Abstract
Background
Recent studies have identified chromosomal regions linked to variation in high density lipoprotein cholesterol (HDL-C), apolipoprotein A-1 (Apo A-1) and triglyceride (TG), although results have been inconsistent and previous studies of American Indian populations are limited
Objective
In an attempt to localize quantitative trait loci (QTLs) influencing HDL-C, Apo A-1 and TG, we conducted genome-wide linkage scans of subjects of the Strong Heart Family Study.
Methods
We implemented analyses in 3484 men and women aged 18 years or older, at three study centers.
Results
With adjustment for age, sex and center, we detected a QTL influencing both HDL-C (LOD = 4.4, genome-wide P = 0.001) and Apo A-1 (LOD = 3.2, genome-wide P = 0.020) nearest marker D6S289 at 6p23 in the Arizona sample. Another QTL influencing Apo A-1 was found nearest marker D9S287 at 9q22.2 (LOD = 3.0, genome-wide P = 0.033) in the North and South Dakotas. We detected a QTL influencing TG nearest marker D15S153 at 15q22.31 (LOD = 4.5 in the overall sample and LOD = 3.8 in the Dakotas sample, genome-wide P = 0.0044) and when additionally adjusted for waist, current smoking, current alcohol, current estrogen, lipid treatment, impaired fasting glucose, and diabetes, nearest marker D10S217 at 10q26.2 (LOD = 3.7, genome-wide P = 0.0058) in the Arizona population.
Conclusions
The replication of QTLs in regions of the genome that harbor well-known candidate genes suggest that chromosomes 6p, 9q and 15q warrant further investigation with fine mapping for causative polymorphisms in American Indians.
Keywords: linkage, genome-wide scan, high density lipoprotein cholesterol, apolipoprotein A-1, triglyceride
INTRODUCTION
High density lipoprotein cholesterol (HDL-C) and triglyceride (TG) concentrations are both independent risk factors for cardiovascular disease (CVD).[1] Apolipoprotein A-1 (Apo A-1), the major apoplipoprotein in HDL-C, is important for biochemical pathways that make HDL-C antiatherogenic. It is estimated that each 1% increase in HDL-C concentration reduces the risk of coronary heart disease by approximately 2% [2], although its etiologic role has been debated, and that each 21% decrease in TG concentration reduces the risk of non-fatal myocardial infarction and CVD death by 40% [3].
According to the Atlas of Heart Disease and Stroke among American Indians and Alaska Natives [4], the age-adjusted prevalence of high blood cholesterol levels (>240 mg/dL) (NCEP [1]) among American Indians living in 35 U.S. states was 30%, notably higher than the approximately 17% prevalence [5] noted among the general United States (U.S.) population. These prevalence measures do not differ significantly between female (29%) and male (31%) American Indians. Similar, as for the prevalence of elevated HDL-C, data from the Strong Heart Study (SHS), prevalence of low HDL-C levels (<40 mg/dL) was 34.2 – 36.7% among American Indian populations, and increased with age [6]. Another study among an urban American Indian population demonstrated a strikingly high prevalence of low HDL-C levels of 54.9% among males and of 54.2% among females [7], higher than the general U.S. population (25.1% for males and 9.1% for females) [5]. This study also reported 32.3% of males and 28.6% of females had high levels of triglycerides (≥ 150 mg/dl) [7], similar to its prevalence in the general U.S. population (30%) [8].
Strong aggregate genetic effects on HDL-C, Apo A-1, and TG variability have been recognized across multiple populations, although findings from candidate gene studies have been conflicting. Nonetheless, recent genome-wide association studies (GWAS) have identified several variants influencing HDL-C, Apo- A-1, and TG. For example, evidence for association between HDL-C and several previously implicated lipid metabolism genes, such as ABCA1 on chromosome (chr) 9 (rs4149268 [9], rs3890182 [10], and rs9282541 [11]) and LIPC on chr 15 (rs261332 and rs11858164 [11]) have been noted. In addition, evidence for association between LIPC rs4775041 and TG and CETP rs1800775 on chr 16 and Apo A-1 concentrations [12] have been reported. Despite these exciting findings, much of the heritable component of HDL-C, Apo-A1, and TG remains to be explained, suggesting that other susceptibility variants are segregating in populations.
Genome-wide linkage scans have been performed to identify novel regions of the genome harboring genes that regulate HDL-C, Apo A-1 and TG levels, but the identification of quantitative trait loci (QTL) has been challenging. As with most common complex phenotypes, several regions of the genome have been implicated, perhaps suggesting locus heterogeneity. A genome-wide scan of HDL-C in a kindred with familial hypercholesterolemia reported genome-wide significant linkage of HDL-C to chromosome 6 (LOD=3.1).[13]. Genome-wide significant linkage of Apo A-1 to chromosome 9q21.21-q33.1 (LOD=3.28), 8p21.1-q13.1 (LOD=3.71) and 10p15.1-p13 (LOD=5.51).[14] Genome wide significant evidence of linkage of TG to chromosome 4q28.3-32.1 (LOD=3.71) [14] and to chromosome 15 (LOD=3.88 at approximately 20–30 cM) [15] has been noted. Few previous gene mapping studies of HDL-C, Apo A-1 and TG in American Indian populations have been conducted.
More studies are necessary to advance our knowledge of the genetic factors influencing HDL-C, Apo A-1 and TG, especially because genome-wide association studies have largely been implemented in Caucasian samples. American Indians are an important United States minority population that can inform knowledge in distinct ways due to their geographic and genetic isolation and high rates of cardiovascular disease risk factors. For instance, because of these qualities we may discover genetic variants that are unique to their genetic and environmental context which can further our understanding of pathways to cardiovascular disease. Further, replication of existing QTLs in independent samples is essential to prioritize regions of the genome for costly comprehensive fine-mapping and ultimately identification of functional variants. Thus, our objectives were to localize new QTLs influencing HDL-C, Apo A-1, and TG as well as to provide new evidence in support of previously identified QTLs by conducting a genome-wide scan in American Indian subjects of the Strong Heart Family Study (SHFS).
MATERIALS AND METHODS
Study population
The SHFS is a multicenter, population-based, family study designed to localize genes that influence risk factors for clinical and subclinical cardiovascular disease, diabetes, and obesity and their progression over time. SHFS data were collected in two phases, the first completed in 1999 and the second initiated in 2001. More than 3600 resident American Indian tribal members aged 18 and older were recruited and examined in three study centers in Arizona (AZ), North and South Dakota (ND/SD), and Oklahoma (OK). The SHFS protocols were approved by the Indian Health Service Institutional Review Board, the institutional review boards of the participating institutions, all participating communities involved, and by all participants. All participants have given informed consent for the genetic study of CVD and associated risk factors, including the present study. Detailed descriptions of the SHFS protocols have been published.[16]
Phenotypic and genotypic data
Phenotypic data
Protocols for the collection of phenotypic data have been described previously [17], and, briefly, are as follows. Fasting blood samples were obtained during a physical examination and assayed at MedStar Research Institute using standard laboratory methods [17]. TG, total cholesterol, HDL-C, and apolipoproteins were measured using enzymatic reagents and the Hitachi 717. Glucose was measured by the hexokinase method (Glucose-HK; BMD, Inc) on the Hitachi 717. Type II diabetes, impaired fasting glucose, and normal glucose tolerance status were defined according to American Diabetes Association criteria [18]. Waist circumference was measured with participants wearing light clothing and without shoes. Information on demographic characteristics was obtained by questionnaire. Ever smoking was defined as having had at least 100 cigarettes during ones lifetime; current smoking was defined when participants answered “yes” to both of the questions: whether they were current smokers and whether they had smoked more than 100 cigarettes in their life. Current and ever alcohol consumption were defined as having had at least 12 alcoholic beverages in the last year (at least one day per month) and/or lifetime, respectively. Reproductive history was queried including parity, gravidity, menopausal status, and estrogen use.
