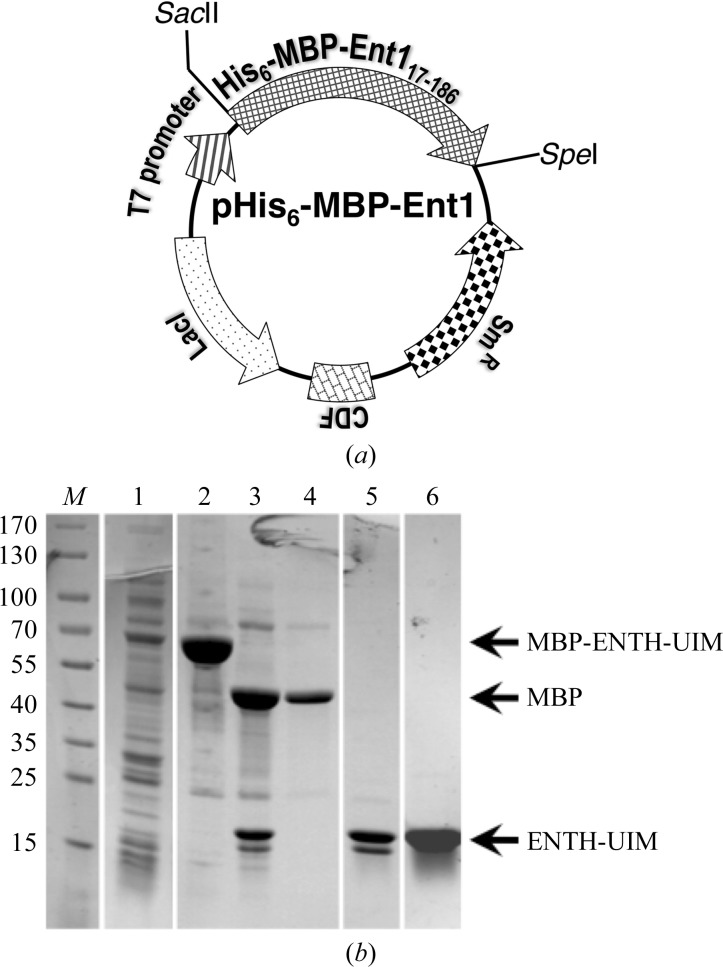
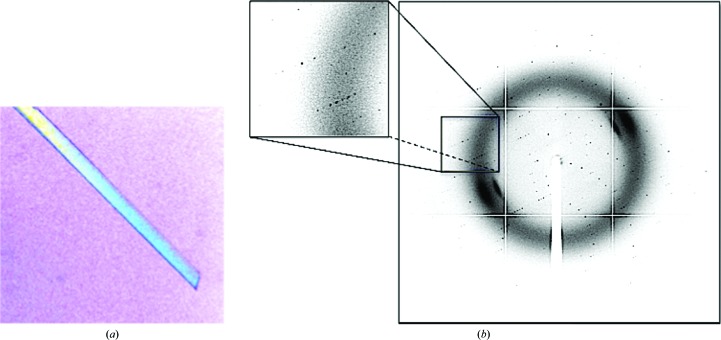
The yeast Epsin-1 (ent1) gene has been cloned and expressed in Escherichia coli. The protein product of a construct containing the ENTH-UIM modules was purified to homogeneity and subjected to crystallization screening using the sitting-drop vapour-diffusion method. Refined conditions containing polyethylene glycol 3350 and Tacsimate yielded thin rod-like crystals.
Keywords: Epsin, ENTH domain, ubiquitylation/clathrin-dependent endocytosis
Abstract
Members of the Epsin protein family regulate the ubiquitin/clathrin-dependent trafficking of transmembrane proteins. The yeast Epsin-1 (ent1) gene was cloned and expressed in Escherichia coli. The protein product of a construct containing the ENTH-UIM modules was purified to homogeneity and subjected to crystallization screening using the sitting-drop vapour-diffusion method. Refined conditions containing polyethylene glycol 3350 and Tacsimate yielded thin rod-like crystals. X-ray analysis revealed that the crystallographic symmetry is primitive orthorhombic, space group P222, with unit-cell parameters a = 32.7, b = 35.5, c = 110.6 Å and a diffraction limit of 2.3 Å. Matthews coefficient calculations suggested that the crystal contained only the ENTH domain. This was corroborated by Coomassie Blue-stained SDS–PAGE analysis of dissolved crystals.
