The JmjC domain-containing histone demethylase NO66 from H. sapiens was overproduced in E. coli, purified and crystallized. Diffraction data were collected to 2.29 Å resolution.
Keywords: histone demethylases, NO66, JmjC domain
Abstract
NO66 is a JmjC domain-containing histone demethylase with specificity towards histone H3 methylated on both Lys4 and Lys36 in vitro and in vivo. A fragment of NO66 lacking the N-terminal 167 amino-acid residues was overexpressed in Escherichia coli, purified and crystallized using the sitting-drop vapour-diffusion method. X-ray diffraction data were collected to a resolution of 2.29 Å. NO66 crystallized in space group P31 or P32, with unit-cell parameters a = 89.35, b = 89.35, c = 304.86 Å, α = β = 90, γ = 120°, and the crystal is likely to contain four molecules in the asymmetric unit.
1. Introduction
Covalent modifications of histone tails, including methylation, acetylation, phosphorylation, ubiquitylation, sumoylation and ADP-ribosylation, are crucial epigenetic markers that are involved in the regulation of diverse cellular processes (Kouzarides, 2007 ▶). Histone lysine methylation is one such modification and plays important roles in transcriptional regulation, X-chromosome inactivation, DNA damage response and heterochromatin formation. The biological outcomes of histone lysine methylation depend on the methylation sites and states. For example, methylated histone H3K9 is usually considered to be a repressive marker of gene transcription, whereas methylated histone H3K4 is enriched in the promoter regions of actively transcribed genes (Martin & Zhang, 2005 ▶; Varier & Timmers, 2011 ▶). Histone lysine methylation is dynamically regulated by histone lysine methyltransferases and histone demethylases. Recently, two families of histone demethylases have been identified: the amine oxidase-related enzymes and the JmjC domain-containing enzymes. The JmjC domain-containing proteins are members of the 2-OG oxygenase family of enzymes that use Fe2+ as a cofactor and oxygen and 2-oxoglutarate as cosubstrates (Hausinger, 2004 ▶; McDonough et al., 2010 ▶; Clifton et al., 2006 ▶).
NO66 is a JmjC domain-containing protein that is highly conserved in eukaryotes and is located in both the nucleolus and the nucleoplasm (Eilbracht et al., 2004 ▶). NO66 has been shown to be a histone demethylase which is specific towards both methylated H3K4 and H3K36 in vitro and in vivo. It has been shown that NO66 lacking the N-terminal 167 amino acids can interact with Osterix (Osx), a key transcription factor controlling osteoblast differentiation. NO66 can inhibit the expression of Osx target genes through the interaction between these two proteins (Sinha et al., 2010 ▶). However, NO66 contains only a JmjC domain; other recognizable modules such as PHD, Tudor and ARID domains that are responsible for interaction with DNA/RNA or proteins are absent. The detailed mechanism of interaction between NO66 and Osx that contributes to the repression of Osx-dependent genes remains unclear. In addition, how NO66 recognizes H3K4 and H3K36 also remains elusive. Here, we cloned, expressed, purified and crystallized human NO66 lacking the N-terminal 167 amino acids (referred to as c-hNO66). Preliminary crystallographic analysis of a c-hNO66 crystal was carried out. Structural and biochemical analysis may lead to an understanding of how NO66 recognizes its substrates and interacts with Osx.
2. Materials and methods
2.1. Cloning
The pET-28a-6×His-SUMO (small ubiquitin-related modifier) vector used below was derived from pET-28a(+) (Novagen) as described by Malakhov et al. (2004 ▶). The nucleotide fragment in pET-28a(+) between NcoI and NdeI was replaced by a DNA fragment encoding a 6×His-SUMO tag, which can be efficiently cleaved by Ulp1 (SUMO protease 1; Malakhov et al., 2004 ▶).
c-hNO66 (Table 1 ▶) was amplified by PCR using the cDNA of human NO66 (a gift from Dr Benoit de Crombrugghe) as template with the primer pair 5′-GAAAGATCTTCGGGGCCGCAGGTGGAT-3′ (forward) and 5′-GAACTCGAGCTAATTTAGGGCTAGAGGCATCTT-3′ (reverse) (Sangon). The PCR product was digested with BglII/XhoI and ligated into the pET-28a-6×His-SUMO vector. The resulting plasmid was confirmed by DNA sequencing (Amphipound Biotech).