Genotypic data
The SHFS genotyping procedures have been documented previously [19]. Specifically, DNA was isolated from fasting blood samples using organic solvents and then amplified in separate PCR reactions with primers specific for short tandem repeat markers using the ABI PRISM Linkage Mapping Set-MD10 version 2.5 (Applied Biosystems, Foster City, CA). Paticipants were genotyped for approximately 400 microsatellite markers spaced at average 10 cM intervals (range, 2.4 to 24.1 cM) across the autosomal chromosomes, with the average heterozygosity of these markers for 0.69 in the Arizon subsample, 0.76 for the Dakotas, and 0.74 for Oklahoma. Analyses and assignment of the marker alleles were done using electrophoresis and computerized algorithms (Applied Biosystems). Sex-averaged chromosomal maps obtained from deCODE Genetics (http://www.decodegenetics.com) were used. All genetic distances are reported in Haldane centimorgans (cM). Pedigree relationships were verified using the PREST (Pedigree Relationship Statistical Tests) package [20]. Mendelian inconsistencies and spurious double recombinants were detected using the SimWalk2 package [21]. The overall blanking rate for both types of errors was <1% of the total number of genotypes for Arizona, North and South Dakota, and Oklahoma. We used web resources of the University of California Santa Cruz (http://genome/ucsc.edu) and Online Mendelian Inheritance in Man (http://www3.ncbi.nlm.nih.gov/entrez/query.fcgi?db 5 OMIM) to determine the cytogenetic location of markers and to search for candidate genes.
Statistical analysis
Fasting plasma HDL-C and TG values were transformed by natural logarithm, and apo A-1 was transformed by dividing 1 by the apo A-1 value, because of their skewed distributions. Multipoint variance component linkage analysis of ln(HDL-C), 1/(apo A-1) and ln(TG) was performed among approximately 3,400 SHFS participants with complete covariate data using SOLAR, version 2.1.4. Details of this model have been described previously.[22] The use of the variance component approach requires an estimate of the identity-by-descent matrices, which were computed using the Loki package [23] with a Markov chain Monte Carlo stochastic procedure. Genome-wide p-values were obtained by the method of Feingold et al. [24]
Stepwise linear regression was used to screen covariates for statistical significance using SAS, version 9.1. Given the sensitivity of variance component analysis to kurtosis, all phenotypic outliers of the trait of interest and all covariate data points (here defined as any value ≥3 standard deviations from the mean) were removed before analysis (the number excluded varies by variable; in all cases n < 30 or approximately 5% of the study observations). To maximize our power to detect genetic effects, we considered two different models of covariate adjustment in each population separately. In model 1, adjustments were made for age, sex, and field center. In model 2, we additionally adjusted for waist circumference, current smoking, current alcohol consumption, current estrogen use, lipid-lowering treatment, diabetes status, and impaired fasting glucose. All analyses were also run, and results reported, stratified by field center. Residuals were generated for both models and used in all subsequent quantitative genetic analyses. Kurtosis values for ln(HDL-C), 1/(apo A-1) and ln(TG) were <0.50 for all analyses.
RESULTS
Descriptive characteristics of the SHFS participants are summarized in Supplementary table 1. The average age ± SD of participants by center was ~37 ± 16, 39 ± 17, and 44 ± 17 years in Arizona, Dakotas, and Oklahoma, respectively. Consistent with the results from our previous studies of a subsample of these family members,[25] a high prevalence of current cigarette smoking and alcohol consumption was observed. As expected, a high prevalence of diabetes was noted, especially in the Arizona center (32%). HDL-C, apo A-1 and TG concentrations varied by center, where HDL-C was lowest in the Arizona population (48.52 ±14 mg/dL), and apo A-1 (143.92 ± 27 mg/dL) and TG (163.59 ± 92 mg/dl) were highest in Oklahoma. In contrast to many similarly-aged U.S. populations, we found a very low prevalence of lipid-lowering therapy (≤ 6%).
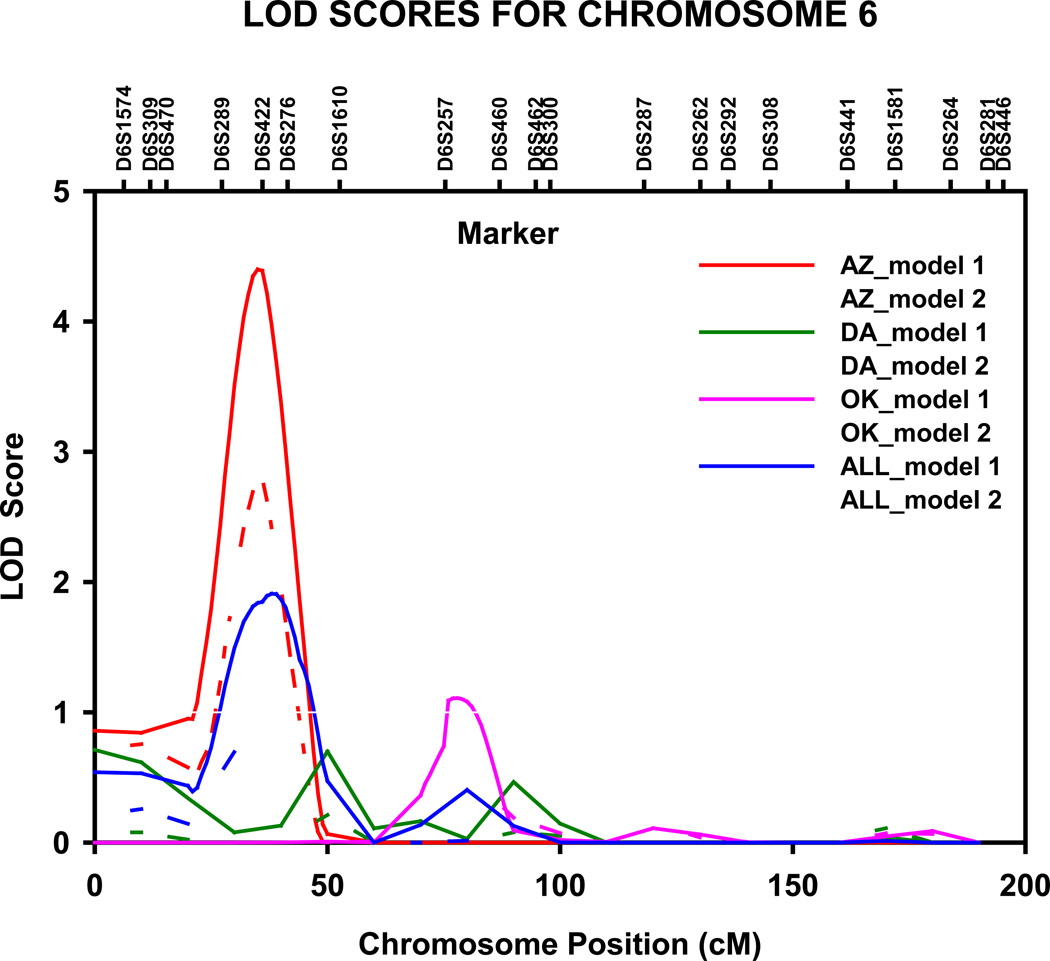
Table 1 presents the multipoint genome-wide adjusted LOD scores and their locations from the variance component linkage analyses of HDL-C, apo A-1 and TG for all linkage peaks with LOD scores ≥1.8 [suggestive evidence for linkage [26]]. A QTL for HDL-C (adjusted LOD=4.4, genome-wide P =0.001) was detected in the Arizona population on chromosome 6p23 at 36 cM, nearest marker D6S289 (fig 1). The 1-cM LOD unit support interval spanned 12 cM, from 29 to 41 cM (10.033–20.578 Mb P terminus), between marker D6S470 and D6S422. Upon adjustment for multiple confounders (model 2), the magnitude of LOD score decreased by 1.6 units, yet suggestive evidence for linkage was still noted (LOD =2.8, genome-wide P =0.054).