1. Introduction
Pivotal accessory factors such as Eps15 and Epsin (Eps15-interacting protein) regulate the function of the endocytic machinery complex (Maldonado-Báez et al., 2008 ▶; McMahon & Boucrot, 2011 ▶). Epsin is composed of several modules that interact with key components of the endocytic machinery complex. The first module is the Epsin N-terminus homology domain (ENTH), which interacts with Cdc42 (Aguilar et al., 2006 ▶) and with phosphatidylinositol 4,5-bisphosphate and also induces membrane curvature (Ford et al., 2002 ▶). The second module is a tandem of several ubiquitin-interaction motifs (UIMs; Young et al., 1998 ▶) that are tethered to the ENTH domain by a flexible hinge and participate in self-ubiquitylation. The third module, the Asn-Pro-Phe (NPF) tripeptide motifs (de Beer et al., 1998 ▶), is located downstream of the UIMs in a large unstructured sequence and promotes binding to Eps15 homology (EH) domains. Finally, a conserved LLDLD-like motif at the carboxyl-terminus binds to a groove in the β-propeller domain of clathrin. This last module recruits clathrin to ubiquitylated transmembrane cargo and thus links the ubiquitylation signal to an endocytic response (ter Haar et al., 2000 ▶). Epsin therefore acts as a ubiquitin receptor that senses and decodes the ubiquitylation signal into cellular responses such as the trafficking of transmembrane ubiquitylated proteins including receptors, transporters and channels. The general architecture of the Epsin protein family is conserved from yeast to human with multiple paralogues in each organism, providing high robustness to the trafficking protein network (Amit et al., 2007 ▶; McMahon & Boucrot, 2011 ▶). Indeed, yeast Ent1 has been shown to complement a quadruple mutant lacking ent1/2 and yap1801/2 (Aguilar et al., 2006 ▶; Dores et al., 2010 ▶). Moreover, removal of the region harbouring the UIM, NPF and LLDLD-like motifs, leaving the protein as the ENTH domain alone, is sufficient for viability and endocytosis in yeast (Aguilar et al., 2006 ▶). The advanced studies provided by yeast genetics have significantly promoted our understanding of the function and interactions of the Epsin protein family. However, while the structures of several ENTH domains from high eukaryotes, including rat, mouse and human, have been determined (PDB entries 1h0a, 1eyh and 1edu for Rattus norvegicus and 1inz for Homo sapiens; Ford et al., 2002 ▶; D. H. Fremont, unpublished work; Hyman et al., 2000 ▶; Koshiba et al., 2001 ▶), no structural information is available for evolutionarily distant proteins such as the yeast ENTH domain. Sequence alignment of ENTH domains revealed only 23–49% identity between the yeast Ent1 ENTH domain and those of known structure. To fill this gap in knowledge, we decided to determine the structure of yeast Ent1. Here, we report the cloning, purification, crystallization and X-ray data collection of the ENTH domain of yeast Ent1.
2. Experimental procedures
2.1. Cloning of the yeast Ent1 gene
The open reading frame (ORF; encoding residues 17–186) for Saccharomyces cerevisiae ent1 was amplified by polymerase chain reaction (PCR) from yeast genomic DNA. The DNA insert was ligated into the SacII–SpeI restriction-endonuclease sites of a modified pCDFDuet vector that harbours His6-MBP (maltose-binding protein) followed by a PreScission protease (human rhinovirus type 14-3C protease) recognition site. The resulting vector expresses a His6-MBP-Ent1 fusion protein that can be affinity-purified sequentially on Ni2+ and amylose affinity matrices. The following oligonucleotide sequences were used to amplify the ent1 ORF: Ent117_SacII_fwd, 5′-TCCCCGCGGTCTTCTACCCAGGTCCTGGTAAGA-3′, and Ent1186_SpeI_rev, 5′-GCGCACTAGTTTAGTCTTCTTCCGCCGTCAACCTGCTA-3′.
