Abstract
Highly active antiretroviral therapy (HAART) treatment for HIV has changed the course of AIDS in societies where the drugs have been readily available. Despite the great success of HAART, drug resistance and toxicity issues still remain a concern for some individuals 1. Thus a number of investigators have been exploring other approaches for inhibiting HIV-1 replication. One of the most potent of these is the use of RNA interference. This review will focus solely on the use of RNA interference for the treatment of HIV-1 infection, including the problems, progress and future prospects.
RNAi as an anti-viral therapeutic
Since the first description of RNA interference (RNAi) in 1998 2, it has rapidly become one of the methods of choice for gene function analyses 3, and is being exploited for therapeutic applications 4–7. Synthetic small interfering RNAs (siRNAs) were first demonstrated to achieve sequence-specific gene knockdown in a mammalian cell line by Tuschl and co-workers 8. Soon thereafter Song et al. successfully showed the first in vivo evidence of siRNA-mediated gene silencing in an animal by demonstrating inhibition of a cell death receptor to recover liver function in a mouse model of hepatitis 9. Because the essential feature of the RNAi mechanism is the sequence-specificity, deriving from complementary Watson-Crick base pairing of a target messenger RNA (mRNA) and the guide strand of the siRNA, RNAi is considered to have some unique therapeutic attributes for the treatment of HIV-1 Infection 10–11. Although there are indications that viruses have evolved ways to escape from the RNAi mechanism12, RNAi-based therapeutics can be multiplexed to prevent escape by combining different siRNAs targeting different mRNAs (viral and host encoded) or by combining siRNAs with other small RNAs (aptamers and ribozymes) or antiviral proteins (Dominant-negative inhibitors such as RevM10). The combinatorial use of antiviral macromolecular drugs could effectively block viral replication and prevent the emergence of resistance variants.
Potential molecular targets for anti-HIV RNAi therapeutics
HIV-1 chronically infects individuals, thus requiring long-term RNAi treatment. In order to achieve potent and durable suppression of viral replication, the viral RNA genome or host cellular transcripts that encode proteins essential for viral replication can be used as anti-HIV targets. Development of viral resistance is a common setback with HIV therapies due to the generation of viral escape mutants. It has been demonstrated that prolonged infection of cells harboring genes encoding anti-HIV siRNAs can result in the evolution of escape variants that are resistant to the expressed siRNA 13–14. A single nucleotide mutation in a critical position within the target sequence relative to the siRNA site of interaction as well as mutations which result in other structural changes or elimination of the target region can diminish and/or eliminate RNAi mediated inhibition 14. Thus, a single siRNA therapy is not sufficient to maintain long-term inhibition of virus replication. One of the strategies for achieving the desired RNAi long term efficacy is mitigating viral escape from RNAi 15–16 through multiple RNAi effectors akin to the HAART approach. It is also important to target sequences that are conserved among different virus strains to reduce the chance of mutant escape and to multiplex siRNAs to several different viral sequences to reduce the probability of viral escape. Host factors that are essential for viral replication can also be targeted, which may reduce the chance of viral escape.
To date, numerous siRNAs targeting a number of HIV-1 or cellular transcripts have been demonstrated to achieve viral inhibition both in vitro and in vivo. Indeed, all the HIV-1 encoded genes (tat, rev, gag, pol, nef, vif, env, vpr and the long terminal repeat (LTR)) are susceptible to RNAi-induced gene silencing in cell lines 17. For example, the Tat and Rev proteins are essential for subsequent expression of HIV-1 structural genes (gag, pol and env) and for the synthesis of full length viral genomic RNA 18. SiRNAs designed to destroy the tat/rev transcripts were found to be highly effective in viral suppression 19.
Several well-known host factors, including NF kappa Beta (NF-κB), the CD4 receptor CD4 and the co-receptors CCR5 (C-C motif receptor 5) and CXCR4 (C-X-C motif receptor 4) have been successfully targeted by siRNAs, thereby suppressing viral replication or entry 11. Recently, three siRNA-screening studies have been conducted to identify hundreds of host factors that are critical for HIV replication 20–22. Brass et al., using a large scale siRNA screen identified 273 genes whose depletion inhibited either p24 production or viral gene activities. There were some surprises among these targets, such as Rab6 (a regulator of retrograde protein transport to the Golgi), Transportin 3-SR2 (TNPO3, a nuclear import factor for serine/arginine-rich (SR) substrates) and Med28 (the Mediator of transcription activation complex component). Specifically, depletion of Rab6 or TNPO3 potently blocked early stages of virus infection. The compilation of hits in the three screening analyses differed due to the conditions (cell types and types of infectious challenges) and readouts of the screens. Nevertheless, these screening approaches have opened up a new landscape of viral-host interactions that are potential targets for RNAi 23. Similar to the conventional HAART strategy, using multiple siRNAs targeting separate, conserved sites in HIV along with targeting HIV host dependency factors (HDFs) has been shown to minimize and even prevent escape while achieving prolonged inhibition 24. In this regard, host factors that are essential for viral entry and replication represent attractive molecular targets. Host genes such as the chemokine receptor CCR5 can be targeted alone or in combination with viral sequences in a tailored fashion to reduce the possibility of viral escape mutants and to avoid toxicity risks associated with long-term HAART treatment 11,16.