Table 1.
Maximum LOD scores suggestive of linkage (LOD ≥ 1.8) for multipoint quantitative trait linkage analyses of HDL-C, apolipoprotein A-1 and triglyceride levels in American Indian Participants of the Strong Heart Family Study
| Phenotype | Center and Model a | Chromosome | Centimorgan | Chromosomal Region |
Maximum LOD Score |
|---|---|---|---|---|---|
| HDL-C | |||||
| Oklahoma, model 2 | 3 | 193 | 3q26.31 – q27.3 | 1.9 | |
| Arizona, model 1, model 2 | 6 | 35–36 | 6p22.3–p24.3 | 4.4, 2.8 | |
| Dakotas, model 1 | 9 | 110 | 9q31.1–q31.3 | 1.9 | |
| Oklahoma, model 2 | 9 | 101 | 9q22.32–q31.1 | 2.0 | |
| apolipoprotein A-1 | |||||
| Dakotas, model 1 | 2 | 214 | 2q33.3–q35 | 2.2 | |
| Arizona, model 1, model 2 | 6 | 36 | 6p22.3–p24.3 | 3.2, 2.1 | |
| Oklahoma, model 2 | 7 | 92 | 7q21.11–q21.13 | 1.9 | |
| Dakotas, model 1 | 9 | 99 | 9q22.2 | 3.0 | |
| Dakotas, model 2 | 9 | 103 | 9q22.2–q31.1 | 2.5 | |
| Dakotas, model 2 | 12 | 162 | 12q24.32 | 2.1 | |
| Triglycerides | |||||
| Arizona, model 1, model 2 | 4 | 168–169 | 4q32.3–q34.1 | 1.9, 3.0 | |
| Arizona, model 1 | 6 | 87 | 6p12.1–q14.1 | 2.9 | |
| Arizona, model 2 | 6 | 75 | 6p12.1–p21.2 | 2.6 | |
| Arizona, model 1, model 2 | 10 | 163–164 | 10q26.2–q26.3 | 2.9, 3.7 | |
| Oklahoma, model 1 | 11 | 67 | 11q12.1–q13.2 | 1.8 | |
| Arizona, model 1 | 15 | 44 | 15q15.1–q21.1 | 2.0 | |
| Dakotas, model 1, model 2 | 15 | 65–68 | 15q22.1–q22.31 | 3.8, 2.2 | |
| Dakotas, model 1 | 18 | 40 | 18p11.21–q12.1 | 1.9 | |
| Oklahoma, model 2 | 21 | 10 | 21q21.1 | 2.0 |
Model 1: adjustment for sex, age, and stratified by field center.
Model 2: additional adjustments for waist circumference, current smoking status, current consumption of alcohol, current estrogen use, lipid treatment status, impaired fasting glucose and diabetes status.
Figure 1.
Multipoint LOD scores on chromosome 6 for HDL cholesterol. Results for combined and stratified analyses shown. Model 1 was adjusted for age, sex, and field center (combined analysis). Model 2 was additionally adjusted for waist circumference, current smoking status, current alcohol consumption, current estrogen use, lipid treatment status, impaired fasting glucose and diabetes status. AZ, Arizona field center; DA, Dakotas field centers; OK, Oklahoma field center; cM, Haldane centimorgans.
Further suggestive evidence of linkage to HDL-C level was observed in the North and South Dakotas population on chromosome 9q31.1-q31.3 at 110 cM (LOD=1.9) between D9S1690 and D9S167, and in the Oklahoma population on chromosome 9q22.2-q31.1 at 101 cM (LOD=2.1), close to marker D9S287. In addition, we found suggestive evidence of linkage to HDL-C in the Oklahoma population at 193 cM on chromosome 3q26.31-q27.3 (LOD=1.9), nearest marker D3S1262.
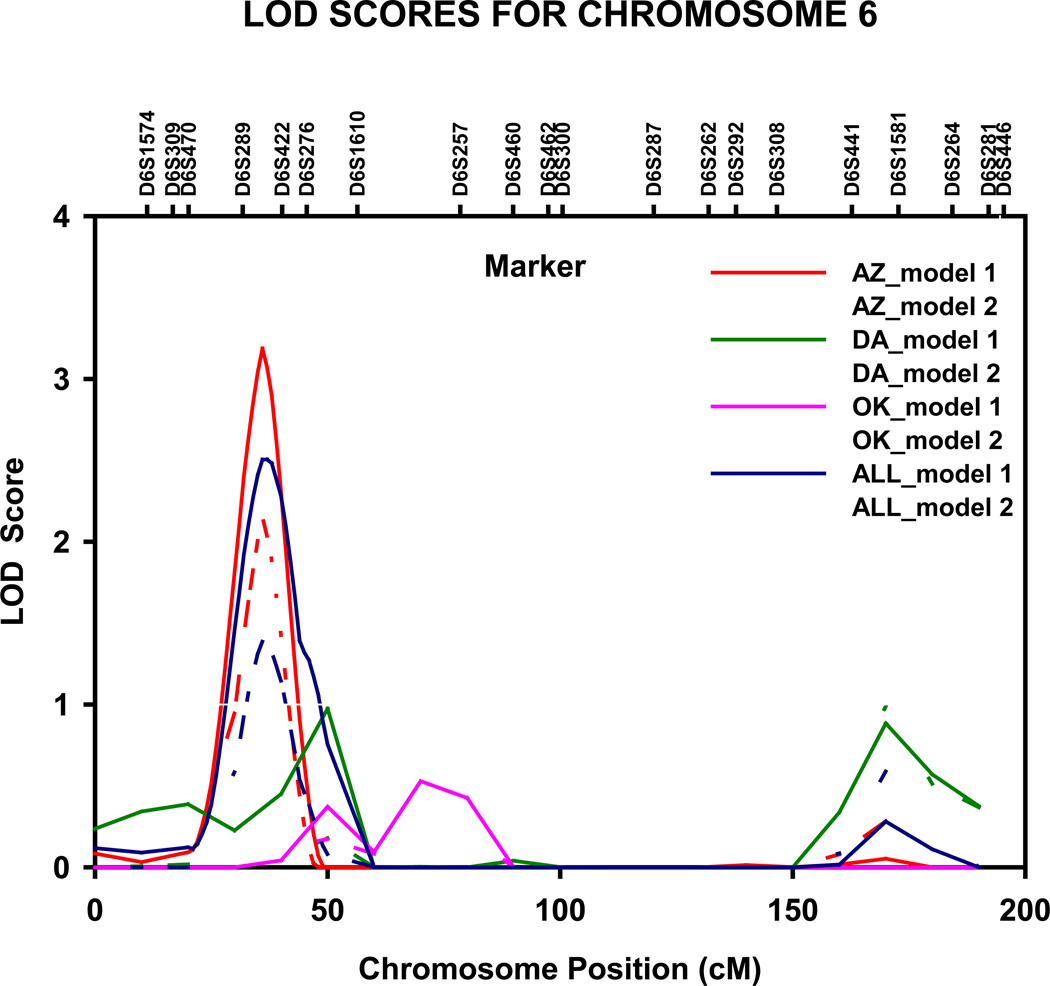
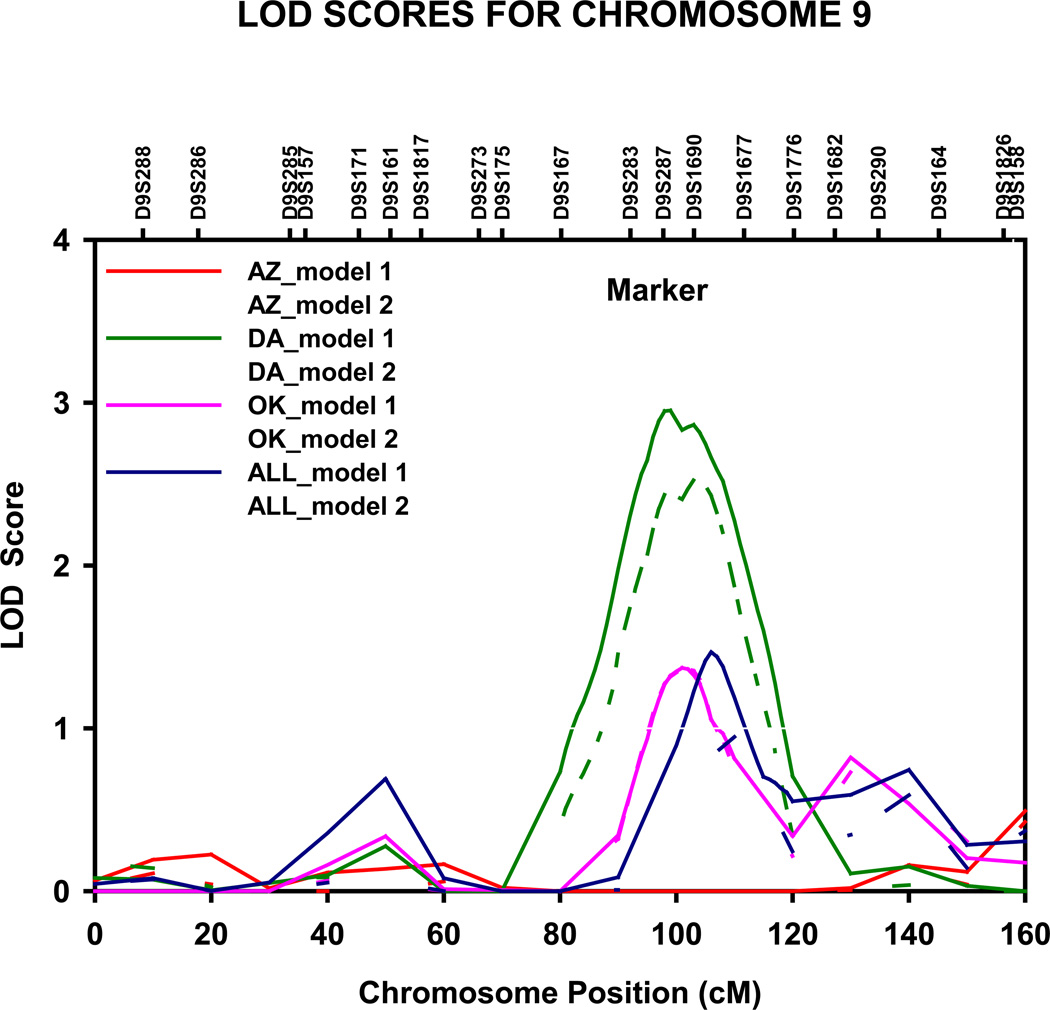