2.2. Protein expression and purification
Escherichia coli Rosetta 2 (λDE3) BL21 cells were transformed with pHis6-MBP-Ent117–186 and grown in Terrific Broth medium supplemented with 50 µg ml−1 streptomycin and 34 µg ml−1 chloramphenicol at 310 K. Expression of Ent1 was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and the bacteria were grown for a further ∼20 h at 289 K. Cells were harvested by centrifugation and resuspended in lysozyme buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.0, 0.1 mg ml−1 lysozyme) supplemented with the protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF). Complete lysis and DNA shearing were achieved by sonication. The soluble fraction was isolated by centrifugation. The protein was purified using nickel–nitrilotriacetic acid (Ni–NTA) affinity chromatography according to the manufacturer’s instructions. The affinity tags were removed by rhinovirus protease during dialysis against 150 mM NaCl, 50 mM Tris–HCl pH 7.0. Subsequently, the sample was diluted to a final concentration of 35 mM NaCl, supplemented with 10% glycerol and loaded onto an ion-exchange column (SP Sepharose Fast Flow, GE Healthcare) pre-equilibrated with the same buffer. The column was washed and a salt gradient (35–1000 mM) was applied. The protein eluted at ∼250 mM NaCl. Finally, size-exclusion chromatography (using a Superdex 75 16/60 HiLoad prep-grade column) was used to further increase the protein purity. The protein sample was concentrated to 10 mg ml−1. 100 µl aliquots were flash-cooled in liquid nitrogen and preserved at 193 K.
2.3. Crystallization of the Ent1 ENTH domain
Initial crystallization screens were performed at 292 K using 96-well sitting-drop plates with various commercially available crystallization kits. Small crystals appeared in 20%(w/v) PEG 3350, 0.1 M HEPES pH 7.5, 2%(v/v) Tacsimate (Hampton Research) pH 7.0. To further refine the crystallization conditions, we varied the Tacsimate concentration. Optimal crystal-growth conditions consisted of a protein concentration of 15 mg ml−1 with a reservoir consisting of 20%(w/v) PEG 3350, 0.1 M HEPES pH 7.4, 3–4% Tacsimate pH 7.0 at 292 K. Crystals appeared after about a week and were harvested for diffraction experiments.
2.4. Screening for cryoprotection conditions
Several cryoprotectants were screened in order to identify one that prevents water icing and does not interfere with X-ray diffraction. Compared with Paratone, glycerol and PEG 400, we found that adding ∼25% ethylene glycol to the crystallization solution generally yielded excellent cryoprotection.
2.5. Data collection
Following a short soak in cryoprotection solution, crystals were cooled by immersion in liquid nitrogen. Using an R-AXIS IV++ image plate and a Rigaku MicroMax-007 rotating-anode X-ray generator operating at 40 kV and 20 mA (Cu Kα radiation; λ = 1.5418 Å), crystals were screened for diffraction quality. Crystals that diffracted with high quality were subjected to a second screen on the ID14-1 beamline, ESRF, Grenoble. A full data set was collected at a wavelength of 0.979 Å. Images were indexed and integrated using MOSFLM (v.7.0.6; Leslie, 1992 ▶) and were scaled using SCALA (Evans, 2006 ▶) from the CCP4 program suite (Winn et al., 2011 ▶).
