To characterize the ISP family of proteins present in apicomplexan parasites, ISP1 from T. gondii was expressed, purified and crystallized. Two crystal forms (cubic and orthorhombic) were analyzed by X-ray diffraction and data were processed to 2.05 and 2.1 Å resolution, respectively.
Keywords: Toxoplasma gondii, inner membrane complex, ISP1
Abstract
The protozoan parasites of the Apicomplexa phylum are devastating global pathogens. Their success is largely due to phylum-specific proteins found in specialized organelles and cellular structures. The inner membrane complex (IMC) is a unique apicomplexan structure that is essential for motility, invasion and replication. The IMC subcompartment proteins (ISP) have recently been identified in Toxoplasma gondii and shown to be critical for replication, although their specific mechanisms are unknown. Structural characterization of TgISP1 was pursued in order to identify the fold adopted by the ISPs and to generate detailed insight into how this family of proteins functions during replication. An N-terminally truncated form of TgISP1 was purified from Escherichia coli, crystallized and subjected to X-ray diffraction analysis. Two crystal forms of TgISP1 belonging to space groups P4132 or P4332 and P212121 diffracted to 2.05 and 2.1 Å resolution, respectively.
1. Introduction
The Apicomplexa are a large phylum of obligate intracellular parasites that represent a major medical and veterinary burden globally (Levine, 1988 ▶). Important human pathogens in this group include Plasmodium spp., the causative agent of malaria, and Toxoplasma gondii, which causes severe disease in immune-compromised patients and congenitally infected neonates (Hill et al., 2005 ▶). The ability of these parasites to infect their mammalian hosts is critically dependent on many unique biological processes and cellular structures, which holds promise for new therapeutic exploitation approaches. One such Apicomplexan-specific structure is a system of flattened vesicles called the inner membrane complex (IMC) that underlies the plasma membrane of the parasite. These vesicles are arranged into a patchwork of rectangular membrane plates with a single cone-shaped plate capping the apex of the structure (Porchet & Torpier, 1977 ▶). The IMC serves as a foundation for a form of gliding motility that is necessary for host-cell invasion and also provides a scaffold for the construction of daughter cells during parasite replication (Keeley & Soldati, 2004 ▶; Striepen et al., 2007 ▶). Despite its key importance in parasite infection and survival, the composition and functions of the protein constituents of the IMC are just beginning to be understood.
Recently, a family of three IMC subcompartment proteins (ISP1, ISP2 and ISP3) has been identified in Toxoplasma (Beck et al., 2010 ▶). The ISP family is conserved throughout the Apicomplexa but is not present outside this phylum, suggesting that these proteins represent unique parasite activities. Indeed, disruption of ISP2 results in major defects during internal budding, indicating a role in coordination of this specialized method of parasite division. The ISP proteins are anchored to the IMC membranes by myristoylation and palmitoylation of conserved N-terminal glycine and cysteine residues, respectively, and are distributed into three distinct subdomains within the organelle. ISP1 localizes exclusively to the cone-shaped apical plate of the IMC, while ISP2 resides in a central IMC compartment that begins at the base of the ISP1 subcompartment and extends about two thirds down the length of the cell. ISP3 spans the central subcompartment and also extends to the far basal end of the IMC. Intriguingly, when ISP1 is genetically ablated, ISP2 and ISP3 are relocalized into the subcompartment normally occupied by ISP1, indicating that a second level of hierarchical targeting exists within these IMC subcompartments. The molecular mechanism of ISP1 gatekeeping activity is unknown, but it depends upon a C-terminal region of ISP1 and this activity cannot be complemented by the homologous C-terminal region of ISP2. The ISPs contain no identifiable domains, hindering the interpretation of the current functional data that has been acquired through genetic approaches. Thus, we took a structural approach in order to identify protein features that may offer important information regarding how these proteins are organized within the IMC and the precise roles that they play.
2. Materials and methods
2.1. Construct design and cloning
The TgISP1 gene was amplified from cDNA of the type II Prugnaud strain of T. gondii and cloned into the NheI–NotI site of a pET28a vector (Novagen) modified to contain an N-terminal hexahistidine tag separated from the TgISP1 sequence by a TEV cleavage site. Two constructs were further subcloned for expression trials: TgISP1-1 and TgISP1-2. TgISP1-1 begins immediately after two N-terminal cysteine residues involved in inner membrane complex targeting and extends through to the C-terminus of the protein. An analysis of the predicted secondary-structure elements of TgISP1, TgISP2 and TgISP3 revealed a core conserved secondary-structure prediction: α-β4-α2-β2-α (McGuffin et al., 2000 ▶). TgISP1-2 encompasses just these ten core predicted elements. Sequence analysis confirmed that no mutations were introduced during amplification procedures.