Genome-wide significant evidence of linkage to apo A-1 levels was detected in the Arizona population on chromosome 6p23 (LOD = 3.2, genome-wide P = 0.020) at 36 cM, nearest marker D6S289 (fig 2). The 1-cM LOD unit support interval spanned 10 cM, from 31 to 41 cM (10.033–20.578 Mb P terminus), between markers D6S470 and D6S422. Upon adjustment for multiple confounders (model 2), the magnitude of the LOD score decreased by 1.1 units, yet suggestive evidence for linkage was still noted (LOD =2.1). We also detected genome wide significant linkage to apo A-1 levels in the North and South Dakotas population on chromosome 9q22.2 at 99 cM (LOD = 3.0, genome-wide P = 0.033), nearest marker D9S287 (fig 3). The 1-cM LOD unit support interval spanned 22 cM from 90 to 112 cM (84.873–111.077 Mb q terminus), between D9S167 and D9S1677.
Figure 2.
Multipoint LOD scores on chromosome 6 for apolipoprotein A-I. Results for combined and stratified analyses shown. Model 1 was adjusted for age, sex, and field center (combined analysis). Model 2 was additionally adjusted for waist circumference, current smoking status, current alcohol consumption, current estrogen use, lipid treatment status, impaired fasting glucose and diabetes status. AZ, Arizona field center; DA, Dakotas field centers; OK, Oklahoma field center; cM, Haldane centimorgans.
Figure 3.
Multipoint LOD scores on chromosome 9 for apolipoprotein A-I. Results for combined and stratified analyses shown. Model 1 was adjusted for age, sex, and field center (combined analysis). Model 2 was additionally adjusted for waist circumference, current smoking status, current alcohol consumption, current estrogen use, lipid treatment status, impaired fasting glucose and diabetes status. AZ, Arizona field center; DA, Dakotas field centers; OK, Oklahoma field center; cM, Haldane centimorgans.
Further suggestive evidence of linkage to apo A-1 level was observed in the North and South Dakotas population on chromosome 2q33.3-q35 at 214 cM (LOD=2.2), nearest marker D2S2382, on chromosome 9q22.2-q31.1 at 103 cM (LOD=2.5) between D9S287 and D9S1690 and on chromosome 12q24.32 at 162 cM (LOD=2.1), close to marker D12S1659, and in the Oklahoma population on chromosome 7q21.11-q21.13 at 92 cM (LOD=1.9), nearest marker D7S669 (table 1).
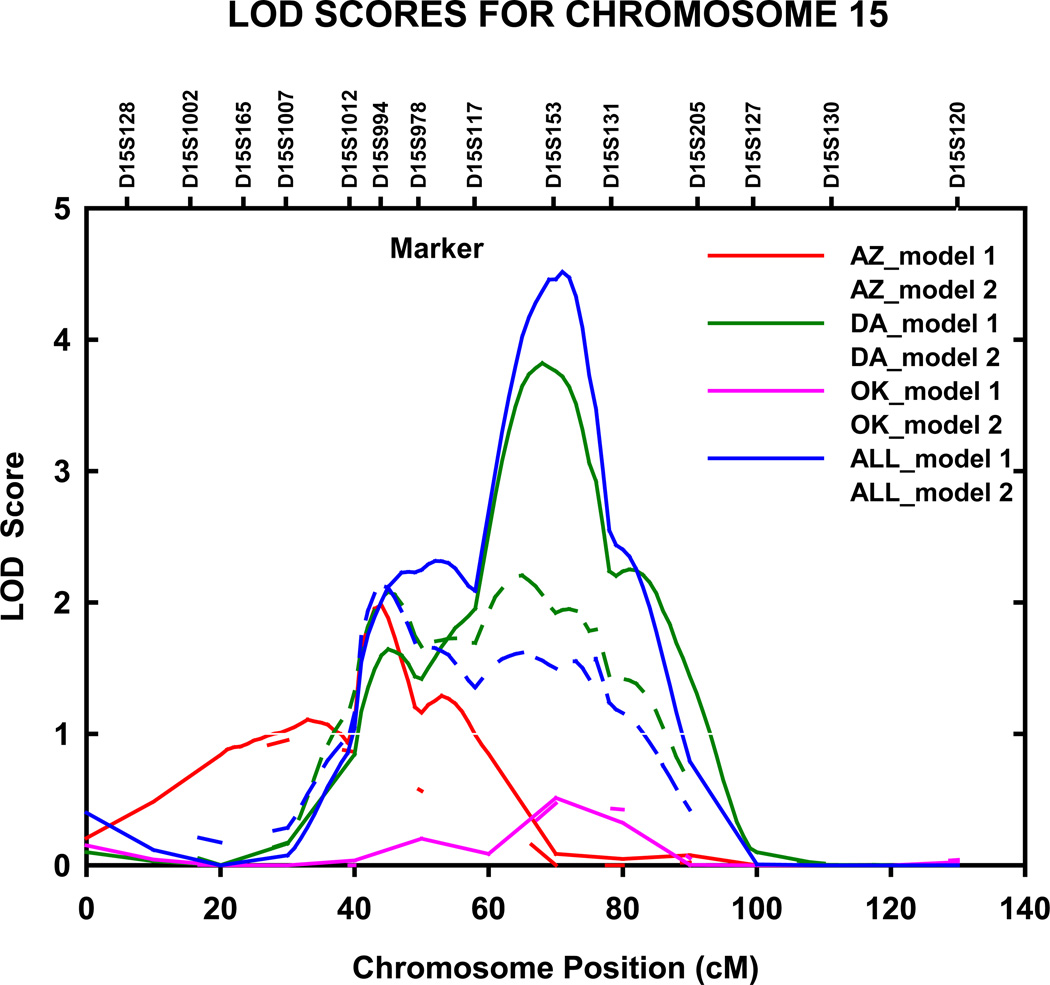
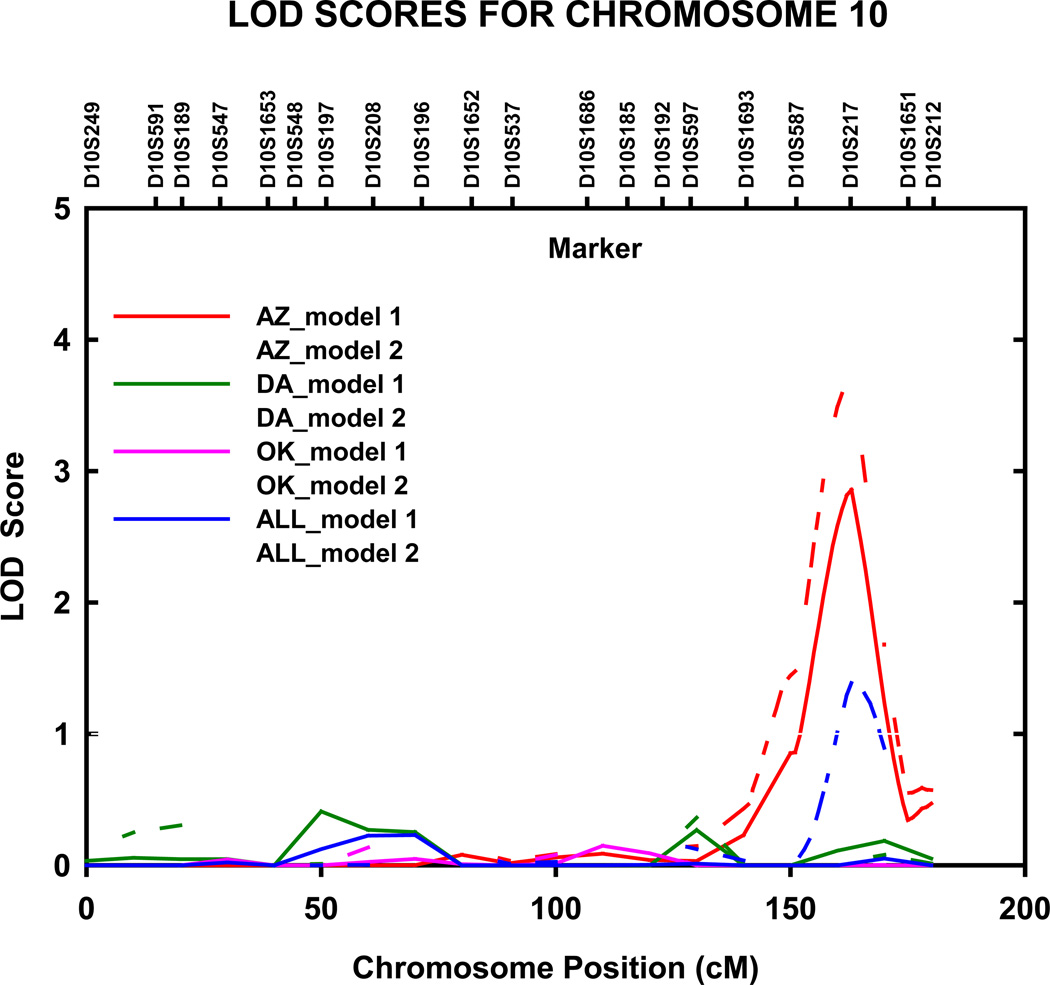
Genome-wide significant evidence of linkage to TG levels was detected in the North and South Dakotas population on chromosome 15q22.31 (LOD = 3.8, genome-wide P =0.0044) at 68 cM, nearest marker D15S153 (fig 4). The 1-cM LOD unit support interval spanned 15 cM from 61 to 76 cM (56.166–69.071 Mb q terminus), between D15S117 and D15S131. When excluding participants on lipid lowering therapy, this LOD score increased to 5.0 (data not shown). We also detected genome wide significant linkage to TG levels in the Arizona population on chromosome 10q26.2 at 164 cM (LOD = 3.7, genome-wide P =0.0058), nearest marker D10S217 (fig 5). This LOD score was slightly lower when adjustments were made for age and sex only (model 1) (LOD = 2.9, genome-wide P =0.042). The 1-cM LOD unit support interval spanned 11 cM from 156 to 167 cM (125.078–129.530 Mb q terminus), between D10S587 and D10S217.
Figure 4.