Systemic delivery of siRNAs for HIV treatment
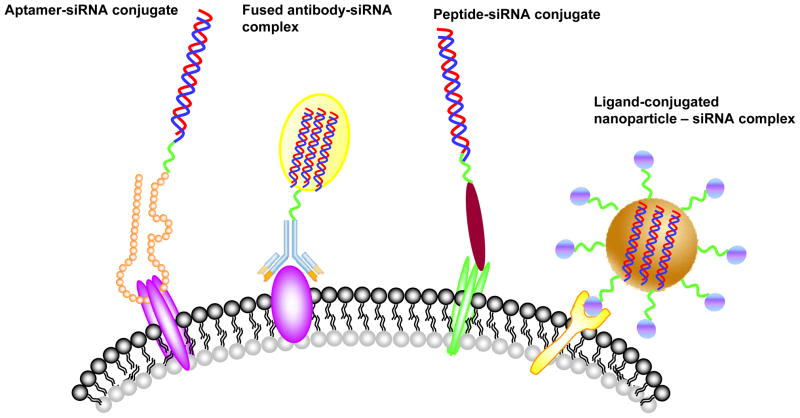
Although siRNAs hold great therapeutic promise for HIV/AIDS, their clinical application has been and still is limited by difficulties of delivering siRNAs in the bloodstream to the right target cells 25–26. In practice, all HIV-susceptible cells such as, human T cells and monocytes are difficult targets for siRNA delivery. Therefore, efficient, systemic delivery of siRNAs in vivo, especially targeted siRNA delivery to HIV-1 susceptible or infected cells, remains a principal challenge for successful anti-HIV therapeutic application. Several recent studies using targeted delivery approaches provide encouragement that this obstacle may soon be overcome (Figure 1).
Figure 1.
Strategies for targeted, systemic delivery of siRNAs for treatment of HIV-1 infection. The various targeting ligands are depicted including aptamers, antibodies, peptides or small molecular ligands. The ligands bind to cell surface antigens or receptors which internalize, delivering the siRNAs to endosomes and ultimately the cytosol for entry into the RNAi pathway.
Single chain antibody-siRNA chimeras targeting either the T-cell CD7 (Cluster of Differentiation 7) receptor 27 or gp120 expressing cells have been shown to functionally deliver anti-HIV siRNAs in vivo28. CD7, a human transmembrane protein on thymocytes and the majority of human T cells, has been documented to be rapidly internalized after antibody binding29. With this in mind an anti-CD7 mAb and its recombinant single-chain (ScFvCD7) fragment have been exploited for targeted delivery. Kumar and coworkers 27 successfully conjugated a Cys-modified scFvCD7 with an oligo-9-arginine peptide to obtain a cell type specific siRNA delivery system (ScFvCD7-9R). In this case, a combination of three siRNAs against two conserved HIV-1 genes (vif and tat) and the cellular CCR5 coreceptor was formulated with scFvCD7-9R system. The complex was systemically administered to HIV-1 infected humanized mice. The weekly injections suppressed endogenous virus and restored CD4+ T cell counts in the viremic mice.
HIV-1 gp120 glycoprotein is exposed on the surface of virus particles and the plasma membrane of HIV-1 infected cells, which thus represents a marker to distinguish HIV-1 infected cells from un-infected cells 30. F105, an anti-HIV-1 gp120 monoclonal antibody, has been demonstrated to bind with high affinity to its ligand gp120 expressed on the surface of a wide range of HIV-1 laboratory strains and primary isolates 31. Song et al. 28 employed a heavy chain fragment of F105 to promote receptor-specific internalization to the cells expressing the HIV-1 envelope protein. In this system, the F105 antibody fragment was fused with protamine. The resulting fusion protein can specifically bind to HIV-1 infected primary T- cells. Additionally, the siRNA targeting the HIV-1 gag capsid gene delivered by the fusion protein suppressed HIV-1 replications only in HIV-1 envelope expressing cells.
