Abstract
To identify lead candidate allele-independent hammerhead ribozymes (hhRz) for the treatment of autosomal dominant mutations in the human rod opsin (RHO) gene, we tested a series of hhRzs for potential to significantly knockdown human RHO gene expression in a human cell expression system. Multiple computational criteria were used to select target mRNA regions likely to be single stranded and accessible to hhRz annealing and cleavage. Target regions are tested for accessibility in a human cell culture expression system where the hhRz RNA and target mRNA and protein are coexpressed. The hhRz RNA is embedded in an adenoviral VAI RNA chimeric RNA of established structure and properties which are critical to the experimental paradigm. The chimeric hhRz-VAI RNA is abundantly transcribed so that the hhRzs are expected to be in great excess over substrate mRNA. HhRz-VAI traffics predominantly to the cytoplasm to colocalize with the RHO mRNA target. Colocalization is essential for second-order annealing reactions. The VAI chimera protects the hhRz RNA from degradation and provides for a long half life. With cell lines chosen for high transfection efficiency and a molar excess of hhRz plasmid over target plasmid, the conditions of this experimental paradigm are specifically designed to evaluate for regions of accessibility of the target mRNA in cellulo. Western analysis was used to measure the impact of hhRz expression on RHO protein expression. Three lead candidate hhRz designs were identified that significantly knockdown target protein expression relative to control (p < 0.05). Successful lead candidates (hhRz CUC↓ 266, hhRz CUC↓ 1411, hhRz AUA↓ 1414) targeted regions of human RHO mRNA that were predicted to be accessible by a bioinformatics approach, whereas regions predicted to be inaccessible supported no knockdown. The maximum opsin protein level knockdown is approximately 30% over a 48 hr paradigm of testing. These results validate a rigorous computational bioinformatics approach to detect accessible regions of target mRNAs in cellulo. The opsin knockdown effect could prove to be clinically significant when integrated over longer periods in photoreceptors. Further optimization and animal testing is the next step in this stratified RNA drug discovery program. A recently developed novel and efficient screening assay based upon expression of a dicistronic mRNA (RHO-IRES-SEAP) containing both RHO and reporter (SEAP) cDNAs was used to compare the hhRz 266 lead candidate to another agent (Rz525/hhRz485) already known to partially rescue retinal degeneration in a rodent model. Lead hhRz 266 CUC↓ proved more efficacious than Rz525/hhRz485 which infers viability for rescue of retinal degeneration in appropriate preclinical models of disease.
Keywords: gene therapy, ribozyme, photoreceptor degeneration, rhodopsin, retinitis pigmentosa
1. Introduction
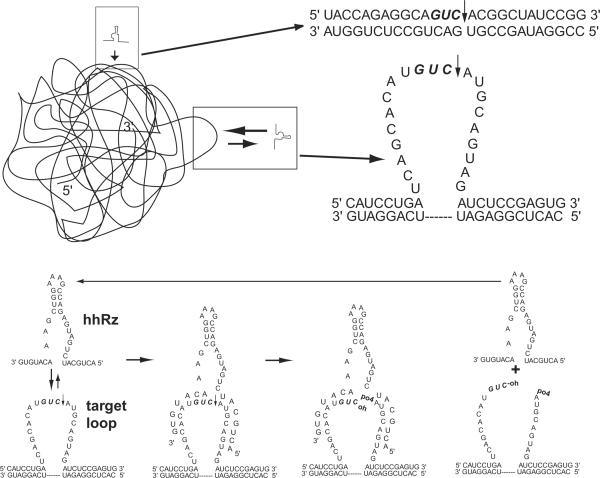
Enzymes are commonly assumed to be proteins. RNA, when of appropriate nucleic acid composition and folded into appropriate structures, can also perform efficient enzymatic catalysis. An RNA-based catalyst is called a ribozyme. Ribozymes are catalytic RNAs that have potential as gene therapy agents for a variety of genetic, degenerative, and infectious diseases of the human eye. The hhRz is a post transcriptional gene silencing agent (PTGS) that can potentially cleave a large number of NUH↓ sites in any given target mRNA (where N is any nucleotide and H is C, U, or A, but not G). The small hhRz has a catalytic core bounded by two flanking antisense (AS) regions (Stems I and III) designed to be complementary by Watson Crick base pairing to the sequence in the mRNA targeted (Fig. 1). The catalytic core of the hhRz is evolutionarily optimized (Nakayama and Eckstein, 1994; Tang and Breaker, 1997; Vaish et al., 1997; Salehi-Ashtiani and Szostak, 2001). Stem II composition, length and capping sequence can be varied and designed for stability (Tuschl and Eckstein, 1993; Homann et al., 1994; Long and Uhlenbeck, 1994). However, the achievement of an active intracellular hhRz therapeutic depends critically upon the accessibility of the region of the target mRNA that contains the NUH↓ hhRz cleavage site, and the composition and length of the AS flanks that guide annealing to that region of the mRNA target. While the hhRz design variable set is limited, this by no means makes for straightforward success in identifying successful lead hhRz candidates against a given target mRNA. The identification of the most accessible target region for annealing and cleavage is a highly non-trivial task because target mRNAs are extensively folded into secondary and tertiary structures, densely coated with heterogeneous protein moieties, dynamically fluctuating on broad time scales, and spatially compartmentalized in the cell at both the macroscopic (nucleus vs. cytoplasm) and microscopic levels (nucleolus, ribosomes, actin bundles). Contemporary structural and biophysical approaches are able to address physical access and conformational dynamics in small RNAs (Higgs, 2000; Doherty and Doudna, 2001; Onoa and Tonoco, 2004; Brauns and Dyer, 2005; Latham et al., 2005), but not in larger mRNA or viral RNAs that are important targets for therapeutic RNA drug development. The biocomplexity of RNA structure is the single major unsolved problem that markedly limits PTGS agent development.
Figure 1.