Multipoint adjusted LOD scores on chromosome 15 for triglycerides. Results for combined and stratified analyses shown. Model 1 was adjusted for age, sex, and field center (combined analysis). Model 2 was additionally adjusted for waist circumference, current smoking status, current alcohol consumption, current estrogen use, lipid treatment status, impaired fasting glucose and diabetes status. AZ, Arizona field center; DA, Dakotas field centers; OK, Oklahoma field center; cM, Haldane centimorgans.
Figure 5.
Multipoint adjusted LOD scores on chromosome 10 for triglycerides. Results for combined and stratified analyses shown. Model 1 was adjusted for age, sex, and field center (combined analysis). Model 2 was additionally adjusted for waist circumference, current smoking status, current alcohol consumption, current estrogen use, lipid treatment status, impaired fasting glucose and diabetes status. AZ, Arizona field center; DA, Dakotas field centers; OK, Oklahoma field center; cM, Haldane centimorgans.
Suggestive evidence of linkage to TG levels was detected in the Arizona population on chromosome 4q32.3-q34.1 at 168 and 169 cM between D4S1597 and D4S1539, on chromosome 6p12.1-q14.1 at 87 cM, between D6S257 and D6S460, on chromosome 15q15.1-q21.1 at 44 cM, close to D15S994, and on chromosome 18p11.21-q12.1 at 40 cM, near D18S53. In the Oklahoma population, suggestive evidence of linkage to TG levels was noted at 67 cM on chromosome 11q12.1-q13.2 between D11S4191 and D11S987, and at 10 cM on chromosome 21q21.1, near D21S1899 (table 1).
DISCUSSION
The present study was conducted to identify regions of the genome harboring genes that influence HDL-C, Apo A-1 and TG levels in large extended American Indian families. Here, we present evidence for a QTL for HDL-C and apo A-1 levels in the Arizona population on chromosome 6p nearest marker D6S289, and for apo A-1 levels in the North and South Dakotas population on chromosome 9 nearest marker D9S287 at 9q22.2. We also detected significant QTLs for TG levels in the North and South Dakotas population on chromosome 15 nearest marker D15S153, and in the Arizona population on chromosome 10, nearest marker D10S217 at 10q26.2.
Chromosome 6p, where we observe our strongest linkage evidence for HDL-C and apo A-1, has previously been implicated in four genome scans of lipid-related traits.[13, 27–29] Genome-wide significant LOD scores were reported in this region for HDL-C in a kindred with familial hypercholesterolemia (LOD=3.1) [13]. Suggestive linkage has also been noted for total apoB in French-Canadian families (LOD=1.35) [29] and in French-Canadian families (LOD=1.32) [28]. The strongest positional candidate genes are located ~4Mb from our highest LOD score, the ELOVL2 and TFAP2A genes. Cell over-expression of ELOVL2 gene, encoding a polyunsaturated fatty acid elongase, enhances TG synthesis and accumulation of lipid droplets [30]. TFAP2A (transcription factor AP-2 alpha) encodes the activator protein (AP)2α and negatively regulates ABCA1 promoter activity [31]. However, to date no previous studies have examined whether ELOVL2 or TFAP2A polymorphisms influence variation in HDL-C or apo A-1. The same QTL detected for both HDL-C and apo A-1 is consistent with the biological structure for apo A-1, the major structural protein of HDL-C.
