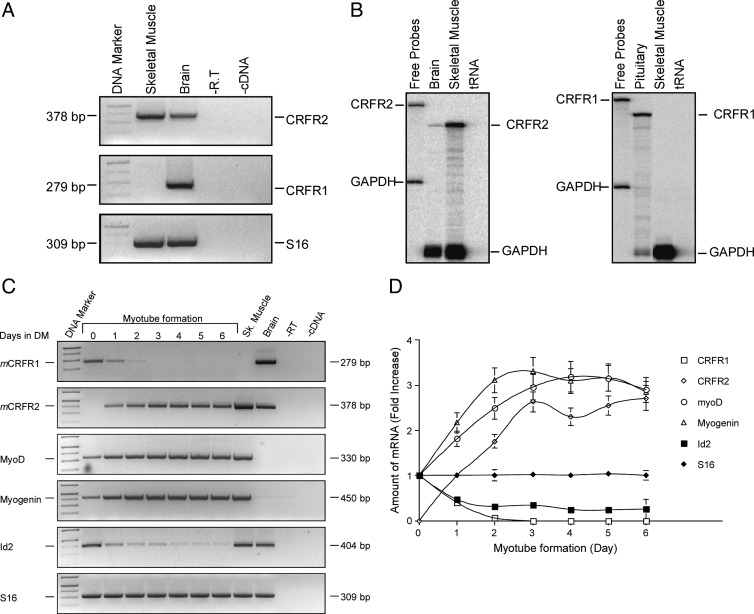
To verify expression of SM CRFRs, total RNA prepared from SM and brain tissues was reverse transcribed to generate cDNAs. The cDNA products were used as templates for specific semiquantitative RT-PCR demonstrating selective CRFR2 expression in SM tissue whereas the brain cDNA served as a positive control for CRFR1 and CRFR2 expression (Fig. 1A). The selective expression of CRFR2, and not CRFR1, in SM tissue was further verified using ribonuclease (RNase) protection assay (Fig. 1B). The multinucleated SM fibers are formed in successive distinct steps involving different types of myoblasts (36). For in vitro investigation of the molecular basis of SM cell differentiation, C2C12 cells, mouse-derived myoblasts that can be propagated as undifferentiated mononuclear cells in serum, serve as a useful experimental model. On serum withdrawal, muscle-specific genes are expressed leading to the formation of differentiated multinucleated myotubes (37,38). To study the CRFR2 expression profile during myogenic differentiation, RNA extracted from C2C12 myoblasts at different time points during myogenic differentiation was reverse transcribed and used as a template for semiquantitative RT-PCR. The myogenic determination factors MyoD and myogenin, as well as the negative regulator of myogenesis, Id2 (39), were used to monitor the differentiation process. Unexpectedly, C2C12 myoblasts were found to exclusively express CRFR1 whereas C2C12 myotubes were found to exclusively express CRFR2 (Fig. 1C). The time-dependent differential expression of the two-receptor forms can be observed during the differentiation process (Fig. 1, C and D). CRFR2 shows expression kinetics similar to MyoD and myogenin expression profiles whereas CRFR1 expression mirrors the expression profile Id2 (Fig. 1, C and D).
Figure 1.
Expression of mCRFR2 mRNA in mouse SM and differential expression of mCRFR1 and mCRFR2 mRNA during C2C12 cells myogenic differentiation. A, Representative image of electrophoretic analysis of the semiquantitative RT-PCR products of mCRFR2 (upper panel), mCRFR1 (middle panel), and the ribosomal protein S16 (lower panel) in the mouse SM. Brain samples served as positive controls for both CRFR1 and CRFR2 gene expression. PCR without reverse transcriptase (RT) enzyme (−R.T) or without cDNA (−cDNA) served as negative controls. B, Representative image of RNase protection assay of mCRFR1 (right panel) and mCRFR2 (left panel) mRNA. SM total RNA was hybridized with the mCRFR1 (right panel), mCRFR2 (left panel), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (both panels) antisense probes. Brain and pituitary gland served as positive controls for CRFR2 and CRFR1 gene expression, respectively. C, Representative image of electrophoretic analysis of the semiquantitative RT-PCR products of mCRFR1, mCRFR2, muscle differentiation markers, MyoD, myogenin, and Id2 and the ribosomal protein S16 in C2C12 myoblasts cultured in differentiation media (DM). RNA extracted from C2C12 myoblasts cultured in DM (containing 2% horse serum) for 0–6 d were reverse transcribed to generate cDNA, which were used as templates to the PCR using specific primers for mCRFR1, mCRFR2, MyoD, myogenin, Id2, and the ribosomal protein S16 that served as an internal control. RNA extracted from mouse SM and brain served as positive controls. PCR without RT enzyme (−R.T) or without cDNA (−cDNA) served as negative controls. D, The bands were quantified, and the normalized values (relative to the control S16 expression) are presented as fold increase. Three independent experiments were conducted and showed similar kinetic of gene expression.
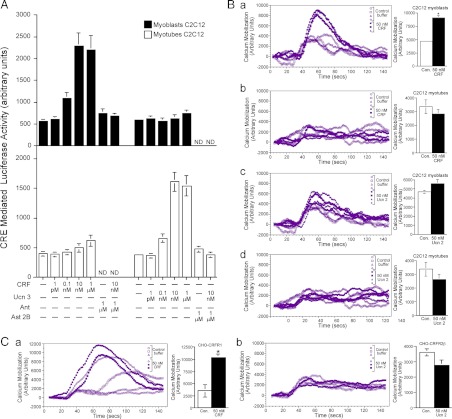
C2C12 cells were further used for demonstrating CRFR1- and CRFR2-selective activation in nondifferentiated (myoblasts) or differentiated (myotubes) state. Receptor functionality was demonstrated by measuring the activation of the cAMP and calcium pathways using CRF or Ucn2/3, which are specific ligands for CRFR1 and CRFR2, respectively (40,41,42). Nondifferentiated C2C12 cells were transfected with a luciferase reporter containing a fragment of the EVX1 gene that contains a cAMP-responsive element (CRE) site. Luciferase activity was used as a measure of receptor activation and was determined after 4 h of treatment with vehicle, and various doses of CRF, or Ucn3 in nondifferentiated, or 48-h differentiated, C2C12 cells. The CRE-luciferase reporter gene was differentially activated in the myoblasts and myotubes after stimulation with CRF or Ucn3, respectively (Fig. 2A). In myoblasts, CRF signaling induced CRE-luciferase activity, which was blocked by the CRFR1-specific antagonist Antalarmin (Ant), whereas in the myotubes, Ucn3 signaling induced CRE-luciferase activity that was blocked by the CRFR2-specific antagonist Astressin 2B (Ast 2B).
Figure 2.
CRFR1 and CRFR2 signaling pathways are differentially activated during the myogenic differentiation. A, Activation of CRE-luciferase reporter by CRF and Ucn3 peptides in C2C12 myoblasts or myotubes, respectively. C2C12 myoblasts were transfected with CRE-luciferase, and luciferase activity was measured after treatment (4 h) with vehicle or 1 pm, 0.1 nm, 10 nm, and 1 μm CRF or Ucn3 in nondifferentiated C2C12 cells (black bars) or 48 h differentiated C2C12 cells (white bars). Assays were normalized to cotransfected β-gal activity. The mean sem of three independent experiments is presented as relative activity. The activation of CRE-luciferase in myoblasts by CRF, or in myotubes by Ucn3, was blocked by the CRFR1-specific antagonist (Ant) or CRFR2-specific antagonist Ast 2B, respectively [*, P < 0.05 vs. vehicle treatment; **, P < 0.05 vs. CRF (10 nm) or Ucn3 (10 nm) treatment]. B. Calcium mobilization in nondifferentiated (myoblasts) or differentiated (myotubes) C2C12 cells by CRF and Ucn2. Nondifferentiated C2C12 cells (a and c) and differentiated C2C12 (b and d) were treated with CRF (a and b) or Ucn2 (c and d), respectively, and the calcium mobilization kinetic was measured using FlexStation (Molecular Devices Corp.). Bar graphs represent the maximum values. Interestingly, CRFR1 but not CRFR2 activation promotes calcium mobilization. Activation of calcium flux was differentially activated in the myoblasts and myotubes after stimulation with CRF. * P < 0.05 vs. control buffer. C, CHO cells stably expressing the CRFR1 (a) or CRFR2 (b) were treated with CRF or Ucn2, respectively, and the calcium mobilization kinetic was measured using FlexStation. Bar graphs represent the maximum values. *, P < 0.05 vs. control buffer. ND, Not determined; Con., control.