Schematic for the structure and function of a hhRz as a catalytic RNA. The structure of a classical hhRz with a 4 bp Stem II region, the catalytic core consensus region, and two antisense flanks that are directed to anneal to the target region of the mRNA. If the targeted region of the mRNA is exposed as a single-stranded platform, full rapid annealing of the hhRz AS flanks can occur. Upon full annealing the core enzyme is in a position to promote hydrolysis of the phosphodiester bond immediately downstream of the NUH↓ cleavage recognition motif. If product release can occur, the hhRz is then able to direct RNA catalysis to other mRNA targets (enzymatic turnover). If the targeted region is not fully exposed the rates of annealing are slowed and efficacy is suppressed.
Rational hhRz design depends upon predetermined knowledge of regions of accessibility, whose identification remains a daunting task. There are hundreds of hhRz NUH↓ cleavage motifs in any moderate sized mRNA target and 236 potential cleavage sites in the dominantly expressed full-length human RHO mRNA (Nathans and Hogness, 1984). While there are many places that might cleave, there are orders of magnitude insufficient resources and time to test them all (Jarvis et al., 1994). Most NUH↓ sites have no cleavage potential because they are buried in secondary or tertiary RNA structure. Highly accessible regions are rare indeed and difficult to identify. The efficacy of hhRz cleavage in vitro does not guarantee function in vivo (Beck and Nassal, 1995), so cellular testing is indicated. Trivial cleavage of small unstructured RNA targets in vitro does not predict kinetic performance in cells where the target mRNA is folded and protein coated. Live cell high throughput screening (HTS) approaches are clearly needed to identify the most accessible regions toward the design of PTGS agents with the greatest clinical impact (Sullivan et al., 2007).
Identifying a highly efficacious hhRz or PTGS agent is one of the more challenging problems in molecular biology today. Our knowledge of target RNA biology is still in its infancy. While there is substantial structural and functional knowledge about the hhRz, identifying the best site to target and then the best agent to use, remains a complex task. There are several bottlenecks in the RNA drug discovery process that can potentially be relieved with high throughput screening technologies (Sullivan et al., 2007). It is critical to embrace all of the known variables of PTGS agent design and use in order to achieve a successful agent (Sullivan et al., in preparation).
To begin to address these challenges in hhRz PTGS for retinal and macular degenerations we chose a model mRNA target, human RHO mRNA. RHO mRNA codes for rod opsin, the apoprotein of the visual pigment in rod photoreceptors that subserves initiation of scotopic range phototransduction. The RHO gene is the site of a large number (>125) of mutations that cause three classes of retinal disease which include autosomal dominant retinitis pigmentosa (RP), autosomal recessive RP, and autosomal dominant congenital stationary night blindness (Gal et al., 1997). Human RHO mRNA is an average size mRNA (1.8 kB dominant photoreceptor transcript), is at least moderately abundant, is slightly GC rich (56.6%), is quite stable in mammalian photoreceptors (τ½ ≈ 12 hrs), and spends most of its lifetime in the cytoplasm (Nathans and Hogness, 1984; Braun and Young, 1986; Korenbrot and Fernald, 1989; McGinnis et al., 1992; Pierce et al., 1993). RHO mRNA codes for a protein that is both highly abundant (>108 copies/rod outer segment) and stable in the photoreceptor (10 days for a mammalian rod, 5–7 weeks for an amphibian rod) or in cultured cells. It is synthesized at a level of approximately107 copies/day to maintain a steady state of visual pigment in the rod outer segment given the daily loss of approximately 10% of pigment by the shedding of distal disks (Young, 1967; Young and Droz, 1968; Sung et al., 1991). Human RHO mRNA is a model target for investigating hhRz design variables that will apply to many mRNA targets for human gene therapy. RHO mRNA is a relatively stable mRNA and thus a good target relative to the catalytic time scale of the intracellular hhRz (minutes to hours). However, RHO protein is incredibly stable in both photoreceptors and cultured cell expression systems. This presents challenges for assessing knockdown (KD) efficacy at the protein level due to long-lived persistence of abundant gene product present at the time of transfection that cannot be impacted by introduction of the PTGS agent. The huge abundance of stable RHO protein decreases the practical dynamic range of measure, for example in stable cell lines expressing RHO. Only cotransfection of RHO and hhRz expression plasmids into naïve cells (no prior levels of RHO expression) can be used to reliably assess KD with acceptable and meaningful linear dynamic range.
Recent computation/experimental studies have substantially cross-validated RNA secondary structure computational approaches to predict accessibility (Sczakiel et al., 1993; Sczakiel and Tabler, 1997; Patzel and Sczakiel, 1998; Scherr and Rossi, 1998; Patzel et al., 1999; Amarzguioui et al., 2000; Scherr et al., 2000). We employed three state-of-the-art RNA computational approaches to predict accessible regions in human RHO mRNA. We recognize the limitations of computational algorithms (only some secondary structures are predicted, and then only to a given level of confidence, while tertiary structures, protein coating, and dynamic fluctuations are not predicted). Nevertheless, these algorithms are useful to identify stable secondary structure motifs that harbor large single-stranded (ss) platforms that are innately and immediately accessible for the second order annealing reaction with a small RNA ligand (the hhRz). This computational approach allowed us to identify accessible regions that supported design of lead KD hhRz expression constructs that suppressed human RHO protein in a human cell expression system. HhRzs were efficacious only when directed to large accessible ss annealing platforms, and not when directed to predicted inaccessible sites. The level of protein KD observed is about 30% in a time-limited (48 hr) coexpression paradigm (cotransfection of both hhRz and target expression plasmids). Optimization of the hhRz AS flanks could improve performance, or as yet unidentified sites might prove as better regions for attack.