Table 1. Amino-acid sequence of NO66.
The sequence of c-hNO66 is shown in bold.
| Protein | Lysine-specific demethylase NO66 (Homo sapiens), gi:106879206, NP_078920.2 |
|---|---|
| Sequence | MDGLQASAGPLRRGRPKRRRKPQPHSGSVLALPLRSRKIRKQLRSVVSRMAALRTQTLPSENSEESRVESTADDLGDALPGGAAVAAVPDAARREPYGHLGPAELLEASPAARSLQTPSARLVPASAPPARLVEVPAAPVRVVETSALLCTAQHLAAVQSSGAPATASGPQVDNTGGEPAWDSPLRRVLAELNRIPSSRRRAARLFEWLIAPMPPDHFYRRLWEREAVLVRRQDHTYYQGLFSTADLDSMLRNEEVQFGQHLDAARYINGRRETLNPPGRALPAAAWSLYQAGCSLRLLCPQAFSTTVWQFLAVLQEQFGSMAGSNVYLTPPNSQGFAPHYDDIEAFVLQLEGRKLWRVYRPRVPTEELALTSSPNFSQDDLGEPVLQTVLEPGDLLYFPRGFIHQAECQDGVHSLHLTLSTYQRNTWGDFLEAILPLAVQAAMEENVEFRRGLPRDFMDYMGAQHSDSKDPRRTAFMEKVRVLVARLGHFAPVDAVADQRAKDFIHDSLPPVLTDRERALSVYGLPIRWEAGEPVNVGAQLTTETEVHMLQDGIARLVGEGGHLFLYYTVENSRVYHLEEPKCLEIYPQQADAMELLLGSYPEFVRVGDLPCDSVEDQLSLATTLYDKGLLLTKMPLALN |
2.2. Overexpression and purification
The expression plasmid was transformed into Escherichia coli Rosetta 2 (DE3) cells. E. coli cells containing the target plasmid were grown in LB medium supplemented with 50 µg ml−1 kanamycin at 310 K until the OD600 nm reached 0.6. Protein overexpression was induced by 0.4 mM isopropyl β-d-1-thiogalactopyranoside for 20 h at 289 K. The cells were harvested by centrifugation at 6000g for 8 min at 277 K. The cell pellets were resuspended in lysis buffer [50 mM Tris–HCl, 500 mM NaCl, 5 mM imidazole, 2 mM β-mercaptoethanol, 5%(v/v) glycerol, pH 7.5]. The cells were lysed by ultrasonication on ice and the lysate was centrifuged at 15 000 rev min−1 for 30 min at 277 K. The supernatant was loaded onto an Ni–NTA nickel-chelating column (Qiagen) pre-equilibrated with lysis buffer. The column was washed with about 20 column volumes of washing buffer [50 mM Tris–HCl, 500 mM NaCl, 40 mM imidazole, 2 mM β-mercaptoethanol, 5%(v/v) glycerol, pH 7.5] to remove contaminants. The resin with target protein was suspended in 10 ml washing buffer. Ulp1, which was expressed and purified as described by Malakhov et al. (2004 ▶), was added to the solution at a molar ratio of 1:100. The mixture was rotated at 277 K overnight to remove the His-SUMO tag. The cleaved protein was again passed through an Ni–NTA nickel-chelating column. The flowthrough fraction was concentrated and applied onto a Superdex 200 10/300 GL size-exclusion column (GE Healthcare) equilibrated with column buffer [20 mM Tris–HCl, 200 mM NaCl, 1 mM dl-dithiothreitol, 1 mM ethylenediaminetetraacetic acid disodium salt, pH 7.5]. All purification steps were performed at 277 K and the result of each step was monitored by SDS–PAGE. The resulting protein was concentrated to 1.8 mg ml−1 by centrifugal ultrafiltration (Millipore; 5 kDa cutoff) for crystallization experiments.
2.3. Crystallization
Initial crystallization trials were carried out using Crystal Screen, Crystal Screen 2, Index (Hampton Research), Wizard I and Wizard II (Emerald BioSystems) at 295 K by the sitting-drop vapour-diffusion method in 48-well plates (XtalQuest Co.). 1 µl protein solution (at 1.8 mg ml−1 in column buffer) was mixed with an equal volume of reservoir solution and equilibrated against 100 µl reservoir solution. After 4 d, microcrystals of c-hNO66 were obtained from several different conditions. The crystallization conditions were optimized and crystals suitable for X-ray diffraction studies were obtained.