3. Results and discussion
The yeast epsin-1 gene (ent1) was cloned into a bacterial expression vector (Fig. 1 ▶ a). The gene was overexpressed in E. coli and its protein product was purified to homogeneity using standard affinity-chromatography techniques followed by ion-exchange and size-exclusion chromatography (Fig. 1 ▶ b). The purified protein was subjected to crystallization trials using the sitting-drop vapour-diffusion method. Thin rod-shaped crystals grew to 300 µm in their longest dimension (Fig. 2 ▶ a). Cryoprotection conditions were screened using our in-house X-ray facility. Diffraction data were collected from a single crystal at the ESRF, Grenoble (Fig. 2 ▶ b). We obtained a complete data set with d min = 2.3 Å. The images were indexed, merged and scaled using the programs MOSFLM and SCALA, respectively (Table 1 ▶). The diffraction pattern suggested the primitive orthorhombic space group P222. Matthews coefficient calculations yielded a V M value of 1.6 A3 Da−1 when considering a single full-length molecule of the protein construct in the asymmetric unit. This suggests that the unit cell cannot accommodate the entire protein construct. Based on this observation, we speculated that the predicted flexible hinge tethering the ubiquitin-interaction motif (UIM) to the ENTH domain was removed during crystallization. Initial phasing derived from the molecular-replacement method using Phaser (McCoy et al., 2007 ▶) supported this assumption. To verify this postulation, crystals were dissolved and the size of their protein content was analyzed by Coomassie Blue-stained SDS–PAGE (Fig. 3 ▶). We found a single band that migrated at ∼14.5 kDa, corresponding to the ENTH domain alone. The latter result corroborates our assumption that the carboxyl-terminus had been cleaved. The alternative possibility is that the missing part of the crystallized protein is the amino-terminal part. This scenario is unlikely since we found that the Ent1 ENTH domain was highly stable. The quality of the data set appears to be sufficient to determine the protein structure. The structure is predicted to provide a high-resolution insight into the structural determinants that bind the GAPs of the small GTPase cell-division cycle 42 protein (Cdc42p), including Rag1, Rag2 and Bem3, and the specific membrane component phosphatidylinositol 4,5-bisphosphate.
Figure 1.
Cloning, expression and purification of yeast Ent117–186. Yeast Ent1 was expressed from the T7 promoter of a modified pMBP-Ent1 vector as a His6-MBP fusion. (a) Essential elements of the expression vector. (b) Coomassie Blue-stained SDS–PAGE of the Ent117–186 purification steps. Lane M, molecular-mass marker (labelled in kDa). Lane 1, total extract of BL21 cells expressing Ent1. Lane 2, purification on Ni2+ affinity matrix. Lane 3, PreScission protease-cleaved protein. Lane 4, flowthrough of the SP column. Lane 5, elution from the SP column. Lane 6, elution from the size-exclusion column.
Figure 2.
Crystal and diffraction of Ent1. (a) A typical rod-shaped crystal of yeast Ent1. (b) Diffraction pattern from a single yeast Ent1 crystal obtained on the ID14-1 beamline, ESRF, Grenoble.
Table 1. Data-collection statistics.