Additionally, calcium mobilization kinetics were measured in nondifferentiated (myoblasts) or differentiated (myotubes) C2C12 cells by CRF or Ucn2 (Fig. 2B). Activation of calcium flux was differentially activated in the myoblasts and myotubes after stimulation with CRF. CRF strongly activated calcium flux in myoblasts, but not in myotubes, (Fig. 2B, a and b). Ucn2 activated calcium flux to a lesser extent in myoblasts, probably due to its low affinity for CRFR1, but did not activate calcium flux in myotubes (Fig. 2B, c and d). To further explore this phenomenon, the calcium mobilization kinetic studies were duplicated in Chinese hamster ovary (CHO) cells stably expressing CRFR1 or CRFR2 treated with CRF or Ucn2, respectively (Fig. 2C). Only CRF activation of CHO cells expressing CRFR1, but not Ucn2 activation of CHO cells expressing CRFR2, promoted calcium mobilization. Demonstrating that CRFR1, but not CRFR2, activation will promote calcium mobilization supports the finding of differential expression of the CRFRs during myogenic differentiation.
Differential activation of mCRFR1 and mCRFR2β promoters during C2C12 myoblast differentiation
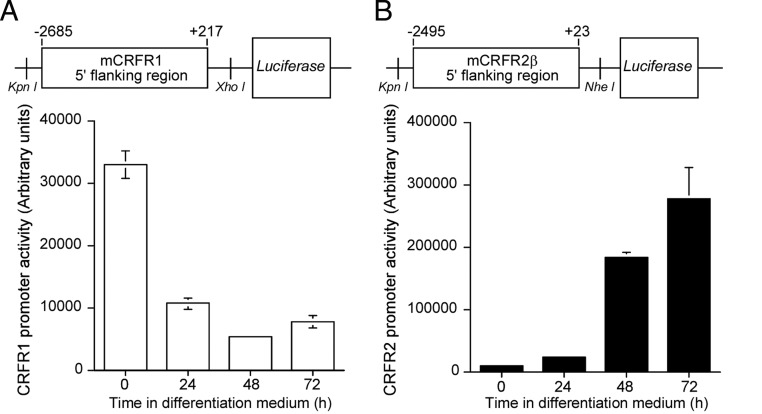
To explore the molecular mechanisms mediating the differential regulation of CRFR1 and CRFR2 during myogenic differentiation, and to examine whether the differential expression is regulated at the promoter level, we isolated the 5′-flanking region of both genes. Subcloning mCRFR1 or mCRFR2β 5′-flanking sequences upstream to a luciferase gene allowed us to study their activity during the myogenic differentiation (Fig. 3). C2C12 cells were transfected with the reporter plasmid DNA, and the luciferase activity over 3 d of myogenic differentiation was determined. Interestingly, myoblast differentiation was accompanied by a significant and rapid inhibition of the mCRFR1 promoter, and time-dependent and robust activation of the mCRFR2β promoter (Fig. 3, A and B, respectively). These sequential changes parallel the decrease and the increase in mRNA level of CRFR1 and CRFR2β, respectively (Fig. 3), and confirm that mCRFR2β expression during differentiation is regulated at the transcriptional level.
Figure 3.
Differential activation of mCRFR1 and mCRFR2β promoters during C2C12 myoblast differentiation. Schematic demonstration of the mCRFR1 (A) and mCRFR2β (B) 5′-flanking region construct fused to the luciferase gene in PGL3 basic vector. C2C12 cells were transfected with the reporter plasmid DNA, and luciferase activity during the myogenic differentiation was determined. The luciferase activity was corrected to β-gal values. (Results are shown as mean ± sem of six independent experiments).
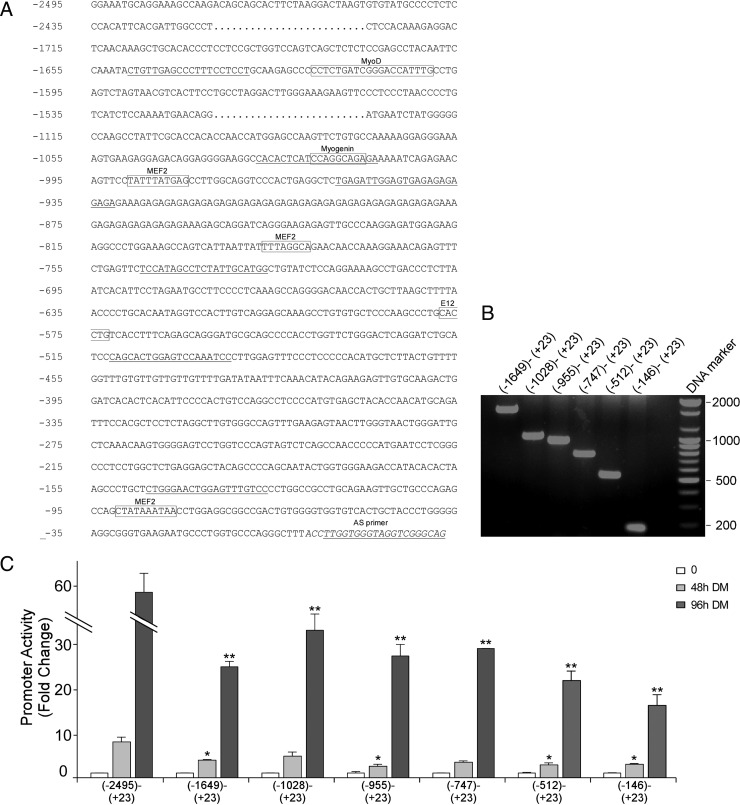
To further study the involvement of putative muscle-specific transcription factors in the activation of the CRFR2β 5′-flanking region, the 5′-flanking region of CRFR2β (−2495 to +23) was analyzed for SM transcription factor consensus sequences using the TESS program (Transcription Element Search System) (Fig. 4A). Six fragments of the 5′-flanking region, with different lengths and numbers of putative muscle-specific transcription factors consensus sequences, were subcloned into a luciferase pGL3 basic vector and used for transfecting C2C12 cells (Fig. 4, B and C). The luciferase activity of each fragment during myogenic differentiation was studied. No basal differences were detected between the different fragments. When differentiation medium (DM) was introduced, the promoter activity increased in a time-dependant manner. The differences between the promoter fragments could be detected as early as 24 h in DM, where the full isolated 5′-flanking region was strongly activated and its activity was significantly higher compared with the truncated fragments, regardless of their length (Fig. 4C). After 96 h in DM, all truncated fragments were robustly activated. However, their activity was significantly lower compared with the full 5′-flanking region (Fig. 4C). Although the activity level of the truncated fragments varied, there was no significant difference between them. Interestingly, even the shortest fragment, consisting of 168 bp, was strongly activated, indicating the importance of the proximal site in mediating CRFR2β transcription (Fig. 4C).
Figure 4.