To further support our results on 6p, we evaluated the P-values from the association between SNP genotypes and HDL-C, apo A-1 and TG concentrations for SNPs within or in LD with biological candidate genes underlying the 1 LOD support interval of our linkage peak on 6p among approximately 2,600 participants of the Diabetes Genetic Initiative (DGI) study (available at http://www.broad.mit.edu/diabetes/). Of the 1,870 SNPs within the 1-cM LOD unit support interval on chromosome 6p, evidence for association with HDL-C was observed for ELOVL2 SNP rs3798712 (P =0.0147), and evidence for association with apo A-1 was observed for ELOVL2 SNP rs3798712 (P =0.0032) and TFAP2A SNP rs12663250 (P =0.0214) (Supplementary table 2).
We also attempted to support our results on 6p using GWAS data from 1087 participants of the offspring cohort of the Framingham Heart Study (FHS) SHARe 100K online resource. All results for autosomal SNPs are available through NCBI dbGaP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?id=phs000007). We examined if any SNP with a P-value ≤ 0.05 was located within a candidate gene for HDL-C, apo A-1 or TG or tagged a candidate gene under our genome wide significant linkage peaks. No evidence for association was observed among the FHS offspring cohort.
Chromosome 9q, where we observe our strongest linkage evidence for Apo A-1 levels, has previously been implicated in six genome scans of lipid-related traits. [9, 14, 32–35] Genome-wide significant LOD scores were reported in this region for Apo A-1 (LOD=3.28) in female sib-pairs [14]. The strongest positional candidate gene is located ~9 Mb from our highest LOD score, the ABCA1 gene. The ATP-binding cassette (ABC) protein encoded by this gene functions as a cholesterol efflux pump in the cellular lipid removal pathway, with cholesterol as its substrate. One study showed an association between apo A-1 levels and ABCA1 14C>T polymorphisms in a Malay population (ORTT/CC=2.92, 95%CI=1.42–6.00; ORCT/CC=1.29, 95%CI=0.81–2.04).[36]
It is notable that ABCA1 is a rate-limiting factor for HDL-C biogenesis. Indeed, GWAS study findings also support an association between ABCA1 variants and HDL-C, for example, rs9282541 (−3.2 mmol/L decrease per T allele, P = 3.2 × 10−5) [11] and rs4149274 (1.51 mg/dL increase per G allele, P ~ 7.4 × 10−8) [9]. On-line GWAS resources further support the association with ABCA1 gene variants. Eight SNPs of the ABCA1 gene within the 1-cM LOD unit support interval of chromosome 9q, were associated with apo A-1 (P≤0.05) in white participants of the DGI study (Supplementary table 2). In contrast, no evidence for this association was observed among the FHS offspring cohort (all P > 0.05).
Chromosome 15q, where we observe our strongest linkage evidence for TG levels, has previously been implicated in four genome scans of lipid-related traits.[37–40] Genome-wide significant LOD scores were reported in this region for TG and HDL-C levels among participants from NHLBI Family Heart study project (LOD=3.0), for HDL-C (LOD=3.3) among Mexican American individuals from the San Antonio Family Heart Study project [40], and for HDL-C among participants from the Genetic Epidemiology of Metabolic Syndrome project (LOD=3.1) [38]. Suggestive linkage has also been noted for TG in family members with familial hypertriglyceridemia (LOD=1.35).[37] Approximately 587 genes underlie the 1 LOD unit support interval (46 cM, 45.2 Mb) of the 15q signal. The strongest positional candidate gene is located ~8Mb from our highest LOD score, the LIPC gene. LIPC (hepatic triglyceride lipase) is a lipolytic enzyme that has dual functions of triglyceride hydrolase and ligand/bridging factor for receptor-mediated lipoprotein uptake [41]. Recent data show inconsistent results in the association between LIPC polymorphisms and TG variation [42, 43] and between LIPC and HDL and LDL concentrations [27, 44]. A recent GWAS study [9] identified a significant association between TG and LIPC SNP rs4775041 (3.62 mg/dL increase per C allele combined P = 1.6 × 10−8). With respect to the on-line DGI GWAS resource, of the 5,844 SNPs on chromosome 15q within the 1-cM LOD unit support interval, nine SNPs within the LIPC gene were associated with TG (P≤0.05) (Supplementary table 2). With respect to the Framingham 100K GWAS resource, LIPC SNPs, rs2899632 (P = 0.0032) and rs8028759 (P = 0.02) were associated with TG measured at FHS Examinations 7 and 4, respectively.
Significant linkage of TG levels to chromosome 10q has also been reported in several other studies. Pajukanta et al [45] studied Finnish families with familial combined hyperlipidemia and found a LOD of 2.6 for cholesterol at the same location on chromosome 10. Approximately 33 known genes underlie the 1 LOD unit support interval (12 cM, 4.5 Mb) of the 10q signal. However, this chromosome 10q locus is in a region lacking any obvious positional candidate genes with known functions related to lipid metabolism, suggesting that this locus may harbor a novel lipid gene.
We also observed several loci with suggestive linkage to HDL-C, apo A-1 and TG, some novel and others supporting positive findings from other studies. Supplementary table 3 summarizes the observed linkage signals from this study that are supported by evidence of linkage to lipid-related traits in other populations on 2q, 3q, 4q, 7q, 11q, 12q, 18, and 21q. Although these signals do not meet the genome-wide significance threshold, these results still offer valuable information in that they may identify regions worthy of further study.
We found that when excluding American Indian participants on lipid lowering therapy (n = 66 participants in the Dakotas center), the magnitude of the LOD score for TG on chromosome 15q increased by 1.2 units (adjusted LOD =5.0). We can offer several possible explanations for these findings. First, as lipid-lowering medications may lower the concentration of TG [46], the TG values of these medicated subjects may not accurately reflect their true values. Secondly, it is possible that by excluding medicated individuals from the analysis, the genetic locus heterogeneity was reduced, thereby strengthening the evidence for linkage at this particular QTL. When additionally adjusting TG for waist circumference, current smoking status, current consumption of alcohol, current estrogen use, lipid-lowering treatment status, and diabetes status, the magnitude of the LOD score on chromosome 10q increased by 1.6 units (LOD =3.7). As the sample of examined individuals remained the same, we infer that by reducing the residual covariate effects (model 1 with only adjustment for sex and age), we had greater power to detect a QTL specific effect at this locus on chromosome 10q. As no other modifications (amplifications or attenuations) of LOD scores were noted upon adjustment for lipid lowering therapies and the percentage of participants taking lipid lowering drugs was low, we report both findings that have been adjusted for lipid lowering therapy and those that did not.
In conclusion, our findings suggest that one or more genes on chromosomes 6 and 9 regulate HDL cholesterol and apopliprotein A-1 concentrations, and on chromosomes 10 and 15 regulate triglyceride concentrations. Excellent candidate genes are implicated in each of these genomic regions and future research should pursue these positional candidate genes, particularly for the ELOV2, the ABCA1 and the LIPC genes, to determine whether polymorphisms in these genes are the source of these highly replicated linkage signals. Our study also provides corroboration of genomic regions that possibly influence interindividual variation in HDL-C, apo A-1 and TG concentrations on chromosome 2, 3, 4, 7, 9, 12, and 18, and new information about genomic regions on 11 and 21. The identification and confirmation of QTLs for HDL-C, apo A-1 and TG concentrations may bring us closer to the identification of the functional genes that influence these phenotypes and aid in the targeting of preventative therapies for individuals with low circulating levels of HDL-C and apo A-1, and high fasting triglyceride concentrations, established risk factors for CVD.