The hhRzs designed in this study are intended for a mutation-independent or KD therapeutic strategy, where one seeks the best site to attack in an mRNA target (Millington-Ward et al., 1997; O'Neill et al., 2000; Sullivan et al., 2002; Gorbatyuk et al., 2005, 2007). This site is present in wild type (WT) mRNA and likely most, if not all, mRNAs harboring mutations in the target gene. A single PTGS therapeutic could potentially be used therapeutically for all known autosomal dominant mutations in a given disease gene that cause toxic mutant (gain-of-function) proteins. The KD hhRzs achieved in this study are capable of cleaving all known human dominant RHO mutations. A KD hhRz suppresses both WT and toxic mutant mRNA and protein. The suppression of WT gene expression by the KD hhRz could create a haploinsufficiency effect. WT expression may need to be reconstituted, through regulated expression of an allelic variant cDNA that transcribes an mRNA that is engineered to resist hhRz cleavage yet translate a WT or near-WT protein. We call this strategy KD-reconstitution (KD-RECON). We have chosen this strategy over any mutation-directed strategy for several reasons: 1) there are typically many autosomal dominant disease mutations in a single gene, and therapeutic use of a single agent for all or most mutations in a given disease gene is pharmaceutically sensible for such orphan diseases, 2) only a small percentage of mutations (~12% for human rod opsin) generate new hhRz cleavage sites that might permit therapy directed to only the mutant mRNA (similar low percentages would be expected in other genes), 3) most single nucleotide mutations occur in regions of primary mRNA sequence that are extensively buried in secondary/tertiary structures that limit or obviate accessibility to hhRz annealing, and 4) antisense effects against WT mRNA will still occur even in a mutation-specific strategy because the hhRz will bind to WT mRNA. Extensive evidence demonstrates that the primary initial challenge to successful development of ribozyme, siRNA, and antisense nucleic acid therapies is to identify rare regions of the target mRNA that are most accessible in live cells to second order kinetic annealing (Patzel et al., 1999; Giddings et al., 2000; Scherr et al., 2000; Zuker, 2000). Only the mutation independent or KD-RECON strategy can embrace these dominant constraints that arise due to target related variables.
The first step in any nucleic acid drug discovery research is to determine the extent to which the candidate drugs can be effective against the target that will be present under in vivo testing in humans. Much of RNA drug screening can be conducted against the human mRNA target in cell culture studies where identification and optimization of lead candidates can be determined under rigorous experimental conditions. Cultured cell expression systems have been the primary source of PTGS development and optimization for many targets. Highly optimized RNA drugs can then progress into preclinical testing in appropriate animal models. What is an appropriate animal model? The ideal animal model is one in which the targets expressed (e.g. WT and mutant) are human mRNAs expressed on a knockout animal background for the homologous gene. Most of the ribozyme or RNAi candidate therapies for retinal degenerations to date have been developed against animal target mRNAs (Drenser et al., 1998; Lewin et al., 1998; LaVail et al., 2000; Gorbatyuk et al., 2005, 2007). The critical factor for ribozyme or RNAi success is accessibility of the target mRNA for rapid annealing by the PTGS ligand. While coding regions in highly conserved proteins such as RHO may yield local mRNA sequences that are totally or partially identical at the primary sequence level, this is no guarantee that the local accessibility for annealing in the secondary and tertiary structure will be similar between the animal and human mRNAs. Small changes in nucleotide sequence can promote substantial changes in local mRNA secondary structure, a primary determinant of mRNA target accessibility. For agents targeting the 5' untranslated region (UTR) or 3' UTR of the target mRNA, which are not as well conserved, lower comparative accessibility is expected. Hence, PTGS agents developed against animal mRNAs for proof-of-principle studies are not arbitrarily expected to perform equivalently well against the human target in clinical trials. This issue raises into question the predictive value of such prior studies for human gene therapy, which is discussed further elsewhere (Sullivan et al., 2007). This is the first study to seek to develop hhRzs designed specifically against a human photoreceptor target (RHO mRNA) with the evaluation conducted in naïve cultured human cells which express abundant levels of the target mRNA and protein. Lead candidate PTGS RNA drug identification and optimization can be carried out efficiently in human cultured cells, as shown in this study, because PTGS operates at the level of cellular housekeeping below any differentiated cellular functions.
We have chosen hhRz technology over siRNA or shRNA because of emerging concerns of off-target RNAi knockdowns that are increasingly documented (Kawasaki et al., 2003; Semizarov et al., 2003; Persengiev et al., 2004; Scacheri et al., 2004; Lin et al., 2005; Shibin et al., 2005; Jackson et al., 2006). The RISC complex of RNAi is mismatch tolerant. RNAi operates in the pathway of microRNA biology which regulates gene expression, differentiation and development. The concept of flooding a natural biological pathway of such importance with therapeutic agents is likely fraught with potential toxicities. It is not surprising that serious untoward effects, including lethality, of RNAi overexpression in mammalian models have been documented (Grimm et al., 2006; Barik, 2006). This should raise concerns for use of RNAi technology in human therapeutic trials. Can RNAi be used therapeutically without inducing cellular and organism toxicity. In contrast the hhRz was originally identified in plants and extensive searches of the human genome have failed to identify human equivalents. The hhRz is thus orthogonal to the human genome, which minimizes its toxicity risk in humans. The hhRz is a self-contained PTGS agent that operates independent of human host cell proteins and pathways, and should be innocuous. Its greatest toxicity risk involves off-site targeting, which is minimized with a total hhRz AS flank of 12–16 nt.
2. Materials and Methods
2.1. Computational Analysis of Human Rod Opsin mRNA Accessibility
Human RHO mRNA construct (GenBank accession number: NM000539) is the full length transcript of rod photoreceptors and begins from transcription start (there are two start sites separated by a single nt- the promoter-proximal site was used) and extends to 21 nt downstream of the initial (dominant) polyadenylation signal (1506–1511): 95 nt 5' UTR (1–95), 1044 nt coding (96–1142), 370 nt 3' UTR (1143–1511) and 21 nt 3' UTR to polyA addition site (1512–1532). The downstream position approximates the point where 3' downstream processing enzymes would cleave the transcript in preparation for terminal polyadenylation to form the resultant 1.8 kB mRNA (Nathans and Hogness, 1984). The polyadenylation component and the remainder of the 3' UTR to the non-dominant polyA signal were not considered in RNA folding analysis.