Sequence-, fragmentation-, and differentiation-induced activation of CRFR2β 5′-flanking region. A, Genomic sequence of mCRFR2β 5′-flanking region. 5′-Untranslated region is shown in italic letters. The six primers used for promoter fragmentation are underlined, and putative sites for muscle-specific transcription factors, recognized using TESS (Transcription Element Search System), are indicated. B, Electrophoretic analysis of pGL3-basic vectors containing truncated mCRFR2β 5′-flanking region. C, Activity of CRFR2β fragmented promoter during myogenic differentiation. C2C12 cells were transfected with the reporter plasmid preceded by the different 5′-flanking region fragments. Luciferase activity during the myogenic differentiation in 2% horse serum containing DM was determined. The relative luciferase activity was corrected to β-gal activity (results are shown as fold increase over the basal activity of each fragment, shown as mean ± sem). AS, Antisense.*, P < 0.05; **, P < 0.001 vs. the full fragment.
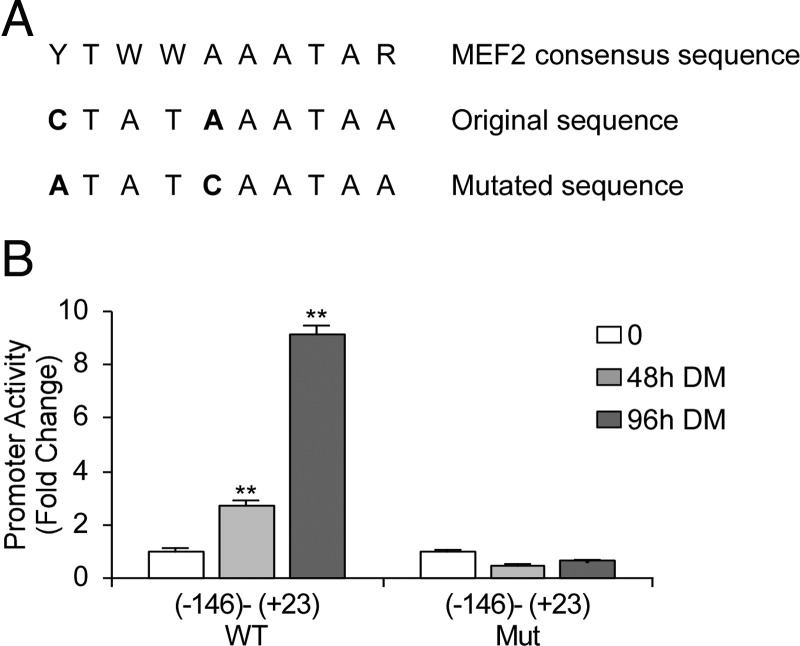
Given the strong potency of the short 5′-flanking fragment (−146 to +23), we examined the importance of the putative myocyte enhancer factor (MEF)2 consensus sequence (located −91 to −82) for this activation. This MEF2 consensus sequence in the minimal 5′-flanking region was mutated and subcloned into pGL3 basic vector. The MEF2 consensus sequence is YTWWAAATAR, where Y stands for T or C, W stands for A or T, and R stands for A or G (43). The mutation included C to A and A to C substitutions (CtatAaataa to AtatCaataa) (Fig. 5A). The mutated sequence is not recognized as MEF2 consensus sequence using the TESS analysis. The WT or mutated fragment were transfected into C2C12 cells, and luciferase activity during myogenic differentiation was measured (Fig. 5B). The mutated fragment was not able to induce transcription as demonstrated by a constant low activation of the luciferase gene throughout the differentiation process, indicating the significance of this MEF2 site for CRFR2β expression during myogenic differentiation.
Figure 5.
Activity of the WT and mutated MEF2 site in the minimal CRFR2β 5′-flanking region during the myogenic differentiation. C2C12 cells were transfected with the WT or the mutated minimal (−146 to +23) 5′-flanking region-luciferase vectors and luciferase activity during myogenic differentiation in 2% HS containing media (DM) was determined. The relative luciferase activity was corrected to β-gal activity (results are shown as mean ± sem). Mut, Mutated.
Regulation of SM CRFR2β expression by stress and its correlation to retinol-binding protein 4 (RBP4) expression level
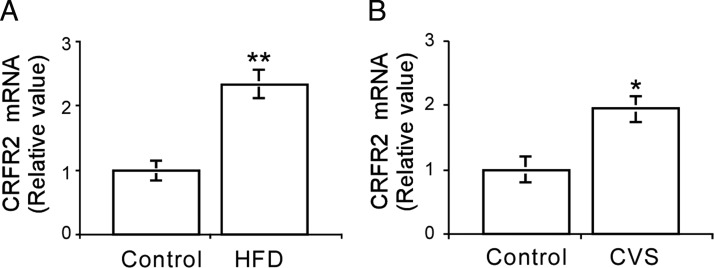
Given the high expression level of CRFR2β and its suggested role in modulating insulin sensitivity and glucose uptake, we further studied its regulation in the mature mouse after exposure to chronic stressors. High-fat diet (HFD) and chronic-variable stress (CVS) paradigms were chosen because they represent prolonged physiological and psychological stressors, respectively. SM RNA obtained from mice maintained for 15 wk on HFD or from mice subjected to CVS protocol, and the respective controls, was reverse transcribed, and CRFR2β expression levels were determined using real-time PCR. Interestingly, both stressors triggered a significant elevation in CRFR2β expression level. HFD induced a 2.3-fold increase (Fig. 6A) whereas CVS induced a 2.0-fold increase (Fig. 6B) in CRFR2β expression level. The expression of Ucn2, the local ligand for SM CRFR2β, did not change significantly under these conditions (data not shown).
Figure 6.
Up-regulation of SM CRFR2β after exposure to chronic stressors. CRFR2β mRNA level determined by real-time PCR in SM obtained from mice kept on HFD compared with control low-fat diet (A) or subjected to CVS (B). CRFR2β expression was corrected by HPRT1 expression level and normalized to control levels (results are shown as mean ± sem). *, P < 0.05 vs. control.
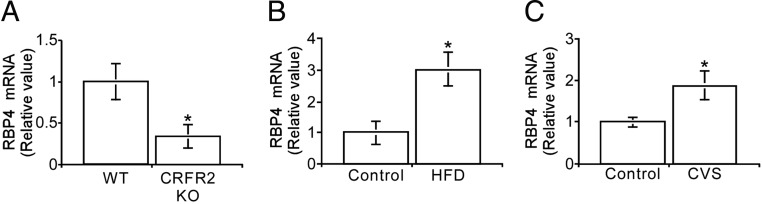
CRFR2 signaling was previously demonstrated to inhibit insulin signaling in SM (8). To further understand the molecular mechanisms mediating the effect of Ucn2/CRFR2 signaling on insulin sensitivity in SM, we compared the gene expression profile of SM obtained from both CRFR2 knockout (KO) and WT littermates using gene expression microarray. The microarray analysis demonstrated a significant reduction of 43.5% in the expression level of RBP4. RBP4 is an adipokine whose serum levels are increased in insulin-resistant subjects, and its administration leads to impaired insulin signaling in muscle (44). To confirm our microarray data, SM was collected from CRFR2-null mice and from their WT littermates, and SM cDNA was used for measuring RBP4 expression level by real-time PCR. The real-time PCR results were in agreement with the microarray findings and showed a significant reduction of 52% in RBP4 expression level (Fig. 7A). Several genes that were found to be up- or down- regulated in the microarray analysis are listed in Supplemental Table 1 (published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). These changes were not further verified using additional quantitative methods. Because CRFR2-KO mice are a developmental KO model and therefore may represent developmental compensatory changes, we further examined RBP4 muscle expression level in conditions that up-regulate CRFR2β expression, namely HFD and CVS, and found a positive correlation between RBP4 and CRFR2β expression. SM RBP4 expression level was significantly elevated in mice subjected to both HFD and CVS manipulations (Fig. 7, B and C).
Figure 7.