Supplementary Material
Key points.
More studies are necessary to advance our knowledge of the genetic factors influencing HDL-C, Apo A-1 and TG among US minority populations, particularly American Indians which can inform knowledge in distinct ways due to their geographic and genetic isolation and high rates of cardiovascular disease risk factors.
In this study, four QTLs influencing HDL-C, apo A-1 , and TG levels were identified in the American Indian participants of the Strong Heart Family Study, on chromosomes 6p, 9q, 15q, and 10q.
These signals implicate excellent candidate genes, particularly for the ELOV2, the ABCA1 and the LIPC genes, and are supported by several linkage and association studies across multiple ethnic groups, suggesting a generalization of lipid related genetic effects across multiple populations.
Acknowledgements
We would first like to thank the Strong Heart Family Study participants. Without their participation, this project would not have been possible. In addition, the cooperation of the Indian Health Service hospitals and clinics, and the directors of the SHS clinics, Betty Jarvis, Dr. Marie Russell, Marcia O’Leary, Dr. Tauqeer Ali, and the many collaborators and staff of the Strong Heart Study that have made this project possible. We would also like to thank Dr. Thomas Welty for his contribution to the Strong Heart Study-Dakota Center. This research was funded by a cooperative agreement that includes grants U01 HL65520, U01 HL41642, U01 HL41652, U01 HL41654, and U01 HL65521 from the National Heart, Lung, and Blood Institute. We would also like to acknowledge US Public Health Service grant MH059490 from the National Institutes of Health. The views expressed in this paper are those of the authors and do not necessarily reflect those of the Indian Health Service. This investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grants C06 RR13556 and C06 RR017515 from the National Center for Research Resources, National Institutes of Health.
Footnotes
Licence for publication:
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd to permit this article (if accepted) to be published in Journal of Medical Genetics and any other BMJPGL products and sublicences such use and exploit all subsidiary rights, as set out in our licence (http://jmg.bmj.com/misc/ifora/licenceform.shtml).
REFERENCES
- 1.Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143–3421. [PubMed] [Google Scholar]
- 2.Gotto AM, Jr, Brinton EA. Assessing low levels of high-density lipoprotein cholesterol as a risk factor in coronary heart disease: a working group report and update. J Am Coll Cardiol. 2004;43(5):717–724. doi: 10.1016/j.jacc.2003.08.061. [DOI] [PubMed] [Google Scholar]
- 3.Secondary prevention by raising HDL cholesterol and reducing triglycerides in patients with coronary artery disease: the Bezafibrate Infarction Prevention (BIP) study. Circulation. 2000;102(1):21–27. doi: 10.1161/01.cir.102.1.21. [DOI] [PubMed] [Google Scholar]
- 4.Casper MLDC, Coolidge JN, Williams GI, Jr, Crowell A, Galloway JM, Cobb N. Atlas of Heart Disease and Stroke Among American Indians and Alaska Natives. 2005. [Google Scholar]
- 5.Rosamond W, Flegal K, Furie K, Go A, Greenlund K, Haase N, Hailpern SM, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O'Donnell C, Roger V, Sorlie P, Steinberger J, Thom T, Wilson M, Hong Y. Heart disease and stroke statistics-2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117(4):e25–e146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- 6.Rhoades DA, Welty TK, Wang W, Yeh F, Devereux RB, Fabsitz RR, Lee ET, Howard BV. Aging and the prevalence of cardiovascular disease risk factors in older American Indians: the Strong Heart Study. J Am Geriatr Soc. 2007;55(1):87–94. doi: 10.1111/j.1532-5415.2006.01018.x. [DOI] [PubMed] [Google Scholar]
- 7.Gupta R, Gupta VP, Sarna M, Bhatnagar S, Thanvi J, Sharma V, Singh AK, Gupta JB, Kaul V. Prevalence of coronary heart disease and risk factors in an urban Indian population: Jaipur Heart Watch-2. Indian Heart J. 2002;54(1):59–66. [PubMed] [Google Scholar]
- 8.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. Jama. 2002;287(3):356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 9.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Gomez Perez FJ, Frazer KA, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40(2):149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 12.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 13.Canizales-Quinteros S, Aguilar-Salinas CA, Reyes-Rodriguez E, Riba L, Rodriguez-Torres M, Ramirez-Jimenez S, Huertas-Vazquez A, Fragoso-Ontiveros V, Zentella-Dehesa A, Ventura-Gallegos JL, Vega-Hernandez G, Lopez-Estrada A, Auron-Gomez M, Gomez-Perez F, Rull J, Cox NJ, Bell GI, Tusie-Luna MT. Locus on chromosome 6p linked to elevated HDL cholesterol serum levels and to protection against premature atherosclerosis in a kindred with familial hypercholesterolemia. Circ Res. 2003;92(5):569–576. doi: 10.1161/01.RES.0000064174.69165.66. [DOI] [PubMed] [Google Scholar]
- 14.Falchi M, Andrew T, Snieder H, Swaminathan R, Surdulescu GL, Spector TD. Identification of QTLs for serum lipid levels in a female sib-pair cohort: a novel application to improve the power of two-locus linkage analysis. Hum Mol Genet. 2005;14(20):2971–2979. doi: 10.1093/hmg/ddi327. [DOI] [PubMed] [Google Scholar]
- 15.Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP. A major susceptibility locus influencing plasma triglyceride concentrations is located on chromosome 15q in Mexican Americans. Am J Hum Genet. 2000;66(4):1237–1245. doi: 10.1086/302849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.North KE, Howard BV, Welty TK, Best LG, Lee ET, Yeh JL, Fabsitz RR, Roman MJ, MacCluer JW. Genetic and environmental contributions to cardiovascular disease risk in American Indians: the strong heart family study. Am J Epidemiol. 2003;157(4):303–314. doi: 10.1093/aje/kwf208. [DOI] [PubMed] [Google Scholar]
- 17.Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, Cucchiara AJ, Savage PJ, Howard BV. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132(6):1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 18.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20(7):1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 19.Diego VP, Goring HH, Cole SA, Almasy L, Dyer TD, Blangero J, Duggirala R, Laston S, Wenger C, Cantu T, Dyke B, North K, Schurr T, Best LG, Devereux RB, Fabsitz RR, Howard BV, MacCluer JW. Fasting insulin and obesity-related phenotypes are linked to chromosome 2p: the Strong Heart Family Study. Diabetes. 2006;55(6):1874–1878. doi: 10.2337/db05-0668. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54(2):99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- 21.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70(2):496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61(3):748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feingold E, Brown PO, Siegmund D. Gaussian models for genetic linkage analysis using complete high-resolution maps of identity by descent. Am J Hum Genet. 1993;53(1):234–251. [PMC free article] [PubMed] [Google Scholar]