The goal of computational analysis was to identify local folding regions with highly probable and stable secondary structures that harbor substantial (≥ 8 nt) ss annealing platforms. HhRz kinetic reactivity (as well as AS and RNAi reactivity) depends upon a second-order annealing reaction between the ligand and target mRNA, which is expected to occur by random coil to double helix transitions following collision and kissing complex formation (Stage-Zimmermann and Uhlenbeck, 1998; Brown et al., 2005). Our hypothesis is that such stable ss platforms constitute suitable sites for hhRz attack as full annealing occurs immediately upon collision without delays due to required melting of local target structure at physiological temperature. From biophysical chemistry principles, any target region that exists in prehybridized state (intramolecular folding) is unavailable for immediate formation of complementary Watson-Crick hydrogen bonds upon intermolecular collision of a PTGS ligand. A kissing complex may form between a part of the ligand and the target over regions of immediately available complementary hydrogen bonding, and this may extend into areas of preformed structure. However, preformed stable secondary (or tertiary) structure(s) require breaking of Watson-Crick hydrogen bonds (positive enthalpy, ΔH > 0) to achieve random coil ss annealing platforms. Prehybridized target structures require a prior spontaneous transition (free energy (ΔG) < 0) of the region at 37°C to form a ss loop to support ligand annealing. This is unlikely to occur unless the entropy (randomness) gained (+ΔS) outweighs the cost of enthalpy input (ΔG = ΔH -TΔS). A preformed and stable secondary structure will always create kinetic delays (Arrhenius) for substantial ss loop formation at physiological temperature, if such a conformational transition is thermodynamically possible. Our rationale is that stable and large preformed ss platforms that allow all or most of the ligand to bind in a single kinetic transition are the best regions to target. The binding energy of the AS flanks must be sufficient for accurate molecular recognition of the target sequence, but not too tight that specificity is lost or product inhibition occurs after cleavage. Product inhibition can occur due to lack of dissociation of upstream and downstream cleavage products from the hhRz AS flanks (Herschlag, 1991; Stage-Zimmermann and Uhlenbeck, 1998).
The set of equilibrium secondary structures (ST) is a geometric scaling related to the number of nucleotides (N) in the RNA (ST = 1.8N) (Zuker and Sankoff, 1984). For a window size of 250 nt the total number of secondary structures is approximately 6.6 × 1063 and would take approximately 6.6 × 1019 seconds (2 × 1012 years) to calculate (Mathews, 2006). The astronomical complexity of potential RNA conformational space cannot be fully appreciated. Nevertheless, this space can be practically sampled. We employed three contemporary algorithms to predict accessible regions in human rod opsin mRNA in a novel approach. These algorithms include MFold (http://mfold.bioinfo.rpi.edu/) (Zuker, 1989, 2000, 2003; Zuker et al., 1999), which is also available on the Genetics Computer Group platform (Accelrys) (UNIX or PC terminal based SeqWeb), SFold (http://sfold.wadsworth.org, (Ding and Lawrence, 2001, 2003; Ding et al., 2004), and OligoWalk (PC-based RNAStructure package, version 4.2, http://rna.urmc.rochester.edu/rnastructure.html, (Mathews et al., 1999a,b; Mathews et al., 2004). MFold uses energy minimization to predict the minimal free energy structure (MFE) and substructures of higher (less stable) free energy over a user stipulated range. We moved windows of 1400 nt or 250 nt along the mRNA in overlapping steps of 100 nt, and output an ensemble of structures (up to 99) for each folding. We chose a window of energy up to 10 kcal/mol above the MFE ΔG and a minimum difference between structures of either 10 bp (1400 nt windows) or 3 bp (250 nt windows). For each ensemble we identified accessible ss platforms defined as greater than or equal to 8 nt and computed the frequency of recurrence for each ensemble and the mean frequency across ensembles that sample the region. The 8 nt ss platform length was chosen because it is sufficient to seed initial annealing after diffusion-limited collisional impact. The MFold output is considered a frequency as opposed to a strict probability because MFold samples cover only a range of secondary structural outputs with folding energies falling in the immediate neighborhood of the MFE structure. Accessible sites were named according to the nearest embracing 10 nt marker. SFolda operates independent of energy minimization, using a Boltzmann-weighted sampling of the total ensemble of all structural states, to directly estimate the true probability of accessibility along an mRNA target. OligoWalk uses the CT file output format from RNAStructure, a Windows version of MFold, to compute several energetic parameters, including local folding energy (LFE) (target break energy), along the mRNA with a user stipulated window size. We used a 15 nt window in OligoWalk to correspond to the antisense platform of the hhRzs tested in this study (7nt/7nt plus one non-annealing nt at site of cleavage). The LFE landscape was calculated by breaking local target secondary structure and including target suboptimal secondary structures in the ΔG calculation. Regions with more positive LFE are less likely to be hybridized into stable intramolecular secondary structures and are likely to be good regions for targeting. To identify likely accessible targeting platforms we sought regions that simultaneously satisfied three conditions: 1) a large stable ss region by MFold, 2) SFold predictions of high probability of access in the same region, and 3) high (less stable) LFE over the same region. Clearly, regions of predicted accessibility must contain available NUH motifs to be subjects for hhRz targeting. We call this approach multiparameter prediction of RNA accessibility (mppRNA), which will be detailed and validated elsewhere (Sullivan et al., manuscript in preparation).