SM RBP4 and CRFR2β expression levels are positively correlated. RBP4 mRNA level determined by real-time PCR in SM obtained from CRFR2 KO mice and their WT littermates (A), mice kept on HFD (B) or mice subjected to CVS (C). RBP4 expression was corrected by HPRT1 expression level and normalized to WT/control (results are shown as mean ± sem). *, P < 0.05 vs. WT/control.
Discussion
SM tissue has been demonstrated to express high levels of CRFR2 transcript (20), which was shown to be associated with controlling glucose transport into the SM. In the present study we demonstrated, using specific mCRFR2 RNase protection assays, RT-PCR, and DNA sequencing, that adult SM tissue expresses CRFR2β, but not the CRFR1, transcripts. Previous reports showed CRFR2β expression in SM to be localized in neural structures, blood vessels, myotendinous junctions, and endomysial/perimysial spaces, but not in myocytes (45).
Here, we showed that C2C12 myoblasts exclusively express CRFR1, whereas the C2C12 myotubes exclusively express CRFR2β. In the myoblast state, serum induces the expression of Id proteins, transcription factors that sequester E12 and E47 into complexes unable to bind DNA (39). Upon serum removal, MyoD family proteins, MyoD, Mrf4, and myogenin, are activated to promote the expression of muscle-specific genes with Mef2 family of transcription factors, which play an important role in this context (46). This sequential expression pattern is also demonstrated in the CRFR expression kinetics, where CRFR1 expression mirrors the expression profile of Id2, and CRFR2β expression kinetics parallels the MyoD and myogenin expression profile. Additional examples of differential expression of CRFRs were reported in other types of muscle tissues. In human nonpregnant myometrium the CRFR1α and CRFR1β-receptor subtypes were found, whereas at term R2α and C variant CRFR subtypes were expressed as well (47). Moreover, we recently reported differential regulation of mCRFR2β by stress in heart myocardium. The mRNA levels of mCRFR2β were down-regulated in hearts of mice that underwent CVS whereas the mRNA levels of a new splice variant of CRFR2β, iv-mCRFR2β, were up-regulated (22). Lipopolysaccharide was also shown to differentially regulate CRFR2β expression in the heart and SM. Systemic injection of lipopolysaccharide up-regulated SM CRFR2 mRNA levels and markedly down-regulated its mRNA heart levels (48).
The pharmacological properties of CRFR1 and CRFR2 activation by different ligands of the CRF peptide family are well established. CRF has relatively lower affinity for CRFR2 compared with its affinity for CRFR1, Ucn1 has equal affinities for both receptors, and Ucn2 and Ucn3 appear to be selective for CRFR2 (40,42,49). Both CRFRs belong to the B1 subfamily of seven-transmembrane-domain receptors that signal by coupling to G proteins (50). CRFR1 and CRFR2 signaling primarily stimulates the adenylyl cyclase/cAMP pathway via coupling and activation of Gαs proteins and protein kinase A activation (13,18,51). In addition, CRFR1 is coupled to activation of plasma membrane calcium channels and CRF signaling and was shown to generate changes in corticotrope cytosolic free calcium concentration (52). The increase in Ca2+ influx involves voltage-gated channels, namely L- and P-type channels (53). That CRFR1 coupling to activation of plasma membrane calcium channels depends on cell type (54) was demonstrated in melanocytes (55). We assessed the differential expression of CRFR signaling in assays based on these similarities and differences. CRF and Ucn3 activated CRE-luciferase reporter gene in myoblasts or myotubes, respectively. The use of selective antagonists and the subsequent activation of the cAMP pathway further emphasized the ligand specificity and functionality of the CRFR subtypes. Furthermore, the ability of CRF to induce calcium mobilization selectively in myoblasts provides an additional level of support to the absence of CRFR1 in differentiated myotubes.
Transient transfection of C2C12 myoblasts with constructs containing the 5′-flanking region of the mCRFR1 or mCRFR2β genes fused to a luciferase reporter showed differential promoter activity during myogenic differentiation. The CRFR1 promoter activity was negatively regulated, whereas CRFR2β promoter activity was positively regulated, during differentiation. This differential regulation is due to the varying responsiveness of the promoters to myogenic transcription factors. Computer-aided sequence analysis revealed the presence of putative muscle-specific transcription factor consensus sequences in the mCRFR2β 5′-flanking region. Different fragments of the 5′-flanking region were cloned into a luciferase vector to identify the crucial area needed for CRFR2β transcription. A robust activation of all the fragmented regions was observed revealing the importance of the 3′-proximal region. The importance of this region was verified by mutating the MEF2 consensus sequence. The mutated fragment was incapable of transcription, as demonstrated by blunted luciferase activity. This short but powerful minimal 5′-flanking region may be further used as a minimal promoter for muscle-specific expression of target genes.
Understanding the regulation of SM CRFR2β may provide further insight into the physiological functions of this receptor. Mice lacking either CRFR2 or Ucn2 demonstrate enhanced glucose tolerance, increased insulin sensitivity, and protection from high fat diet-induced insulin resistance (6,8). In Ucn2 KO mice, systemic Ucn2 administration before glucose tolerance test or insulin tolerance test impaired glucose clearance and reduced insulin sensitivity, respectively (8), showing that this phenotype is mediated by peripheral CRFR2. Both obesity and high stress, hallmarks of a modern lifestyle, are correlated with insulin resistance (56,57). We showed that both a physiological stressor (chronic consumption of a HFD) and a psychological stressor (CVS) share the same consequence of elevated SM CRFR2β expression level. Consequently, the increased SM CRFR2β expression may contribute to the reduced insulin sensitivity, which characterizes these conditions. Elucidation of this phenomenon is essential for better management of the metabolic consequences that coincide with both HFD and chronic psychological stress. Interestingly, a similar CVS protocol mediated a reduction in CRFR2β mRNA levels in the hearts of mice (22); however, these tissue-specific differences might be attributed to the up-regulation of the dominant-negative iv-mCRFR2β isoform.
A positive correlation between leptin serum levels and CRFR2α mRNA levels in the ventromedial hypothalamus has been shown (30). Because leptin is produced in proportion to fat stores (58), and full-length leptin receptor is expressed by SM (59), it is intriguing to hypothesize that the increased CRFR2β expression under HFD is regulated by leptin. However, the association with leptin does not explain the increased CRFR2β expression in mice subjected to CVS, because CVS was demonstrated to reduce serum leptin levels (60). Both HFD consumption and chronic stress lead to elevated glucocorticoids (60,61,62), which may regulate CRFR2β expression under these conditions. HFD and CVS may be considered representatives of modern lifestyle characteristics, which include high stress load and increased intake of high-fat foods. The indication that mice lacking Ucn2 exhibited increased insulin sensitivity and better glucose tolerance (8) implies that Ucn2 is endogenously secreted under hypoglycemic and hyperglycemic states. Glucose serves as the primary fuel molecule in the fight or flight response and is crucial for the organism’s survival (4,63). Therefore, the inhibitory effect of SM CRFR2β on insulin signaling may function to regulate the stress-induced elevation in blood glucose levels and to allow availability of glucose to other tissues. Whereas this function is beneficial under normal conditions, it may be maladaptive under chronic stress conditions, under which SM CRFR2β expression is elevated and consequently insulin sensitivity is reduced.
Whole-genome microarray expression data comparing the expression profile of SM obtained from CRFR2 KO or WT littermates showed a robust reduction in RBP4 expression, which was further confirmed by real-time PCR. RBP4 is mainly expressed in liver and adipose tissue (64). RBP4 serum levels are increased in insulin-resistant mice and in humans with type 2 diabetes (65), and weight loss in morbidly obese patients reduces RBP4 serum level (66). Adipose-specific glucose transporter 4 KO mice demonstrated elevated serum RBP4 levels and secondary insulin resistance in the muscle and the liver (65), a metabolic phenotype that mirrors the observed phenotype of the CRFR2 and Ucn2 KO mice.