2.2. Expression and purification of TgISP1-1 and TgISP1-2
TgISP1-1 and TgISP1-2 were recombinantly produced in Escherichia coli BL21 Codon Plus cells (Stratagene) grown in Overnight Express autoinduction medium with 35 µg ml−1 chloramphenicol and 50 µg ml−1 ampicillin (Novagen) from a 5% inoculum. Following 4 h of growth at 310 K and 16 h at 303 K, the cells were harvested by centrifugation and the pellets were resuspended in 20 mM HEPES pH 8.0, 750 mM NaCl, 30 mM imidazole, 2 mM β-mercaptoethanol. Cells in suspension were lysed using a French press, insoluble material was removed by centrifugation and the soluble fraction was applied onto a HisTrap FF 5 ml column (GE Healthcare). Bound TgISP1-1 and TgISP1-2 were eluted with an increasing concentration of imidazole and the His6 tag was subsequently removed from each protein by TEV cleavage overnight at 291 K in HBS (20 mM HEPES pH 7.0, 150 mM NaCl) with 1 mM dithiothreitol (DTT; Sigma) and 0.5 mM EDTA. The TgISP1-1 and TgISP2-2 samples were further purified by gel filtration on a Superdex 75 16/60 HiLoad column (GE Healthcare) in HBS with 1 mM DTT. The elution volumes of TgISP1-1 and TgISP1-2 were compared with the elution profile of a set of globular protein standards to assess the multimeric state. Fractions containing pure sample were pooled and concentrated to a final concentration of 12 mg ml−1 using Amicon Ultra (Millipore) centrifugal filter devices for crystallization trials. The purity of TgISP1-1 and TgISP1-2 was assessed by SDS–PAGE at each stage and protein concentrations were determined by the Bradford assay owing to the absence of tryptophan residues.
2.3. Crystallization, data collection and processing
Initial crystallization trials for TgISP1-1 and TgISP1-2 were set up using commercial screens (JCSG+ from Molecular Dimensions and Index and SaltRx from Hampton Research) in 96-well plates (Emerald BioSystems). The final sitting drops consisted of 1.2 µl protein solution (12 mg ml−1 in HBS with 1 mM DTT) and 1.2 µl reservoir solution and were equilibrated against 100 µl reservoir solution. While TgISP1-1 was refractory to crystallization, two crystal forms of TgISP1-2 were observed. The first form (crystal form 1) was identified in multiple high ammonium sulfate conditions, such as 2.0 M ammonium sulfate with 0.1 M sodium acetate pH 4.6, after 1 d at 291 K. The second crystal form (crystal form 2) was identified only in 0.2 M ammonium sulfate, 0.1 M bis-Tris pH 5.5, 25% PEG 3350 after 7 d at 291 K.
Single TgISP1-2 crystals were looped, stepped into a final cryoprotectant consisting of reservoir solution supplemented with saturated lithium sulfate for crystal form 1 or with 25% glycerol for crystal form 2 and flash-cooled directly in the cryostream (100 K). Diffraction data for TgISP1-2 crystal form 1 were collected on beamline 9-1 at the Stanford Synchrotron Radiation Laboratory (SSRL) using a PILATUS detector and were processed to 2.05 Å resolution. Diffraction data for TgISP1-2 crystal form 2 were collected on a Rigaku R-AXIS IV++ area detector coupled to a Rigaku MicroMax-002 X-ray generator with Osmic Blue optics and an Oxford Cryostream 700 and were processed to 2.1 Å resolution. Diffraction data for both crystal forms were processed using iMOSFLM (Battye et al., 2011 ▶) and SCALA (Evans, 2006 ▶) from the CCP4 suite of programs (Winn et al., 2011 ▶).
Data-collection and processing statistics are presented in Table 1 ▶.
Table 1. Data-collection and processing statistics for TgISP1-2.