2.4. X-ray data collection and processing
The crystals of c-hNO66 were soaked in cryoprotectant solution consisting of 10% PEG 8000, 200 mM ammonium sulfate, 0.1 M Tris–HCl pH 7.0, 20% glycerol for several seconds. The crystals were flash-cooled in liquid nitrogen and subjected to X-ray diffraction data collection at 100 K on beamline BL17U1 at Shanghai Synchrotron Radiation Facility (SSRF). 230 diffraction images were recorded from a single crystal with an oscillation angle of 0.5° per image. Diffraction data were processed using the HKL-2000 program package (v.0.98.699; Otwinowski & Minor, 1997 ▶). The statistics of diffraction data are shown in Table 2 ▶.
Table 2. Crystal parameters and data-collection statistics for the crystal of c-hNO66.
Values in parentheses are for the highest resolution shell.
| No. of crystals | 1 |
| Beamline | BL17U, SSRF |
| Wavelength (Å) | 0.9999 |
| Detector | ADSC Quantum 315r |
| Crystal-to-detector distance (mm) | 200 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 115 |
| Exposure time per image (s) | 1.6 |
| Resolution range (Å) | 50–2.29 (2.33–2.29) |
| Space group | P31 or P32 |
| Unit-cell parameters (Å, °) | a = 89.35, b = 89.35, c = 304.86, α = β = 90, γ = 120 |
| Mosaicity (°) | 0.33 |
| Total No. of measured intensities | 1602752 |
| Unique reflections | 123682 |
| Multiplicity | 3.3 (2.7) |
| Average I/σ(I) | 11.0 (2.0) |
| Completeness (%) | 98.9 (99.9) |
| R merge † (%) | 10.8 (55.1) |
| Overall B factor from Wilson plot (Å2) | 44.7 |
R
merge = 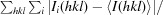
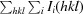 , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity of all observations i of reflection hkl.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity of all observations i of reflection hkl.
3. Results and discussion
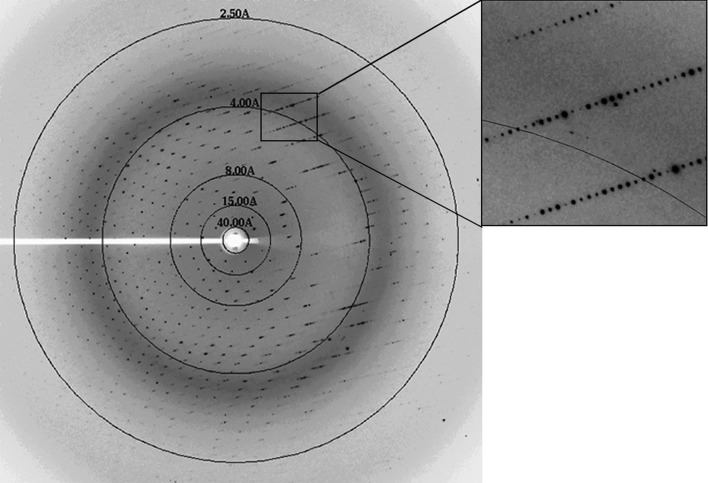
The purified c-hNO66 was concentrated to 1.8 mg ml−1 for initial crystallization experiments. 4 d after the initial crystal screening experiment was set up, microcrystals were observed in condition No. 10 of Wizard I (20% PEG 2000 MME, 0.1 M Tris–HCl pH 7.0), No. 32 of Wizard II (20% PEG 1000, 0.1 M Tris–HCl pH 8.5), No. 34 of Wizard II (10% PEG 8000, 0.1 M imidazole pH 8.0) and No. 69 of Index (25% PEG 3350, 0.2 M ammonium sulfate, 0.1 M Tris–HCl pH 8.5). To obtain crystals suitable for X-ray diffraction studies, the initial crystallization conditions were optimized. A linear gradient of PEG 8000, PEG 2000, PEG 1000, PEG 3350 and pH was screened and different salts were used as additives. These experiments were performed at 281 or 295 K. Finally, large single crystals were obtained using 10% PEG 8000, 200 mM ammonium sulfate, 0.1 M Tris–HCl pH 7.0 at 295 K. Typical crystal dimensions were 0.2 × 0.1 × 0.05 mm (Fig. 1 ▶). The crystals diffracted to 2.29 Å resolution (Fig. 2 ▶) and belonged to space group P31 or P32. The calculated Matthews coefficient (V M) was 3.25 Å3 Da−1, suggesting that there may be four molecules in the asymmetric unit (Matthews, 1968 ▶).