RBP4 and CRFR2 signaling disrupt components of SM insulin signaling that play a role in the control of glucose transporter 4 translocation. RBP4 reduces both phosphoinositide3-kinase activity and insulin-stimulated tyrosine phosphorylation of insulin receptor substrate-1 at tyrosine residue 612, a docking site for the p85 subunit of phosphoinositide3-kinase (65), whereas Ucn2 signaling inhibits insulin-induced Akt phosphorylation and reduces ERK1/2 phosphorylation (8). It was demonstrated that RBP expressed ectopically in mice muscle can elevate serum RBP levels (65). Here, we demonstrate that HFD and CVS conditions mediate an increase both in SM CRFR2β and RBP4 expression levels. This dual increased expression may synergistically act in an autocrine fashion to inhibit insulin signaling and magnify metabolic complications.
The current findings further position the SM-CRFR2 pathways as a relevant physiological system that may affect the known reciprocal relationship between psychological and physiological challenges and the metabolic syndrome. A better understanding of SM CRFR2β pathway, its physiological roles, and its regulation may provide benefits in related pathological conditions, such as obesity and type 2 diabetes.
Materials and Methods
Animals
Mice were housed and handled in a pathogen-free temperature-controlled (22 C ±1) mouse facility on a 12-h light, 12-h dark cycle (lights on from 1900 h–0700 h), with food and water given ad libitum, according to institutional guidelines. Adult C57BL/6 male mice were used in all experiments. CRFR2-null (129×C57Bl/6 mixed background) mice were used for microarray and real-time PCR studies. All experimental protocols were approved by the Institutional Animal Care and Use Committee of The Weizmann Institute of Science.
Cell lines
C2C12 myoblasts were grown to 50% confluence in DMEM (Invitrogen Life Technologies, Carlsbad, CA), containing 10% FBS supplemented with 100 μg/ml of penicillin/streptomycin (Invitrogen Life Technologies) (normal growth medium). For differentiation, C2C12 were grown to 90% confluency and washed with serum-free medium, and their medium was replaced with DMEM containing 2% horse serum. CHO cells were grown in normal growth medium as previously described (22).
RNA and cDNA preparation
RNA was extracted from brain, pituitary, gastrocnemius muscle, or C2C12 cells using Tri-Reagent RNA isolation reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s recommendations. To avoid false-positive results caused by DNA contamination, a deoxyribonuclease treatment was performed for 30 min at 37 C using the RQ1 RNase-free deoxyribonuclease (Promega Corp., Madison, WI). RNA preparations were reverse transcribed to generate cDNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Inc., Foster city, CA). The cDNA products were used as templates for semiquantitative and quantitative PCR analysis.
Semiquantitative RT-PCR
Semiquantitative RT-PCR was used to amplify the levels of endogenous mCRFR2 and mCRFR1 present in the mouse SM and brain. The expression of mCRFR1, mCRFR2 as well as muscle differentiation markers MyoD, myogenin and Id2 levels were studied during C2C12 differentiation. The cDNA products were used as templates for semiquantitative RT-PCR analysis using specific primers for mCRFR2, mCRFR1, MyoD, myogenin, and Id2 and the ribosomal protein S16 (for sequences see Table 1).
Table 1.
Sequence of PCR primers
| Gene | Primer sequence (5′ to 3′) | GenBank accession no. |
|---|---|---|
| CRFR1 | NM_007762 | |
| Sense | GGT GTG CCT TTC CCC ATC ATT | |
| Antisense | CAA CAT GTA GGT GAT GCC CAG | |
| CRFR2 | NM_009953 | |
| Sense | GGC AAG GAA GT GGT GAT TTG | |
| Antisense | GGC GTG GTG GTC CTG CCA GCG | |
| MyoD1 | NM_010866 | |
| Sense | GAG CAA AGT GAA TGA GGC CTT | |
| Antisense | CAC TGT AGT AGG CGG TGT CGT | |
| Myogenin | NM_031189 | |
| Sense | TCA GAA GAG GAT GCT CTC TGC | |
| Antisense | TCA GAA GAG GAT GCT CTC TGC | |
| Id2 | NM_010496 | |
| Sense | ATG AAA GCC TTC AGT CCG GTG | |
| Antisense | TTA GCC ACA GAG TAC TTT GCT | |
| S16 | M11408 | |
| Sense | TGC GGT GTG GAG CTC GTG CTT GT | |
| Antisense | GCT ACC AGG CCT TTG AGA TGG A |
PCR without reverse transcriptase enzyme (−R.T) or without cDNA (−cDNA) served as negative control. The expression of ribosomal protein S16 served as internal control. The PCR conditions were as follows: cDNA equivalent to 200 ng of total RNA was amplified by PCR for 35 cycles at an annealing temperature of 62 C. The final MgCl2 concentration was 3 mm, and each reaction contained 2.5 U of Taq DNA polymerase (BIO-X-ACT DNA polymerase; Bioline UK Ltd., London, UK).
RNase protection assay
SM total RNA was hybridized with the mCRFR1, mCRFR2, and glyceraldehyde-3-phosphate dehydrogenase antisense probes. Brain and pituitary gland served as positive control for CRFR2 and CRFR1 gene expression, respectively. RNase protection assay was performed as previously described (23).
Transient transfections and luciferase assay
C2C12 were used for the CRE activation and for the promoter studies. All transfections were carried out in 12-well plates using Lipofectamine 2000 Transfection Reagent (Invitrogen Life Technologies) according to manufacturer’s instructions. For CRE-luciferase activation, C2C12 myoblasts were plated to 90% confluency and transfected with 1.5 μg of the luciferase reporter containing a fragment of the EVX1 gene, which contains a potent CRE site (kindly provided by Marc Montminy, The Salk Institute) and 50 ng β-gal expression plasmid. Cells were treated for 4 h with vehicle or 1 pm, 0.1 nm, 10 nm, and 1 μm CRF or Ucn3 in nondifferentiated C2C12 cells, or 48 h differentiated C2C12 cells with or without the presence of CRFR1- and CRFR2-selective antagonists (Ant and Ast 2B, respectively). For promoter studies, C2C12 myoblasts were plated to 90% confluency and transfected with 1.5 μg of the luciferase reporter plasmid or empty pGL3 vector and 50 ng β-gal expression plasmid. After 24 h the medium was replaced with DM. The promoter activity was monitored at the basal state and after the indicated times (24–96 h) in DM. The cells were harvested, and the luciferase reporter activity was assayed as previously described (20). Transfections were performed at least three times (in triplicate) for each construct or treatment tested. To correct for variations in transfection efficiencies, luciferase activities were normalized to β-gal activity. Results were corrected by the activity of the promoterless pGL3 vector.
Calcium-mobilization assay
Calcium-mobilization kinetics in nondifferentiated or differentiated C2C12 cells or in CHO cells stably transfected with either mCRFR1 or mCRFR2 after treatment of CRF or Ucn2 (50 nm) were measured using FlexStation (Molecular Devices, Sunnyvale, CA) as previously described (67).