Values in parentheses are for the highest resolution shell.
| Crystal form 1 | Crystal form 2 | |
|---|---|---|
| Space group | P4132 or P4332 | P212121 |
| Unit-cell parameters (Å, °) | a = b = c = 129.21, α = β = γ = 90 | a = 58.11, b = 81.09, c = 120.07, α = β = γ = 90 |
| Wavelength | 0.9795 | 1.54 |
| Resolution range (Å) | 43.07–2.05 (2.16–2.05) | 29.09–2.10 (2.21–2.10) |
| Measured reflections | 362703 | 160946 |
| Unique reflections | 23759 | 33884 |
| Multiplicity | 15.3 (13.3) | 4.7 (4.8) |
| Completeness (%) | 100.0 (100.0) | 99.9 (100.0) |
| 〈I/σ(I)〉 | 15.1 (4.8) | 16.3 (4.5) |
| R merge † | 0.102 (0.469) | 0.069 (0.380) |
R
merge = 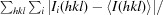
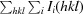 , where 〈I(hkl)〉 is the average of symmetry-related observations of a unique reflection.
, where 〈I(hkl)〉 is the average of symmetry-related observations of a unique reflection.
3. Results and discussion
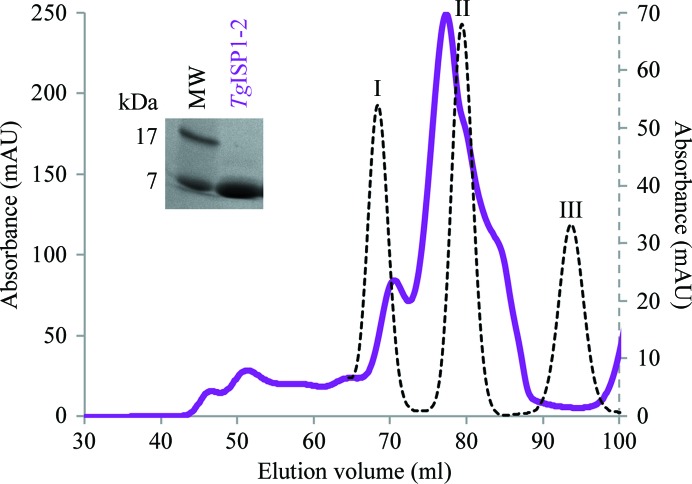
Two constructs of TgISP1 were cloned, produced in E. coli and purified to homogeneity. While the near-complete sequence of TgISP1 (TgISP1-1) was refractory to crystallization, the core domain (TgISP1-2) designed based on secondary-structure prediction analysis of the ISP family was successfully crystallized. Despite the small size of TgISP1 (176 amino acids), seven cysteine residues are present, with two N-terminal cysteines involved in IMC targeting (Beck et al., 2010 ▶). The presence of five cysteines in the small core domain (119 amino acids) suggests a mixture of free cysteines and disulfide bonds, which may explain the multiple closely related species of TgISP1-2 that were reproducibly observed during gel filtration (Fig. 1 ▶). The low theoretical pI of TgISP1-2 (5.8) is likely to be the cause of its fast migration during SDS–PAGE (expected molecular weight of 13.9 kDa; Fig. 1 ▶, inset).
Figure 1.
Purification of TgISP1-2 by gel-filtration chromatography on a Superdex 75 16/60 HiLoad column. Standards (plotted against secondary axis, dotted curve): peak I, carbonic anhydrase (29 kDa); peak II, ribonuclease A (13.7 kDa); peak III, aprotinin (6.5 kDa). TgISP1-2 (solid purple curve) elutes as a monomer with conformational impurity. Inset: SDS–PAGE analysis of a representative fraction showing a low apparent molecular weight of 7 kDa (the expected molecular weight is 13.9 kDa).