2.2. Expression Constructs
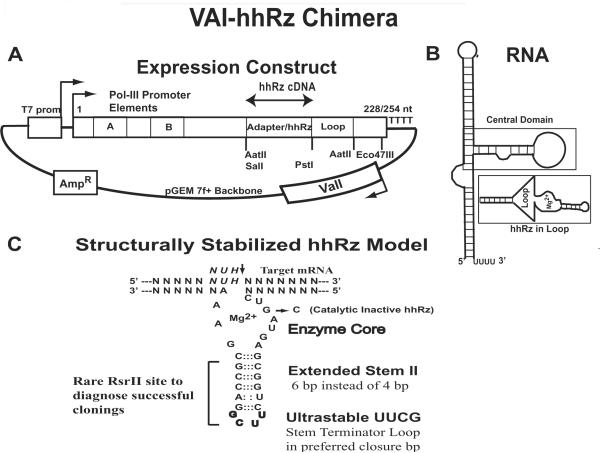
The human RHO expression vector used in this study contains a cDNA harvested from plasmid pCIS-hRHO (Nathans and Hogness, 1984; Nathans et al., 1989) and cloned downstream of the CMV promoter in pCDNA3 to form pCDNA3-WT-RHO (Sullivan and Satchwell, 2000). This construct expresses abundant WT or mutant opsin proteins in HEK293S cells (Sullivan and Satchwell, 2000). In this construct the first 74 nt (of 95 nt) of the 5' UTR are replaced with vector sequence. No hhRzs were targeted at the 5' UTR and predicted accessible structures were not influenced by the replacement of this element of the 5' UTR by the specific vector sequence The polyadenylated mRNA transcribed by Pol-II in human cells is approximately 1.8 kB. Chimeric VA1-hhRz vectors were generated by first annealing two fully complementary synthetic oligonucleotides with cohesive overhanging ends and ligating the hhRz cDNA between the SalI and PstI sites in pGVAL (Lieber and Strauss, 1995). We modified pGVAL with an adapter between the SalI and PstI sites to allow highly efficient cloning of hhRz cDNAs into the expected loop region by a process of restriction endonuclease mediated genetic selection against parental plasmid background. The adapter separated the SalI and PstI sites to allow more efficient hhRz cDNA cloning when compared to the original construct in which the sites were immediately adjacent (see Fig. 7A). This adapter contained EcoRV, XbaI, and AflII restriction sites that are unique to the pGVAL-ad vector. These restriction sites allow post-ligation (by EcoRV) selection to increase the frequency of positive clones by minimizing parental background. All hhRz cDNAs insert a rare and unique 7 bp RsrII restriction site in pGVAL-mod that allowed simple diagnosis of successful clones. A hhRz catalytic core mutation was used to promote inactivation in order to test the source of KD (Sheldon and Symons, 1984).
Figure 7.
VAI-hhRz chimeric expression construct. (A) The VAI-hhRz chimera RNA is expressed from a gene with an intragenic Pol-III promoter (A and B boxes) and terminates after a polyuridine tetramer. The loop sequence into which the hhRz cDNA is cloned between the SalI and PstI sites is stabilized by a long (19 bp) GC rich stem. An adapter sequence inserted between and separating the SalI and PstI sites in the loop makes hhRz cDNA cloning much more efficient. Other than this adapter element, the overall sequence of the pGVAL vector was not altered from the vector which has been used successfully in prior studies (Lieber and Strauss, 1995). (B) The hhRz was cloned into an expected loop region engineered into the adenoviral VAI RNA (Lieber and Strauss, 1995). This VAI RNA has stable apical and basal stems and its central domain (functional) (box) was obviated by the engineered stem loop structure to hold the hhRz and create a flexible steric environment where it could function (box). (C). Stabilized hhRz design. A 7 nt/7 nt symmetrical antisense flank design was used centered on the intended NUH↓ cleavage motif. The catalytic core is shown using the established numbering scheme (Hertel et al., 1992). The nt in the core enzyme that is mutated to form a catalytically inactivated hhRz is also shown (G5C). Stem II was extended by 2 bp to add six hydrogen bonds to the helical stack in order to stabilize folding. This extended stem was capped with a single stranded 5' UUCG 3' loop that also promotes stabilization of the overlying stem structure. Stable folding of Stem II protects the catalytic domain from misfolding. Stem II is engineered with a rare 7 bp RsrII site that allows straightforward identification of successful hhRz cDNA ligation into pGVAL-ad.
2.3. Cell Transfections and Target Protein Analysis
HEK293S cells are an environmental suspension-adapted version of the ATCC line (CRL 1573), which are transformed and immortalized with adenovirus E1A and E1B proteins (Stillman and Gluzman, 1985). HEK293E cells are a stably transformed derivative of HEK293S cells that constitutively express the Epstein Barr virus nuclear antigen. Resuspended HEK293 cells were quantified by hemocytometer-calibrated OD800 measurements (Mohler et al., 1996). Equivalent numbers of cells (6 × 106 cells) were grown in 10 cm plates at 37°C in DMEM/F12 supplemented with 10% (v/v) heat inactivated calf serum, 2 mM L-glutamine, and penicillin/streptomycin (Sullivan and Satchwell, 2000). Cells were cotransfected at 60–70% confluence with pCDNA3-human rod opsin cDNA (7181 bp) (2 μg, 4.21 × 10−13 moles plasmid, 1.28 × 1013 moles of RHO expression construct), pGVAL-hhRz (3533 bp) (5 μg, 2.14 × 10−12 moles plasmid, 1.62 × 10–13 moles of VAI-hhRz expression construct) (or 5 μg pGVAL, control) and pEGFP (2 μg) expression plasmids by Lipofectamine Plus (InVitrogen). The molar ratio of hhRz/target template expression constructs in the transfection mixes was 1.27. High transfection efficiencies (≥ 80–90%) were routinely achieved. Plates were screened to insure uniformity of transfection efficiency through measures of cellular EGFP fluorescence directly from the 10 cm culture plates on a Storm 860 fluorimeter (Molecular Dynamics-Amersham) in blue diode mode (with a thin layer of water between the glass and the polystyrene plates) and also in some Western blots by probing with an anti-EGFP antibody. Western blots acceptable for analysis were those that originated from experiments with uniform transfection efficiency and readily assayed control sample (pGVAL with no hhRz) opsin counts.