Construction of luciferase reporter plasmids
The mCRFR1 and mCRFR2β 5′-flanking region constructs were cloned by PCR using mouse genomic DNA. The primers used for the construct were designed to include artificial restriction sites (KpnI and XhoI for mCRFR1; KpnI and NheI for mCRFR2β). The primer sequences were as follows: for mCRFR1 sense primer (−2685 to 2663): 5′-TTG GGT TAC GTA TGC TGC TCC TT-3′ and antisense primer (+196 to +217): 5′-CCT CGG GCT CGC TCT GTC AGC-3′. For mCRFR2β sense primer (−2495 to 2473): 5′-GGA AAT GCA GGA AAG CCA AGA CA-3′ and antisense primer (+4 to +23): 5′-CTG CCC GAC CTA CCC ACC AA-3′. Fragmentation of the mCRFR2β 5′-flanking region was done using the above mentioned antisense primer along with six sense primers located at: (−1649 to −1630), (−1028 to −1009), (−955 to −932), (−747 to −726), (−512 to −492), (−146 to −127). The primers sequences are indicated in Fig. 4; all primers contained an artificial KpnI restriction site. For mutating the MEF2 recognition site located at −91 to −82, the proximal 5′-flanking area was amplified using the above-mentioned antisense primer with the following sense primer, which contains an endogenous PstI restriction site: 5′-CTGCAGAAGTTGCTGCCCAGAGCCAGATATCAATAACCTGG-3′. The mutation introduces a unique EcoRV restriction site (in italics), which was later used for identifying mutated clones. The PCR products were analyzed by agarose gel electrophoresis and eluted from the gel. After digestion by the appropriate restriction enzymes, the DNA fragments were cloned into the luciferase reporter plasmid pGL3 (Promega Corp.), and the sequences were verified using automated direct DNA sequencing.
HFD
Mice were fed ad libitum a high-fat (60% of calories) (n = 13) or low-fat (10% of calories) (n = 5) diet (D12492 and D12450B, respectively; Research Diets, Inc., New Brunswick, NJ) for 15 wk.
CVS
CVS mice (n = 5) were housed in a temperature-controlled room (22 C ±1) and were subjected to the CVS protocol for a period of 4 wk as previously described (22).
Real-Time PCR
SM cDNA products were used as templates for real-time PCR analysis. Sense and antisense primers were selected to be located on different exons to avoid false-positive results caused by DNA contamination. The following specific primers were designed using Primer Express software (Applied Biosystems, PerkinElmer, Foster City, CA). For mCRFR2: 5′-TACCGAATCGCCCTCATTGT-3′ and 5′-CCACGCGATGTTTCTCAGAAT-3′ corresponding to nucleotides 479-498 and 640-620, respectively (GenBank accession no. AY445512); for mRBP4: 5′-GCTTCCGAGTCAAGGAGAACTTC-3′ and 5′-TCCACAGAAAACTCAGCGATGA-3′ corresponding to nucleotides 479-498 and 640-620, respectively (GenBank accession no. NM_011255). For mouse hypoxanthine guanine phosphoribosyl transferase 1 (HPRT1), which served as an internal control: 5′-GCAGTACAGCCCCAAAATGG-3′ and 5′-GGTCCTTTTCACCAGCAAGCT-3′ corresponding to nucleotides 389-411 and 509-488, respectively (GenBank accession no. NM_013556). Real-time PCRs were carried out on a 7500 Real-Time PCR system (Applied Biosystems, Inc.), using fluorescent SYBR Green technology (Abgene; Epsom, Surrey, UK). Reaction protocols had the following format: 15 min at 95 C for enzyme activation, followed by 45 cycles of 15 sec at 94 C and 60 sec at 60 C. The specificity of the amplification products was checked by melting curve analysis. All reactions contained the same amount of cDNA, 10 μl Master Mix, and 250 nm primers to a final volume of 20 μl.
Microarray preparation and data are described under Gene Expression Omnibus accession number GSE25045. Briefly, total RNA was extracted from adult skeletal muscle obtained from four CRFR2 KO mice and four WT littermates, using Tri-Reagent RNA isolation reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s protocol. The RNA was pooled such that each sample consisted of two muscles from each genotype (a total of four samples). Total RNA (100 μg) was further cleaned using Qiagen RNA purification kit (QIAGEN Inc., Valencia, CA), and RNA integrity was verified using gel electrophoresis and 260/280 ratios. cRNA synthesis and hybridization to Affymetrix Murine Genome-U74Av2 array (Affymetrix, Santa Clara, CA) was performed by the UCSD Biological Services Unit. Data was analyzed using Affymetrix Microarray Analysis Suite 5.1.
Supplementary Material
Acknowledgments
We thank Mr. S. Ovadia for his devoted assistance with animal care.
This research was supported in part by the Clayton Medical Research Foundation, Inc. A.C. was supported by the following: Roberto and Renata Ruhman, Brazil; Mark Besen and the Pratt Foundation, Australia; the Israel Science Foundation; the Legacy Heritage Biomedical Science Partnership D-Cure Fellowship; Nella and Leon Benoziyo Center for Neurosciences; Nella and Leon Benoziyo Center for Neurological Diseases; Carl and Micaela Einhorn-Dominic Brain Research Institute; Irwin Green Alzheimer’s Research Fund; Gerhard and Hannah Bacharach (Fort Lee, NJ) and is incumbent of the Philip Harris and Gerald Ronson Career Development Chair. W.V. was supported by Award No. 5P01DK026741-30 from the National Institute of Diabetes and Digestive and Kidney Diseases and is a Clayton Medical Research Foundation, Inc. Senior Investigator and the Helen McLoraine Professor of Molecular Neurobiology.
Footnotes
Disclosure Summary: W.V. is a cofounder, consultant, equity holder, and member of the Board of Directors of Neurocrine Biosciences, Inc., and Acceleron Pharma, Inc. These companies are developing products that are related to some of the topics discussed. However, none if these products are as yet on the market. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health. Y.K., O.I., J.V., L.B., and A.C. have nothing to disclose.
Abbreviations: Ant, Antalarmin; Ast 2b, Astressin 2B; CHO, Chinese hamster ovary; CRE, cAMP-responsive element; CRF, corticotropin-releasing factor; CRFR2, corticotropin-releasing factor receptor type 2; CVS, chronic variable stress; DM, differentiation medium; HFD, high-fat diet; HPRT1, hypoxanthine guanine phosphoribosyl transferase 1; iv, insertion variant; KO, knockout; MEF, myocyte enhancer factor; RBP4, retinol-binding protein 4; RNase, ribonuclease; SM, skeletal muscle; Ucn, urocortin; WT, wild type.