Values in parentheses are for the last shell.
| Wavelength (Å) | 0.979 |
| Space group | P222 |
| Unit-cell parameters (Å, °) | a = 32.69, b = 35.46, c = 110.62, α = β = γ = 90 |
| Resolution range (Å) | 48.0–2.3 (2.4–2.3) |
| Observed reflections | 22787 (3323) |
| Unique reflections | 6033 (859) |
| Multiplicity | 3.8 (3.9) |
| Completeness (%) | 98.9 (99.6) |
| 〈I/σ(I)〉 | 9.7 (4.5) |
| R merge † (%) | 10.3 (28.1) |
R
merge = 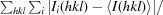
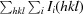 , where Ii(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the average intensity of reflection hkl.
, where Ii(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the average intensity of reflection hkl.
Figure 3.
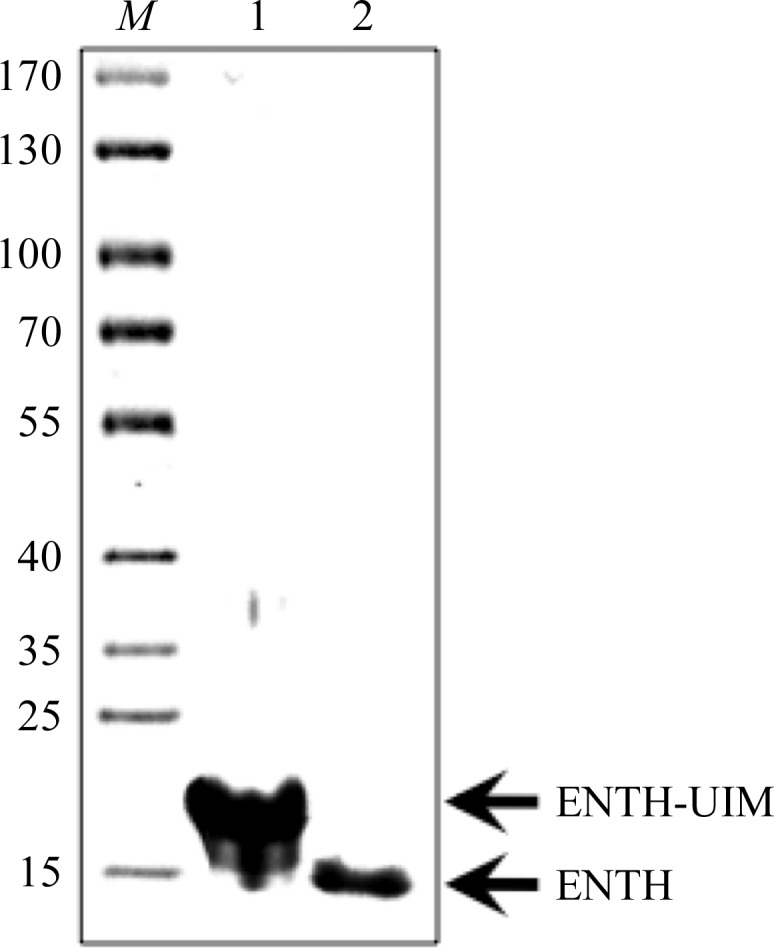
Ent1 crystal content. Coomassie Blue-stained SDS–PAGE showing the content of the crystals. Lane M, molecular-mass marker (labelled in kDa). Lane 1 contains the protein used in the drop. Lane 2 contains crystals extensively washed in mother liquor and dissolved. Faster migration of the protein from the dissolved crystal by about 4 kDa is clearly evident.
Acknowledgments
We thank the staff of the ESRF, Grenoble, France for their exceptional help and for maintaining and upgrading the facility. This work was supported by grants from the Israeli Science Foundation (Nos. 1695/08 and 464/11), from the EC FP7 Marie Curie International Reintegration Grant (No. PIRG03-GA-2008-231079) and from the Israeli Ministry of Health (No. 3000005108) to GP.
References
- Aguilar, R. C., Longhi, S. A., Shaw, J. D., Yeh, L.-Y., Kim, S., Schön, A., Freire, E., Hsu, A., McCormick, W. K., Watson, H. A. & Wendland, B. (2006). Proc. Natl Acad. Sci. USA, 103, 4116–4121. [DOI] [PMC free article] [PubMed]
- Amit, I., Wides, R. & Yarden, Y. (2007). Mol. Syst. Biol. 3, 151. [DOI] [PMC free article] [PubMed]
- Beer, T. de, Carter, R. E., Lobel-Rice, K. E., Sorkin, A. & Overduin, M. (1998). Science, 281, 1357–1360. [DOI] [PubMed]
- Dores, M. R., Schnell, J. D., Maldonado-Baez, L., Wendland, B. & Hicke, L. (2010). Traffic, 11, 151–160. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Ford, M. G., Mills, I. G., Peter, B. J., Vallis, Y., Praefcke, G. J., Evans, P. R. & McMahon, H. T. (2002). Nature (London), 419, 361–366. [DOI] [PubMed]
- Haar, E. ter, Harrison, S. C. & Kirchhausen, T. (2000). Proc. Natl Acad. Sci. USA, 97, 1096–1100. [DOI] [PMC free article] [PubMed]
- Hyman, J., Chen, H., Di Fiore, P. P., De Camilli, P. & Brunger, A. T. (2000). J. Cell Biol. 149, 537–546. [DOI] [PMC free article] [PubMed]
- Koshiba, S., Kigawa, T., Kikuchi, A. & Yokoyama, S. (2001). J. Struct. Funct. Genomics, 2, 1–8. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACMB Newsl. Protein Crystallogr. 26
- Maldonado-Báez, L., Dores, M. R., Perkins, E. M., Drivas, T. G., Hicke, L. & Wendland, B. (2008). Mol. Biol. Cell, 19, 2936–2948. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McMahon, H. T. & Boucrot, E. (2011). Nature Rev. Mol. Cell Biol. 12, 517–533. [DOI] [PubMed]
- Winn, M. D. et al. (2010). Acta Cryst. D67, 235–242.
- Young, P., Deveraux, Q., Beal, R. E., Pickart, C. M. & Rechsteiner, M. (1998). J. Biol. Chem. 273, 5461–5467. [DOI] [PubMed]