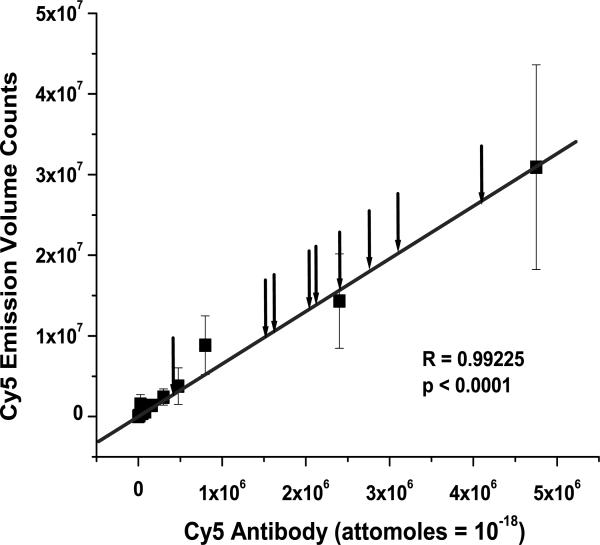
Cells for western analysis were harvested at 48 hours post transfection, and counted in 1× PBS by OD800. At the time of harvest there were 1–2 × 107 cells per plate. Cells were pelleted and resuspended and extracted overnight with 1% (w/v) n-dodecyl-β-maltoside in 1× PBS at 4°C with nutation. Nuclei were sedimented by centrifugation (14000g) and total cytoplasmic protein was quantified by modified detergent-insensitive Lowry assay (DC Assay, BioRad). Equivalent amounts of total cellular protein extract from each sample (10–40 μg) were loaded into adjacent wells of PAGE-SDS gels (0.75 mm thick, 4% stacking, 12% resolving) and electrophoresed at 200 volts for 45–60 minutes in standard electrophoresis buffer. Western electrotransfer onto nitrocellulose membranes occurred at 30 volts for 1 hour in ice-cooled transfer buffer (25 mM Tris, pH 8.0, 0.15M glycine, 20% methanol). Laemmli sample buffer was modified with 8M urea in order to better denature opsin protein from the detergent micelles and suppress aggregation which otherwise occurs in standard cracking buffer. With this buffer the dimer and trimer opsin bands were markedly suppressed compared to extraction with standard Laemmli buffer. Blots were processed as reported previously (Sullivan and Satchwell, 2000) using excess 1D4 mouse anti-bovine opsin monoclonal antibody (same epitope in human rod opsin), except that a Cy5-labeled goat-anti-mouse-IgG polyclonal antibody (Jackson ImmunoResearch Labs, West Grove, PA, #115-175-003, lot 46983) (peak absorption 650 nm, peak emission, 670 nm) was used as the secondary antibody to quantitate opsin protein on a Storm 860 platform (800V PMT, 200 um resolution) in red laser diode mode (635 ± 5 nm excitation) (Fradelizi et al., 1999). In this assay there is a 1:1 relationship expected between the numbers of opsin molecules bound on the nitrocellulose surface and the number of excess 1D4 antibodies bound. The number of Cy5 labeled secondary antibodies bound is expected to be proportional (not necessarily linearly) to the number of 1D4 antibodies bound because of varying affinities of the antibodies within the mixture, but with clear bias to higher affinity antibodies that remain bound to RHO during washing. Each 1D4 primary and Cy5-labeled secondary antibodies are expected to bind statistically independently. By spotting dilutions of Cy5 labeled secondary antibody onto the glass scanning platform of the Storm 860 fluorimeter we determined that the assay has a linear dynamic range of measurement in red laser diode mode over several log orders (see Fig. 8A). The mean raw Cy5 volume counts (not normalized to controls) obtained from opsin measures on western blots used to obtain Figs 8B and 8C were all within the linear range of direct Cy5 labeled antibody measures on the Storm 860 platform. In addition, given the expected level of opsin expression in transfected HEK293S cells (~5 × 106 opsins/cell; Sullivan and Satchwell, 2000; Sullivan and Shukla, 1999; Shukla and Sullivan, 1999), and assuming a 1:1:1 relationship between opsin molecules on the membrane, 1D4 molecules bound to opsin, and Cy5 secondary molecules bound to 1D4, the measured slope of the Cy5 secondary antibody measure curve (6.52 volume counts/attomole Cy5 or 1.08 × 10−5 volume counts/molecule) we estimate a fluorescent output for RHO that is on the experimentally-determined linear dynamic range of Cy5 antibody measure. To minimize dominant sources of variation in western analyses, all experiments were conducted by the same individual (HA), and a single stable fluorescent secondary antibody staining solution preparation was used throughout the study (Koller and Watzig, 2005; Watzig, 2005).
Figure 8.