First Published Online November 17, 2010
References
- Petersen KF, Shulman GI 2002 Pathogenesis of skeletal muscle insulin resistance in type 2 diabetes mellitus. Am J Cardiol 90:11G–18G [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Savage DB, Bilz S, Solomon G, Yonemitsu S, Cline GW, Befroy D, Zemany L, Kahn BB, Papademetris X, Rothman DL, Shulman GI 2007 The role of skeletal muscle insulin resistance in the pathogenesis of the metabolic syndrome. Proc Natl Acad Sci USA 104:12587–12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GI 2004 Unraveling the cellular mechanism of insulin resistance in humans: new insights from magnetic resonance spectroscopy. Physiology (Bethesda) 19:183–190 [DOI] [PubMed] [Google Scholar]
- Brown MR, Fisher LA, Spiess J, Rivier C, Rivier J, Vale W 1982 Corticotropin-releasing factor: actions on the sympathetic nervous system and metabolism. Endocrinology 111:928–931 [DOI] [PubMed] [Google Scholar]
- Dallman MF, Akana SF, Strack AM, Hanson ES, Sebastian RJ 1995 The neural network that regulates energy balance is responsive to glucocorticoids and insulin and also regulates HPA axis responsivity at a site proximal to CRF neurons. Ann NY Acad Sci 771:730–742 [DOI] [PubMed] [Google Scholar]
- Bale TL, Anderson KR, Roberts AJ, Lee KF, Nagy TR, Vale WW 2003 Corticotropin-releasing factor receptor-2-deficient mice display abnormal homeostatic responses to challenges of increased dietary fat and cold. Endocrinology 144:2580–2587 [DOI] [PubMed] [Google Scholar]
- Carlin KM, Vale WW, Bale TL 2006 Vital functions of corticotropin-releasing factor (CRF) pathways in maintenance and regulation of energy homeostasis. Proc Natl Acad Sci USA 103:3462–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Brar B, Choi CS, Rousso D, Vaughan J, Kuperman Y, Kim SN, Donaldson C, Smith SM, Jamieson P, Li C, Nagy TR, Shulman GI, Lee KF, Vale W 2006 Urocortin 2 modulates glucose utilization and insulin sensitivity in skeletal muscle. Proc Natl Acad Sci USA 103:16580–16585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperman Y, Chen A 2008 Urocortins: emerging metabolic and energy homeostasis perspectives. Trends Endocrinol Metab 19:122–129 [DOI] [PubMed] [Google Scholar]
- Huising MO, van der Meulen T, Vaughan JM, Matsumoto M, Donaldson CJ, Park H, Billestrup N, Vale WW 2010 CRFR1 is expressed on pancreatic β cells, promotes β cell proliferation, and potentiates insulin secretion in a glucose-dependent manner. Proc Natl Acad Sci USA 107:912–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperman Y, Issler O, Regev L, Musseri I, Navon I, Neufeld-Cohen A, Gil S, Chen A 2010 Perifornical urocortin-3 mediates the link between stress-induced anxiety and energy homeostasis. Proc Natl Acad Sci USA 107:8393–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita N, Laurent P, Lefort S, Chalon P, Lelias JM, Kaghad M, Le Fur G, Caput D, Ferrara P 1993 Primary structure and functional expression of mouse pituitary and human brain corticotrophin releasing factor receptors. FEBS Lett 335:1–5 [DOI] [PubMed] [Google Scholar]
- Chen R, Lewis KA, Perrin MH, Vale WW 1993 Expression cloning of a human corticotropin-releasing-factor receptor. Proc Natl Acad Sci USA 90:8967–8971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Pearse II RV, O'Connell S, Rosenfeld MG 1993 Identification of a seven transmembrane helix receptor for corticotropin-releasing factor and sauvagine in mammalian brain. Neuron 11:1187–1195 [DOI] [PubMed] [Google Scholar]
- Stenzel P, Kesterson R, Yeung W, Cone RD, Rittenberg MB, Stenzel-Poore MP 1995 Identification of a novel murine receptor for corticotropin-releasing hormone expressed in the heart. Mol Endocrinol 9:637–645 [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Pearse II RV, Lin CR, Rosenfeld MG 1995 A sauvagine/corticotropin-releasing factor receptor expressed in heart and skeletal muscle. Proc Natl Acad Sci USA 92:1108–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin M, Donaldson C, Chen R, Blount A, Berggren T, Bilezikjian L, Sawchenko P, Vale W 1995 Identification of a second corticotropin-releasing factor receptor gene and characterization of a cDNA expressed in heart. Proc Natl Acad Sci USA 92:2969–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovenberg TW, Liaw CW, Grigoriadis DE, Clevenger W, Chalmers DT, De Souza EB, Oltersdorf T 1995 Cloning and characterization of a functionally distinct corticotropin-releasing factor receptor subtype from rat brain. Proc Natl Acad Sci USA 92:836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pett K, Viau V, Bittencourt JC, Chan RK, Li HY, Arias C, Prins GS, Perrin M, Vale W, Sawchenko PE 2000 Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. J Comp Neurol 428:191–212 [DOI] [PubMed] [Google Scholar]
- Chen A, Perrin M, Brar B, Li C, Jamieson P, Digruccio M, Lewis K, Vale W 2005 Mouse corticotropin-releasing factor receptor type 2alpha gene: isolation, distribution, pharmacological characterization and regulation by stress and glucocorticoids. Mol Endocrinol 19:441–458 [DOI] [PubMed] [Google Scholar]
- Lovenberg TW, Chalmers DT, Liu C, De Souza EB 1995 CRF2 α and CRF2 β receptor mRNAs are differentially distributed between the rat central nervous system and peripheral tissues. Endocrinology 136:4139–4142 [DOI] [PubMed] [Google Scholar]
- Sztainberg Y, Kuperman Y, Issler O, Gil S, Vaughan J, Rivier J, Vale W, Chen A 2009 A novel corticotropin-releasing factor receptor splice variant exhibits dominant negative activity: a putative link to stress-induced heart disease. FASEB J 23:2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Blount A, Vaughan J, Brar B, Vale W 2004 Urocortin II gene is highly expressed in mouse skin and skeletal muscle tissues: localization, basal expression in corticotropin-releasing factor receptor (CRFR) 1- and CRFR2-null mice, and regulation by glucocorticoids. Endocrinology 145:2445–2457 [DOI] [PubMed] [Google Scholar]
- Hall JE, Kaczor JJ, Hettinga BP, Isfort RJ, Tarnopolsky MA 2007 Effects of a CRF2R agonist and exercise on mdx and wildtype skeletal muscle. Muscle Nerve 36:336–341 [DOI] [PubMed] [Google Scholar]
- Hinkle RT, Donnelly E, Cody DB, Bauer MB, Isfort RJ 2003 Urocortin II treatment reduces skeletal muscle mass and function loss during atrophy and increases nonatrophying skeletal muscle mass and function. Endocrinology 144:4939–4946 [DOI] [PubMed] [Google Scholar]
- Brar BK, Jonassen AK, Egorina EM, Chen A, Negro A, Perrin MH, Mjos OD, Latchman DS, Lee KF, Vale W 2004 Urocortin-II and urocortin-III are cardioprotective against ischemia reperfusion injury: an essential endogenous cardioprotective role for corticotropin releasing factor receptor type 2 in the murine heart. Endocrinology 145:24–35; discussion 21–23 [DOI] [PubMed] [Google Scholar]
- Makino S, Asaba K, Takao T, Hashimoto K 1998 Type 2 corticotropin-releasing hormone receptor mRNA expression in the heart in hypertensive rats. Life Sci 62:515–523 [DOI] [PubMed] [Google Scholar]
- Makino S, Nishiyama M, Asaba K, Gold PW, Hashimoto K 1998 Altered expression of type 2 CRH receptor mRNA in the VMH by glucocorticoids and starvation. Am J Physiol 275:R1138–R1145 [DOI] [PubMed] [Google Scholar]
- Makino S, Asaba K, Nishiyama M, Hashimoto K 1999 Decreased type 2 corticotropin-releasing hormone receptor mRNA expression in the ventromedial hypothalamus during repeated immobilization stress. Neuroendocrinology 70:160–167 [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Makino S, Asaba K, Hashimoto K 1999 Leptin effects on the expression of type-2 CRH receptor mRNA in the ventromedial hypothalamus in the rat. J Neuroendocrinol 11:307–314 [DOI] [PubMed] [Google Scholar]