Selective targeting and delivery of an anti-CCR5 siRNA to lymphocytes in vivo has been demonstrated using nanoparticles harboring the lymphocyte function associated antigen-1 (LFA-1) 32. LFA-1, the predominant integrin present on all leukocytes has been used to selectively deliver siRNAs to multiple immune cell types, including T cells, macrophages, and dendritic cells that play key roles in HIV infection and pathogenesis. In vivo administration of the LFA-1 integrin-targeted immunoliposome - anti-CCR5 siRNA nanoparticles resulted in selective uptake of siRNA and specific gene silencing by T-cells and macrophages, further protecting humanized mice from HIV challenge with an enhanced resistance to infection.
An RNA aptamer that binds to the HIV envelope protein gp120 expressed on the surface of HIV infected cells has also been used for functional siRNA delivery in vivo 33. These studies demonstrated that 2’-F modified RNA aptamers that specifically bind HIV-1 gp120 and are rapidly internalized into HIV-1 infected cells can neutralize HIV-1 replication 34–35. Recently, our group has successfully used an anti-HIV-1 gp120 aptamer for cell type-specific delivery of anti-HIV dsiRNAs in HIV-1 infected, humanized Rag-hu mice 33. The anti-gp120 aptamer has dual functions, serving as an HIV neutralizing agent and as a targeted delivery vehicle for siRNAs. The gp120 aptamer-siRNA chimera was systemically administered once a week into viremic animals for several weeks. The treatments resulted in several logs of suppression of HIV-1 viral loads. As expected, the anti HIV-1 tat/rev dsiRNA delivered by the aptamer was processed by Dicer leading to incorporation into the RNA induced silencing complex and target mRNA cleavage in vivo.
As mentioned above, the rapid emergence of viral escape mutants often abrogates RNAi efficacy. Therefore, continued efforts also have been made to develop an aptamer-mediated delivery of combinatorial RNAi therapeutics 34. A single HIV-1 gp120 aptamer was tightly complexed with three different dsiRNAs targeting HIV-1 tat/rev or the host dependency factors (CD4 and Transportin-3 (TNPO3)). Such a combination of aptamer and cocktailed siRNAs has the added benefits of mitigating the emergence of viral escape mutants.
A different aptamer that binds to the CD4 receptor and internalizes was shown to be an effective mucosal microbicide 36. CD4 is well-known as the primary receptor for the HIV and plays an important role in HIV entry into host T-cells 37–39. CD4 targeting RNA aptamers have blocked HIV-1 and CD4 interactions and proven to be useful anti-HIV agents 40. CD4 aptamer-siRNA chimeric RNAs (CD4-AsiCs) were used to specifically deliver siRNAs to CD4+ cells and inhibit target gene expression in vitro, as well as in cervicovaginal tissue explants 36. When applied intravaginally in a humanized mouse model, the CD4-AsiCs bearing siRNAs targeting HIV gag and vif or host CCR5 significantly prevented viral transmission to cervicovaginal explants and to the mice.
Thus far in humanized mice at least, cell type specific delivery of siRNAs for the treatment of HIV-1 infection seems to be working quite well. The next challenges will be to use these approaches in non-human primates to validate safety and efficacy.