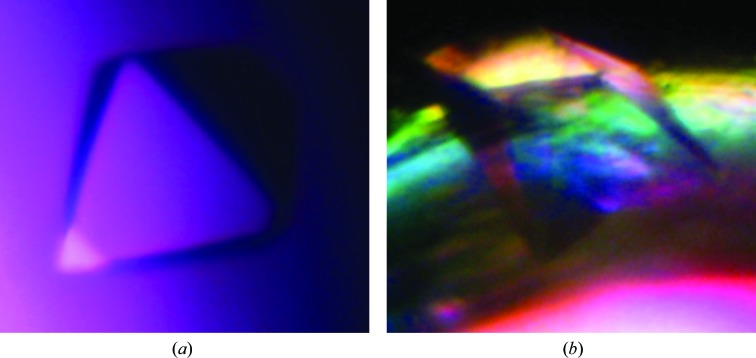
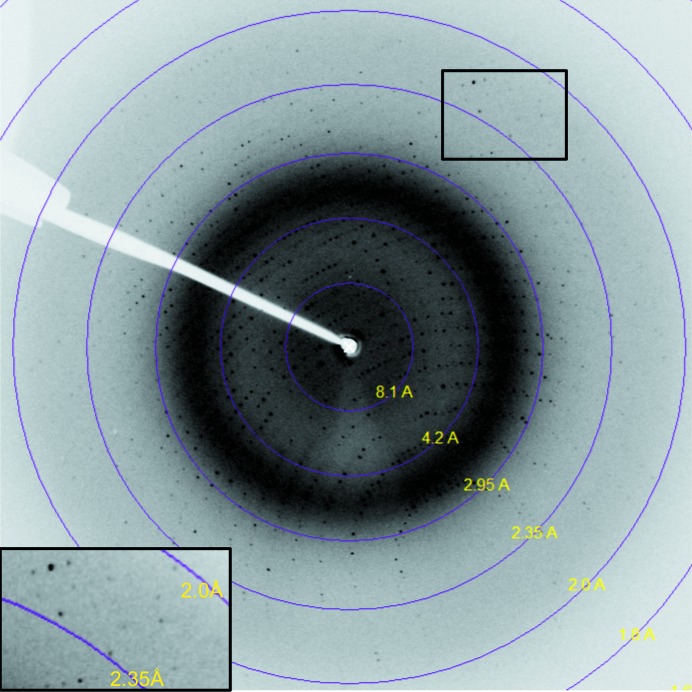
Crystallization trials with the redox-sensitive TgISP1-2 led to two high-quality crystal forms (Fig. 2 ▶). The first crystal form diffracted to 2.05 Å resolution on SSRL beamline 9-1. Processing of these data suggested that TgISP1-2 crystallized with three molecules in the asymmetric unit of the cubic unit cell, with a solvent content of 43.05% and a Matthews coefficient of 2.16 Å3 Da−1 (Matthews, 1968 ▶). The second crystal form diffracted to 2.1 Å resolution using an in-house X-ray generator (Fig. 3 ▶). Processing of these data suggested that TgISP1-2 crystallized with four molecules in the asymmetric unit of the orthorhombic unit cell, with a solvent content of 49.56% and a Matthews coefficient of 2.44 Å3 Da−1 (Matthews, 1968 ▶). Attempts to solve the structure of TgISP1-2 in both crystal forms by molecular replacement have thus far been unsuccessful. Therefore, we are currently pursuing selenomethionine and bromide phasing strategies.
Figure 2.
Crystallization of TgISP1-2. (a) TgISP1-2 crystal form 1 grown in a sitting drop with 2.0 M ammonium sulfate, 0.1 M sodium acetate pH 4.6 as the reservoir solution. (b) TgISP1-2 crystal form 2 nucleated from the wall of a sitting-drop well with 0.2 M ammonium sulfate, 0.1 M bis-tris pH 5.5, 25% PEG 3350 as the reservoir solution.
Figure 3.
X-ray diffraction image from TgISP1-2 crystal form 2. Inset: high-resolution area; data were processed to 2.1 Å resolution.
Acknowledgments
This work was supported by research grant MOP82915 from the Canadian Institutes for Health Research (CIHR) to MJB. MJB is a Canada Research Chair and a Michael Smith Foundation for Health Research (MSFHR) scholar. MLT is supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Alexander Graham Bell Canada Graduate Scholarship (CGSD3).
References
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Beck, J. R., Rodriguez-Fernandez, I. A., Cruz de Leon, J., Huynh, M.-H., Carruthers, V. B., Morrissette, N. S. & Bradley, P. J. (2010). PLoS Pathog. 6, e1001094. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Hill, D. E., Chirukandoth, S. & Dubey, J. P. (2005). Anim. Health Res. Rev. 6, 41–61. [DOI] [PubMed]
- Keeley, A. & Soldati, D. (2004). Trends Cell Biol. 14, 528–532. [DOI] [PubMed]
- Levine, N. D. (1988). J. Protozool. 35, 518–520. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McGuffin, L. J., Bryson, K. & Jones, D. T. (2000). Bioinformatics, 16, 404–405. [DOI] [PubMed]
- Porchet, E. & Torpier, G. (1977). Z. Parasitenkd. 54, 101–124. [DOI] [PubMed]
- Striepen, B., Jordan, C. N., Reiff, S. & van Dooren, G. G. (2007). PLoS Pathog. 3, e78. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.