- Eghbal-Ahmadi M, Avishai-Eliner S, Hatalski CG, Baram TZ 1999 Differential regulation of the expression of corticotropin-releasing factor receptor type 2 (CRF2) in hypothalamus and amygdala of the immature rat by sensory input and food intake. J Neurosci 19:3982–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama K, Gaudriault GE, Bradbury MJ, Vale WW 2000 Regulation of corticotropin-releasing factor receptor type 2 β messenger ribonucleic acid in the rat cardiovascular system by urocortin, glucocorticoids, and cytokines. Endocrinology 141:2285–2293 [DOI] [PubMed] [Google Scholar]
- Kageyama K, Gaudriault GE, Suda T, Vale WW 2003 Regulation of corticotropin-releasing factor receptor type 2β mRNA via cyclic AMP pathway in A7r5 aortic smooth muscle cells. Cell Signal 15:17–25 [DOI] [PubMed] [Google Scholar]
- Coste SC, Heldwein KA, Stevens SL, Tobar-Dupres E, Stenzel-Poore MP 2001 IL-1α and TNFα down-regulate CRH receptor-2 mRNA expression in the mouse heart. Endocrinology 142:3537–3545 [DOI] [PubMed] [Google Scholar]
- Pournajafi-Nazarloo H, Partoo L, Sanzenbacher L, Azizi F, Carter CS 2007 Modulation of corticotropin-releasing hormone type 2 receptor and urocortin 1 and urocortin 2 mRNA expression in the cardiovascular system of prairie voles following acute or chronic stress. Neuroendocrinology 86:17–25 [DOI] [PubMed] [Google Scholar]
- Biressi S, Molinaro M, Cossu G 2007 Cellular heterogeneity during vertebrate skeletal muscle development. Dev Biol 308:281–293 [DOI] [PubMed] [Google Scholar]
- Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY 2002 A myogenic differentiation checkpoint activated by genotoxic stress. Nat Genet 32:585–593 [DOI] [PubMed] [Google Scholar]
- Kislinger T, Gramolini AO, Pan Y, Rahman K, MacLennan DH, Emili A 2005 Proteome dynamics during C2C12 myoblast differentiation. Mol Cell Proteomics 4:887–901 [DOI] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V 2000 Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol 185:155–173 [DOI] [PubMed] [Google Scholar]
- Reyes TM, Lewis K, Perrin MH, Kunitake KS, Vaughan J, Arias CA, Hogenesch JB, Gulyas J, Rivier J, Vale WW, Sawchenko PE 2001 Urocortin II: a member of the corticotropin-releasing factor (CRF) neuropeptide family that is selectively bound by type 2 CRF receptors. Proc Natl Acad Sci USA 98:2843–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SY, Hsueh AJ 2001 Human stresscopin and stresscopin-related peptide are selective ligands for the type 2 corticotropin-releasing hormone receptor. Nat Med 7:605–611 [DOI] [PubMed] [Google Scholar]
- Lewis K, Li C, Perrin MH, Blount A, Kunitake K, Donaldson C, Vaughan J, Reyes TM, Gulyas J, Fischer W, Bilezikjian L, Rivier J, Sawchenko PE, Vale WW 2001 Identification of urocortin III, an additional member of the corticotropin-releasing factor (CRF) family with high affinity for the CRF2 receptor. Proc Natl Acad Sci USA 98:7570–7575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V, Nadal-Ginard B 1992 Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev 6:1783–1798 [DOI] [PubMed] [Google Scholar]
- Graham TE, Yang Q, Blüher M, Hammarstedt A, Ciaraldi TP, Henry RR, Wason CJ, Oberbach A, Jansson PA, Smith U, Kahn BB 2006 Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med 354:2552–2563 [DOI] [PubMed] [Google Scholar]
- Samuelsson S, Lange JS, Hinkle RT, Tarnopolsky M, Isfort RJ 2004 Corticotropin-releasing factor 2 receptor localization in skeletal muscle. J Histochem Cytochem 52:967–977 [DOI] [PubMed] [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F 2003 The formation of skeletal muscle: from somite to limb. J Anat 202:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatopoulos D, Dai Y, Chen J, Karteris E, Papadopoulou N, Easton AJ, Hillhouse EW 1998 Human corticotropin-releasing hormone receptor: differences in subtype expression between pregnant and nonpregnant myometria. J Clin Endocrinol Metab 83:2539–2544 [DOI] [PubMed] [Google Scholar]
- Heldwein KA, Duncan JE, Stenzel P, Rittenberg MB, Stenzel-Poore MP 1997 Endotoxin regulates corticotropin-releasing hormone receptor 2 in heart and skeletal muscle. Mol Cell Endocrinol 131:167–172 [DOI] [PubMed] [Google Scholar]
- Vaughan J, Donaldson C, Bittencourt J, Perrin MH, Lewis K, Sutton S, Chan R, Turnbull AV, Lovejoy D, Rivier C, et al. 1995 Urocortin, a mammalian neuropeptide related to fish urotensin I and to corticotropin-releasing factor. Nature 378:287–292 [DOI] [PubMed] [Google Scholar]
- Bale TL, Vale WW 2004 CRF and CRF receptors: role in stress responsivity and other behaviors. Annu Rev Pharmacol Toxicol 44:525–557 [DOI] [PubMed] [Google Scholar]
- Perrin MH, Vale WW 1999 Corticotropin releasing factor receptors and their ligand family. Ann NY Acad Sci 885:312–328 [DOI] [PubMed] [Google Scholar]
- Guérineau N, Corcuff JB, Tabarin A, Mollard P 1991 Spontaneous and corticotropin-releasing factor-induced cytosolic calcium transients in corticotrophs. Endocrinology 129:409–420 [DOI] [PubMed] [Google Scholar]
- Kuryshev YA, Childs GV, Ritchie AK 1996 Corticotropin-releasing hormone stimulates Ca2+ entry through L- and P-type Ca2+ channels in rat corticotropes. Endocrinology 137:2269–2277 [DOI] [PubMed] [Google Scholar]
- Hauger RL, Olivares-Reyes JA, Braun S, Catt KJ, Dautzenberg FM 2003 Mediation of corticotropin releasing factor type 1 receptor phosphorylation and desensitization by protein kinase C: a possible role in stress adaptation. J Pharmacol Exp Ther 306:794–803 [DOI] [PubMed] [Google Scholar]
- Fazal N, Slominski A, Choudhry MA, Wei ET, Sayeed MM 1998 Effect of CRF and related peptides on calcium signaling in human and rodent melanoma cells. FEBS Lett 435:187–190 [DOI] [PubMed] [Google Scholar]
- Kyrou I, Tsigos C 2007 Stress mechanisms and metabolic complications. Horm Metab Res 39:430–438 [DOI] [PubMed] [Google Scholar]
- James WP, Rigby N, Leach R 2006 Obesity and the metabolic syndrome: the stress on society. Ann NY Acad Sci 1083:1–10 [DOI] [PubMed] [Google Scholar]
- Münzberg H, Myers Jr MG 2005 Molecular and anatomical determinants of central leptin resistance. Nat Neurosci 8:566–570 [DOI] [PubMed] [Google Scholar]
- Liu YL, Emilsson V, Cawthorne MA 1997 Leptin inhibits glycogen synthesis in the isolated soleus muscle of obese (ob/ob) mice. FEBS Lett 411:351–355 [DOI] [PubMed] [Google Scholar]
- Lu XY, Kim CS, Frazer A, Zhang W 2006 Leptin: a potential novel antidepressant. Proc Natl Acad Sci USA 103:1593–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallman MF, Pecoraro N, Akana SF, La Fleur SE, Gomez F, Houshyar H, Bell ME, Bhatnagar S, Laugero KD, Manalo S 2003 Chronic stress and obesity: a new view of “comfort food.” Proc Natl Acad Sci USA 100:11696–11701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallman MF, Akana SF, Strack AM, Scribner KS, Pecoraro N, La Fleur SE, Houshyar H, Gomez F 2004 Chronic stress-induced effects of corticosterone on brain: direct and indirect. Ann NY Acad Sci 1018:141–150 [DOI] [PubMed] [Google Scholar]
- Ulrich-Lai YM, Herman JP 2009 Neural regulation of endocrine and autonomic stress responses. Nat Rev Neurosci 10:397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaner WS 1989 Retinol-binding protein: the serum transport protein for vitamin A. Endocr Rev 10:308–316 [DOI] [PubMed] [Google Scholar]
- Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB 2005 Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 436:356–362 [DOI] [PubMed] [Google Scholar]
- Haider DG, Schindler K, Prager G, Bohdjalian A, Luger A, Wolzt M, Ludvik B 2007 Serum retinol-binding protein 4 is reduced after weight loss in morbidly obese subjects. J Clin Endocrinol Metab 92:1168–1171 [DOI] [PubMed] [Google Scholar]
- McDermott DH, Fong AM, Yang Q, Sechler JM, Cupples LA, Merrell MN, Wilson PW, D'Agostino RB, O'Donnell CJ, Patel DD, Murphy PM 2003 Chemokine receptor mutant CX3CR1-M280 has impaired adhesive function and correlates with protection from cardiovascular disease in humans. J Clin Invest 111:1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.