Gene therapy mediated delivery of shRNAs
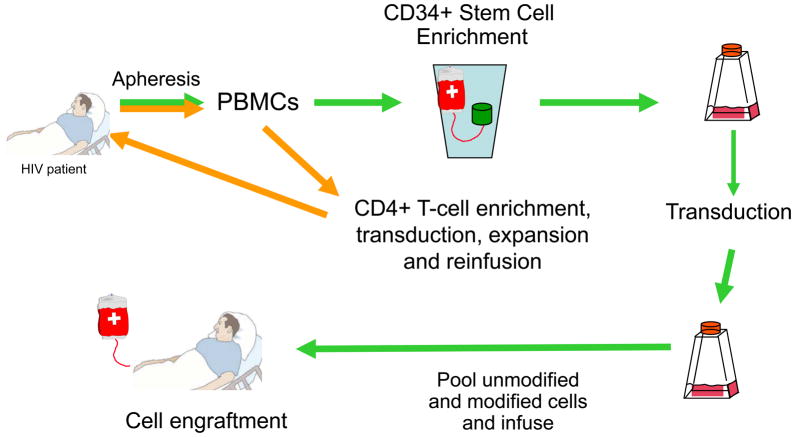
The alternative to systemic siRNA delivery is to use ex vivo gene therapy to deliver integrating or non-integrating viral vectors harboring transcription units that express short hairpin RNAs (shRNAs) targeting HIV-1 or cellular sequences required for HIV-1 infection (Figure 2). The concept of gene therapy for the treatment of HIV-1 infection received a huge boost with the demonstration that HIV-1 infection was eradicated from an AIDS/leukemia patient who received an allogeneic hematopoietic stem cell transplant from a donor who had a homozygous deletion in the chemokine receptor gene CCR5 41. It is known that CCR5 is a primary co-receptor for HIV-1 entry, so in effect the transplant patient was given a protective therapy. Although this is a single patient result, the idea that repopulation of the hematopoietic system with cells resistant to HIV-1 infection can have a major impact on the disease is now a reality. Finding CCR5−\− compatible donors is a major challenge and as such this cannot be considered to be a treatment that will have broad applications. In addition, virus that mutates to CXCR4 tropism can infect the CCR5−\cells and reactivate the infection. Thus, it is of importance to consider additional anti-viral approaches in a combination gene therapy setting 19,42. The use of autologous, gene modified hematopoietic stem cells is an attractive alternative for treatment of HIV-1 infection. For this purpose, HIV-1 derived VSV-G pseudotyped lentiviral vectors have received the most attention.
Figure 2.
Representation of autologous stem and T-cell gene therapy for treatment of HIV-1 infection. CD34+ progenitor cells are mobilized, collected, transduced and infused into patients. This process requires some myeloablation to create space for stem cell engraftment. The T-cell approach requires isolation of PBMCs, removal of CD8+ cells and transduction of CD4+ cells followed by ex vivo expansion and infusion into the patients.
Currently, lentiviral vectors represent one of the most popular choices due to their ability to transduce non-dividing hematopoietic stem cells. We have shown that a single lentiviral vector harboring a triple combination of an shRNA targeting HIV-1 tat/rev mRNAs combined with two HIV-specific RNA-based inhibitors (a CCR5 targeting hammerhead ribozyme and a nucleolar-localizing TAR element that serves as a Tat decoy) efficiently suppressed HIV-1 replicated over 42 days and was more effective than a single anti-tat/rev shRNA or double combinations of the shRNA/ribozyme or decoy 19. This lentiviral vector harboring the three RNA-based anti-HIV moieties has been used for ex vivo gene delivery to hematopoietic stem cells in a first in man clinical trial 43. In this case, after autologous hematopoietic progenitor cells (HPCs) were programmed with the lentiviral vector, transfected cells were successfully engrafted in all four infused patients by day 11 without unexpected infusion-related toxicities. A long-term (up to 24 months) expression of an ectopically expressed siRNA and ribozyme in multiple peripheral blood cell lineages of two of the transplanted patients was observed. These results support the development of a multiple RNA-based cell therapy platform for HIV.
Other investigators are developing multiple HIV inhibitory encoding vectors for future use in gene therapy trials 44–46. A foamy virus vector has been engineered with a combination of anti-HIV shRNAs and the C46 fusion inhibiting peptide and shown to provide potent protection from HIV infection in a humanized mouse model. The use of a non-integrating viral vector such as AAV may in the future prove to be useful for transduction of shRNAs into T-lymphocytes for ex vivo applications.
Concluding remarks
Within the past decade, RNA interference has show great potential as a therapeutic modality for various diseases. HIV-1 became one of the first infectious agents targeted by RNAi due to its well-understood life cycle and pattern of gene expression. However, two major obstacles for the long-term use of RNAi against chronic HIV-1 infection are viral escape and poor cellular uptake/stability of siRNA.
Currently, significant progress has been made to identify therapeutic targets and in the development of efficient and safe siRNA delivery approaches. Efforts have been undertaken to identify hundreds of cellular targets and to develop combination therapies. Moreover, several examples discussed in this article demonstrate cell type-specific, non-viral mediated siRNA delivery into HIV-1 infected and target cells. Moreover viral vector mediated multi-RNAi therapeutic approaches may provide a complementary mechanism for combining the power of RNAi with other nucleic acid therapeutics, thereby providing a versatile technology platform for the treatment of various diseases.