- 25.North KE, Almasy L, Goring HH, Cole SA, Diego VP, Laston S, Cantu T, Williams JT, Howard BV, Lee ET, Best LG, Fabsitz RR, MacCluer JW. Linkage analysis of factors underlying insulin resistance: Strong Heart Family Study. Obes Res. 2005;13(11):1877–1884. doi: 10.1038/oby.2005.230. [DOI] [PubMed] [Google Scholar]
- 26.Rao DC, Gu C. False positives and false negatives in genome scans. Adv Genet. 2001;42:487–498. doi: 10.1016/s0065-2660(01)42038-4. [DOI] [PubMed] [Google Scholar]
- 27.Malhotra A, Coon H, Feitosa MF, Li WD, North KE, Price RA, Bouchard C, Hunt SC, Wolford JK. Meta-analysis of genome-wide linkage studies for quantitative lipid traits in African Americans. Hum Mol Genet. 2005;14(24):3955–3962. doi: 10.1093/hmg/ddi419. [DOI] [PubMed] [Google Scholar]
- 28.Bosse Y, Chagnon YC, Despres JP, Rice T, Rao DC, Bouchard C, Perusse L, Vohl MC. Compendium of genome-wide scans of lipid-related phenotypes: adding a new genome-wide search of apolipoprotein levels. J Lipid Res. 2004;45(12):2174–2184. doi: 10.1194/jlr.R400008-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Yang H, Quinones MJ, Bulnes-Enriquez I, Jimenez X, De La Rosa R, Modilevsky T, Yu K, Li Y, Taylor KD, Hsueh WA, Hodis HN, Rotter JI. A genome-wide scan for carotid artery intima-media thickness: the Mexican-American Coronary Artery Disease family study. Stroke. 2005;36(3):540–545. doi: 10.1161/01.STR.0000155746.65185.4e. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi T, Zadravec D, Jacobsson A. ELOVL2 overexpression enhances triacylglycerol synthesis in 3T3-L1 and F442A cells. FEBS Lett. 2007;581(17):3157–3163. doi: 10.1016/j.febslet.2007.05.081. [DOI] [PubMed] [Google Scholar]
- 31.Iwamoto N, Abe-Dohmae S, Ayaori M, Tanaka N, Kusuhara M, Ohsuzu F, Yokoyama S. ATP-binding cassette transporter A1 gene transcription is downregulated by activator protein 2alpha. Doxazosin inhibits activator protein 2alpha and increases high-density lipoprotein biogenesis independent of alpha1-adrenoceptor blockade. Circ Res. 2007;101(2):156–165. doi: 10.1161/CIRCRESAHA.107.151746. [DOI] [PubMed] [Google Scholar]
- 32.Hsiao CF, Chiu YF, Chiang FT, Ho LT, Lee WJ, Hung YJ, Chen YD, Donlon TA, Jorgenson E, Curb D, Risch N, Hsiung CA. Genome-wide linkage analysis of lipids in nondiabetic Chinese and Japanese from the SAPPHIRe family study. Am J Hypertens. 2006;19(12):1270–1277. doi: 10.1016/j.amjhyper.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Arya R, Duggirala R, Almasy L, Rainwater DL, Mahaney MC, Cole S, Dyer TD, Williams K, Leach RJ, Hixson JE, MacCluer JW, O'Connell P, Stern MP, Blangero J. Linkage of high-density lipoprotein-cholesterol concentrations to a locus on chromosome 9p in Mexican Americans. Nat Genet. 2002;30(1):102–105. doi: 10.1038/ng810. [DOI] [PubMed] [Google Scholar]
- 34.Rust S, Walter M, Funke H, von Eckardstein A, Cullen P, Kroes HY, Hordijk R, Geisel J, Kastelein J, Molhuizen HO, Schreiner M, Mischke A, Hahmann HW, Assmann G. Assignment of Tangier disease to chromosome 9q31 by a graphical linkage exclusion strategy. Nat Genet. 1998;20(1):96–98. doi: 10.1038/1770. [DOI] [PubMed] [Google Scholar]
- 35.Li WD, Dong C, Li D, Garrigan C, Price RA. A genome scan for serum triglyceride in obese nuclear families. J Lipid Res. 2005;46(3):432–438. doi: 10.1194/jlr.M400391-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Tan JH, Low PS, Tan YS, Tong MC, Saha N, Yang H, Heng CK. ABCA1 gene polymorphisms and their associations with coronary artery disease and plasma lipids in males from three ethnic populations in Singapore. Hum Genet. 2003;113(2):106–117. doi: 10.1007/s00439-003-0943-3. [DOI] [PubMed] [Google Scholar]
- 37.Austin MA, Edwards KL, Monks SA, Koprowicz KM, Brunzell JD, Motulsky AG, Mahaney MC, Hixson JE. Genome-wide scan for quantitative trait loci influencing LDL size and plasma triglyceride in familial hypertriglyceridemia. J Lipid Res. 2003;44(11):2161–2168. doi: 10.1194/jlr.M300272-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Yu Y, Wyszynski DF, Waterworth DM, Wilton SD, Barter PJ, Kesaniemi YA, Mahley RW, McPherson R, Waeber G, Bersot TP, Ma Q, Sharma SS, Montgomery DS, Middleton LT, Sundseth SS, Mooser V, Grundy SM, Farrer LA. Multiple QTLs influencing triglyceride and HDL and total cholesterol levels identified in families with atherogenic dyslipidemia. J Lipid Res. 2005;46(10):2202–2213. doi: 10.1194/jlr.M500137-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Arnett DK, Miller MB, Coon H, Ellison RC, North KE, Province M, Leppert M, Eckfeldt JH. Genome-wide linkage analysis replicates susceptibility locus for fasting plasma triglycerides: NHLBI Family Heart Study. Hum Genet. 2004;115(6):468–474. doi: 10.1007/s00439-004-1182-y. [DOI] [PubMed] [Google Scholar]
- 40.Almasy L, Hixson JE, Rainwater DL, Cole S, Williams JT, Mahaney MC, VandeBerg JL, Stern MP, MacCluer JW, Blangero J. Human pedigree-based quantitative-trait-locus mapping: localization of two genes influencing HDL-cholesterol metabolism. Am J Hum Genet. 1999;64(6):1686–1693. doi: 10.1086/302425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai SJ, Wong DM, Chen SH, Chan L. Structure of the human hepatic triglyceride lipase gene. Biochemistry. 1989;28(23):8966–8971. doi: 10.1021/bi00449a002. [DOI] [PubMed] [Google Scholar]
- 42.Baroni MG, Berni A, Romeo S, Arca M, Tesorio T, Sorropago G, Di Mario U, Galton DJ. Genetic study of common variants at the Apo E, Apo AI, Apo CIII, Apo B, lipoprotein lipase (LPL) and hepatic lipase (LIPC) genes and coronary artery disease (CAD): variation in LIPC gene associates with clinical outcomes in patients with established CAD. BMC Med Genet. 2003;4:8. doi: 10.1186/1471-2350-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moennig G, Wiebusch H, Enbergs A, Dorszewski A, Kerber S, Schulte H, Vielhauer C, Haverkamp W, Assmann G, Breithardt G, Funke H. Detection of missense mutations in the genes for lipoprotein lipase and hepatic triglyceride lipase in patients with dyslipidemia undergoing coronary angiography. Atherosclerosis. 2000;149(2):395–401. doi: 10.1016/s0021-9150(99)00330-5. [DOI] [PubMed] [Google Scholar]
- 44.Rainwater DL, Almasy L, Blangero J, Cole SA, VandeBerg JL, MacCluer JW, Hixson JE. A genome search identifies major quantitative trait loci on human chromosomes 3 and 4 that influence cholesterol concentrations in small LDL particles. Arterioscler Thromb Vasc Biol. 1999;19(3):777–783. doi: 10.1161/01.atv.19.3.777. [DOI] [PubMed] [Google Scholar]
- 45.Pajukanta P, Terwilliger JD, Perola M, Hiekkalinna T, Nuotio I, Ellonen P, Parkkonen M, Hartiala J, Ylitalo K, Pihlajamaki J, Porkka K, Laakso M, Viikari J, Ehnholm C, Taskinen MR, Peltonen L. Genomewide scan for familial combined hyperlipidemia genes in finnish families, suggesting multiple susceptibility loci influencing triglyceride, cholesterol, and apolipoprotein B levels. Am J Hum Genet. 1999;64(5):1453–1463. doi: 10.1086/302365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaefer EJ, McNamara JR, Tayler T, Daly JA, Gleason JL, Seman LJ, Ferrari A, Rubenstein JJ. Comparisons of effects of statins (atorvastatin, fluvastatin, lovastatin, pravastatin, and simvastatin) on fasting and postprandial lipoproteins in patients with coronary heart disease versus control subjects. Am J Cardiol. 2004;93(1):31–39. doi: 10.1016/j.amjcard.2003.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.