Figure 1.
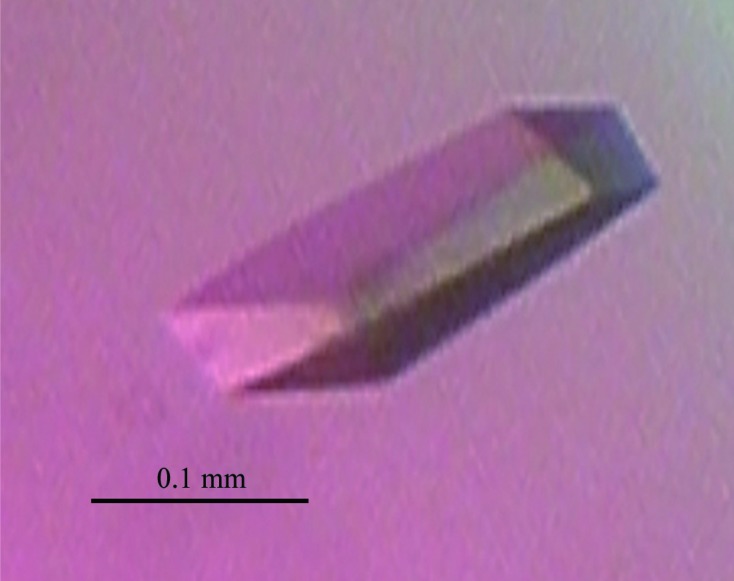
Crystal of c-hNO66.
Figure 2.
X-ray diffraction image from the crystal of c-hNO66. A section of the image is enlarged in the box.
Acknowledgments
We thank Dr Benoit de Crombrugghe for providing the cDNA of human NO66. We thank the staff at beamline BL17U1 of SSRF for assistance with data collection. This work was supported by grants from the National Natural Science Foundation of China (Nos. 31171241 and 30970576), grants from the Chinese Ministry of Science and Technology (Nos. 2009CB825502 and 2012CB917200), the PhD Programs Foundation of the Ministry of Education of China (No. 20113402110033) and the ‘100 Talents Program’ of the Chinese Academy of Sciences to JZ.
References
- Clifton, I. J., McDonough, M. A., Ehrismann, D., Kershaw, N. J., Granatino, N. & Schofield, C. J. (2006). J. Inorg. Biochem. 100, 644–669. [DOI] [PubMed]
- Eilbracht, J., Reichenzeller, M., Hergt, M., Schnölzer, M., Heid, H., Stöhr, M., Franke, W. W. & Schmidt-Zachmann, M. S. (2004). Mol. Biol. Cell, 15, 1816–1832. [DOI] [PMC free article] [PubMed]
- Hausinger, R. P. (2004). Crit. Rev. Biochem. Mol. Biol. 39, 21–68. [DOI] [PubMed]
- Kouzarides, T. (2007). Cell, 128, 693–705. [DOI] [PubMed]
- Malakhov, M. P., Mattern, M. R., Malakhova, O. A., Drinker, M., Weeks, S. D. & Butt, T. R. (2004). J. Struct. Funct. Genomics, 5, 75–86. [DOI] [PubMed]
- Martin, C. & Zhang, Y. (2005). Nature Rev. Mol. Cell Biol. 6, 838–849. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McDonough, M. A., Loenarz, C., Chowdhury, R., Clifton, I. J. & Schofield, C. J. (2010). Curr. Opin. Struct. Biol. 20, 659–672. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sinha, K. M., Yasuda, H., Coombes, M. M., Dent, S. Y. & de Crombrugghe, B. (2010). EMBO J. 29, 68–79. [DOI] [PMC free article] [PubMed]
- Varier, R. A. & Timmers, H. T. (2011). Biochim. Biophys. Acta, 1815, 75–89. [DOI] [PubMed]