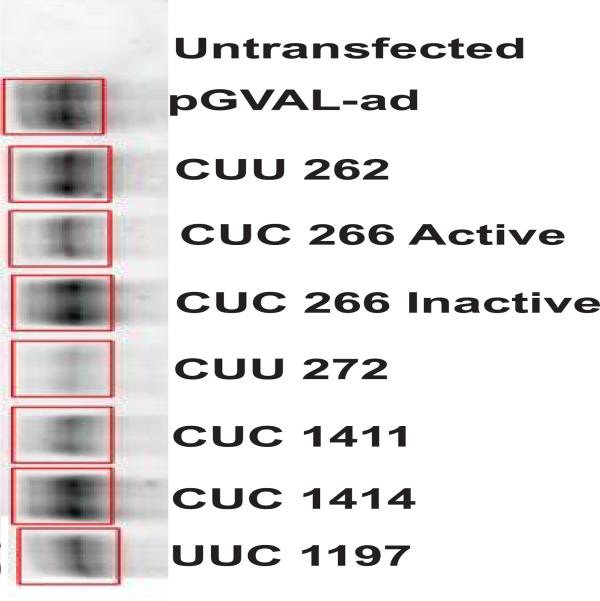
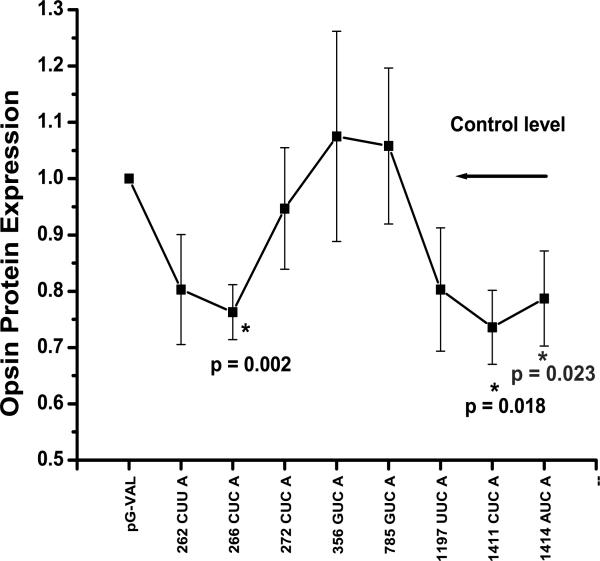
Quantitative analysis of hhRz knockdown of opsin protein expression. (A) Broad linear dynamic range of measure. We determined the dynamic range of Cy5-labeled antibody measure for western blotting. Lyophilized Cy5-labeled goat anti-mouse-IgG antibody was prepared with water and 50% glycerol to a final concentration of 1.33 mg/ml (9.5 μM). This stock was diluted serially in water (concentration range: 0.19 μM (1/50 dil) – 19 pM (1/500,000 dil)) and 25 μl aliquots were spotted in triplicate directly onto the window of a Storm 860 fluorimeter platform and scanned at 200 μm resolution at 800 Volts in the red laser diode excitation mode. Cy5 fluorescence volume counts were acquired in ImageQuant. Mean values and SEM are plotted vs. the number of moles of antibody spotted (1 attomole = 10−18 moles). A linear regression line was fit through the origin (Slope = 6.51977 ± 0.21479 volume counts/attomole Cy5 (1.08E-5 volume counts/Cy5 molecule); R = 0.99225, p ≤ 0.0001). Arrows pointing to the fitted line show the mean values (unnormalized) of Cy5 fluorescence obtained from each sample of the RHO KD analysis. All values fall within the linear dynamic range of Cy5 measure. (B) Western blot analysis of opsin protein KD by pGVAL-hhRz constructs. Transfected HEK293E cells were harvested at 48 hrs post transfection and cell number was determined prior to nonionic detergent extraction. After nuclei removal total cellular protein (cytoplasmic, microsomal) was measured prior to Western analysis with equal amounts of total protein added to each well on a single gel. Data from a representative immunoblot is shown from cells cotransfected with pGVAL-ad (without hhRz) (control) or with pGVAL-hhRz (specific constructs), and pCDNA3-WT-RHO. pEGFP-N1 was used to assess uniformity of transfection efficiency across samples (data not shown). Membranes were probed with 1D4 mouse opsin monoclonal IgG primary and then a Cy5-labelled goat anti-mouse IgG. Volume fluorescence counts of RHO in each region of interest box were obtained and output to spreadsheets for statistical analysis. (C) Ribozyme efficacy analysis. The data set was assembled from a large number of experiments (Numbers of experiments (N) for each condition: Control: 29; hhRz 262: 18; hhRz 266: 23; hhRz 272: 20; hhRz 356: 11; hhRz 780: 11; hhRz 1197: 19; hhRz 1411: 19; hhRz 1414: 16). Mean levels of opsin remaining (REM) after pGVAL-hhRz transfections were normalized to opsin REM in pGVAL (no hhRz) control. The mean and SEM is shown for each experimental factor. pGVAL (no hhRz) is always normalized to itself so the mean is 1.0 and variance zero. ANOVA and post-hoc statistical tests were used (see text) to determine that the 266, 1411, and 1414 pGVAL-hhRz constructs promote statistically significant (p < 0.05) KD of opsin protein relative to pGVAL. The 356 and 785 control pGVAL-hhRz constructs do not promote any KD relative to pGVAL.
Fluorescent images of western blots were acquired on the Storm 860 platform and analyzed with ImageQuant software (Molecular Dynamics-Amersham) by volume count processing. The heterogeneously glycosylated monomer band of human rod opsin (~43–68 kD) was subjected to volume count analysis by drawing a quantitation box around the band of the control sample and using the identical box to quantitate each experimental sample from the same blot. The box was moved to compensate for slight variation in RHO band mobility across different lanes on each gel. Opsin protein levels in samples containing hhRz expression constructs were ratio-normalized by opsin expression levels in the control sample(s) that received the empty expression vector (pGVAL) without an embedded hhRz cDNA.
2.4. High Throughput Ribozyme Screening Assay
The methods for this approach have been previously reported (Yau and Sullivan, 2006, 2007; Yau et al., 2008; Yau and Sullivan, manuscript submitted). In brief, the full length human opsin cDNA was cloned upstream of an IRES (internal ribosome entry site) element and a secreted alkaline phosphatase (SEAP) reporter cDNA sequence. The dicistronic mRNA was expressed in HEK293S cells from a strong CMV promoter after plasmid transfections. Both opsin and SEAP proteins are translated from the dicistronic mRNA. While the opsin is an integral membrane protein, SEAP is secreted in bulk into the culture medium where it can be readily assayed. Ribozyme cleavage anywhere within the dicistronic mRNA promotes more rapid degradation of the dicistronic mRNA, a shorter half-life, and suppression of both opsin and SEAP translation and SEAP secretion. SEAP levels in extracellular fluid were measured by a high throughput enzyme assay based upon conversion of the nonfluorescent SEAP substrate (4-methyl-umbelliferyl-phosphate) to a fluorescent product (4-methyl-umbelliferone). Fluorescence was measured on a 96-well plate reader. The maximum level of suppression in this screening assay is approximately 50%.
2.5. Statistical and Quantitative Analysis
Statistical and graphical analysis was conducted in SPSS (Version 11.5, SPSS Inc., Chicago, IL), and Origin (Version 6.1, MicroCal, Northampton MA) using one-way analysis of variance and post-hoc tests for between condition comparisons with a previously chosen significance level of 0.05. Nonparametric tests were chosen whenever the assumptions of parametric tests were unsatisfied (Zar, 1984). RNA secondary structure figures were output as PostScript files for detailing.