RNAi technology still requires refinements before its full potential can be utilized for the routine clinical treatment of HIV. For example when targeting host dependency factors the safety and toxicity profiles of down-regulating these host targets must be carefully investigated in long-term knockdown studies. In addition, further efforts should be conducted to refine siRNA delivery schemes and to reduce or avoid unwanted immunogenicity and unwanted side-effects.
Acknowledgments
This work was supported by grants from the NIH to JJR: AI29329, AI42552 and HL07470
References
- 1.Richman DD, et al. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Zamore PD. RNA interference: big applause for silencing in Stockholm. Cell. 2006;127:1083–1086. doi: 10.1016/j.cell.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 6.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurreck J. RNA interference: from basic research to therapeutic applications. Angew Chem Int Ed Engl. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 9.Song E, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 10.Scherer L, Rossi JJ, Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- 11.Singh SK, Gaur RK. Progress towards therapeutic application of RNA interference for HIV infection. BioDrugs. 2009;23:269–276. doi: 10.2165/11317120-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 12.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das AT, et al. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pusch O, et al. Nucleotide sequence homology requirements of HIV-1-specific short hairpin RNA. Nucleic Acids Res. 2003;31:6444–6449. doi: 10.1093/nar/gkg876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsygankov AY. Current developments in anti-HIV/AIDS gene therapy. Curr Opin Investig Drugs. 2009;10:137–149. [PubMed] [Google Scholar]
- 18.Podlekareva D, et al. Factors associated with the development of opportunistic infections in HIV-1-infected adults with high CD4+ cell counts: a EuroSIDA study. J Infect Dis. 2006;194:633–641. doi: 10.1086/506366. [DOI] [PubMed] [Google Scholar]
- 19.Li MJ, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–09. doi: 10.1016/j.ymthe.2005.07.524. S1525-0016(05)01112-3 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 21.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Goff SP. Knockdown screens to knockout HIV-1. Cell. 2008;135:417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ter Brake O, et al. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 25.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkhout B, ter Brake O. Towards a durable RNAi gene therapy for HIV-AIDS. Expert Opin Biol Ther. 2009;9:161–170. doi: 10.1517/14712590802653619. [DOI] [PubMed] [Google Scholar]
- 27.Kumar P, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 29.Peipp M, et al. A recombinant CD7-specific single-chain immunotoxin is a potent inducer of apoptosis in acute leukemic T cells. Cancer Res. 2002;62:2848–2855. [PubMed] [Google Scholar]
- 30.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posner MR, et al. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J Immunol. 1991;146:4325–4332. [PubMed] [Google Scholar]
- 32.Kim SS, et al. RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice. Mol Ther. 2010;18:370–376. doi: 10.1038/mt.2009.271. mt2009271 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neff CP, et al. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med. 2011;3:66ra66. doi: 10.1126/scitranslmed.3001581. 3/66/66ra6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler LA, et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J Clin Invest. 2011;121:2401–2412. doi: 10.1172/JCI45876. 45876 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussey RE, et al. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature. 1988;331:78–81. doi: 10.1038/331078a0. [DOI] [PubMed] [Google Scholar]
- 38.Sattentau QJ, Moore JP. The role of CD4 in HIV binding and entry. Philos Trans R Soc Lond B Biol Sci. 1993;342:59–66. doi: 10.1098/rstb.1993.0136. [DOI] [PubMed] [Google Scholar]
- 39.Dalgleish AG, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 40.Kraus E, James W, Barclay AN. Cutting edge: novel RNA ligands able to bind CD4 antigen and inhibit CD4+ T lymphocyte function. J Immunol. 1998;160:5209–5212. [PubMed] [Google Scholar]
- 41.Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. 360/7/692 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Robbins MA, et al. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nat Biotechnol. 2006;24:566–571. doi: 10.1038/nbt1206. nbt1206 [pii] [DOI] [PubMed] [Google Scholar]
- 43.DiGiusto DL, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. 2/36/36ra43 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berkhout B. Toward a durable anti-HIV gene therapy based on RNA interference. Ann N Y Acad Sci. 2009;1175:3–14. doi: 10.1111/j.1749-6632.2009.04972.x. NYAS4972 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkhout B, Sanders RW. Molecular strategies to design an escape-proof antiviral therapy. Antiviral Res. 2011 doi: 10.1016/j.antiviral.2011.04.002. S0166-3542(11)00281-6 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Liu YP, Berkhout B. Lentiviral delivery of RNAi effectors against HIV-1. Curr Top Med Chem. 2009;9:1130–1143. doi: 10.2174/156802609789630866. [DOI] [PubMed] [Google Scholar]