3. Results
3.1. In Silico Bioinformatics Approach to Target Site Selection
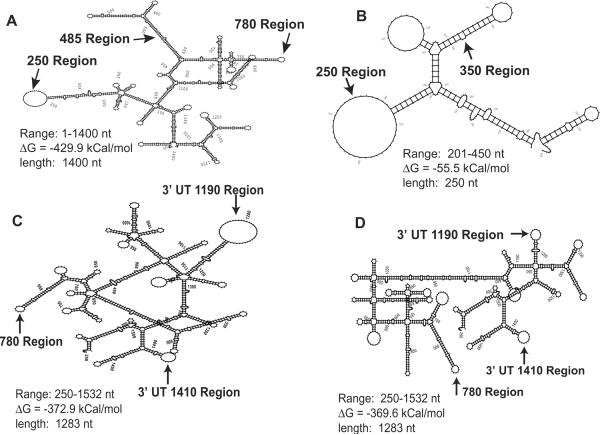
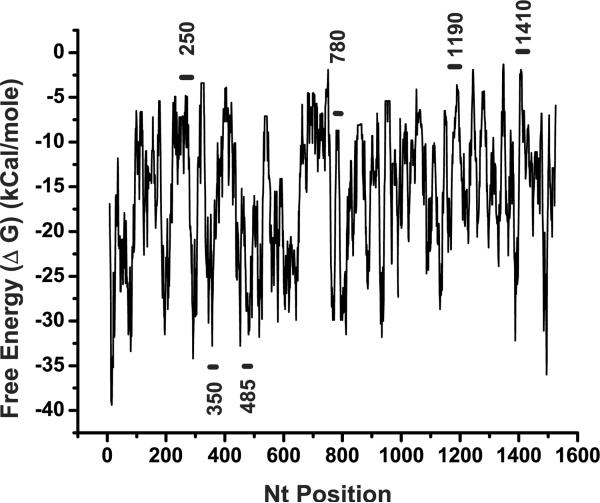
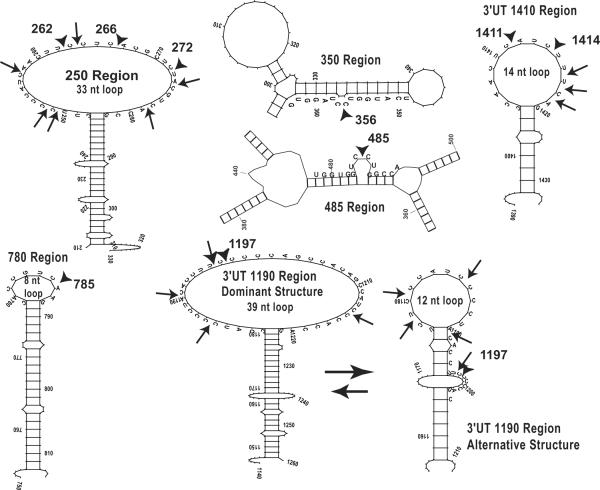
Most of the nucleotides in any RNA are hybridized into intramolecular secondary structures. Given the potential complexity, one expects to find both multiple stable conformational states of a given RNA target and dynamic fluctuations among these states. Our working model assumes that an accessible region in a target mRNA is one that simultaneously has: 1) a stable and large ss region in MFold, 2) a high probability of access in SFold, and 3) a less stable LFE in OligoWalk. Such a predicted ss region is expected to permit rapid annealing of the AS flanks of a colliding hhRz, the essential first step in RNA catalysis. We tabulated regions of RHO mRNA that contain ss platforms greater than or equal to 8 nt. In this study we searched for stable stem loop structures where the loop was expected to be accessible. Examples of discrete structural state outputs from MFold (there were 1088 images analyzed) that show large ss loops or hybridized stem structures (controls) are shown (Fig. 2). Initial MFold local structural state analysis suggested large stable ss loops in regions embracing 250, 1190, and 1410. A smaller stable loop was identified at region 780. The region around the target site at site 485 in the MFE shows a small bulge loop (4 nt) containing the GUC↓ site in an otherwise strong secondary structure. A hybridized stem structure at 350 was selected from many possibilities as a control. In different regions there were substantial numbers of different substates that contained ss platforms such that a frequency of the dominant substate could be determined. The frequency of the dominant substate in these regions was estimated by MFold analysis (Table 1). SFold output (true probability of accessibility) was also estimated in the regions identified by Mfold, and also varies extensively across the entire mRNA (Fig. 3). The regions presenting stable ss loops in local secondary structures had substantial access probabilities by SFold, varying over primary sequence spans consistent with the size of the ss loops seen in MFold analysis, whereas the control hybridized region had low probability. OligoWalk output across the entire mRNA shows regions of high (relatively unstable) and low (relative stable) LFE (Fig. 4). All regions with predicted ss loops had relatively positive LFE, whereas the control region at 350 was one of the most stable regions within the fold. Note that the LFE map in the 780 region has a tiny positive plateau surrounded by deep negative regions. This represents the tiny 8 nt ss loop capping a stable hybridized stem structure.
Figure 2.
Examples of MFold search for sterically accessible regions in human RHO mRNA. MFold was used with 250 nt overlapping windows or 1400 nt overlapping windows. Each fold of each overlapping segment of the target mRNA generated a set or ensemble of structures. Output from each ensemble (up to 99 structural images) was carefully examined for large ss regions. In this study these accessible sites sought were loops capping stable stems. The frequency of recurrence of ss site in a single ensemble was taken as the number of times the ss element occurred over the total number of images in the ensemble. Estimates of potentially accessible regions were averaged over multiple ensembles from folding windows containing the region. The frequency of recurrence of the dominant substate conformation was also tabulated. Shown here are minimal folding energy structures from 1400 nt or 250 nt windows (Panel A: 1400 nt wide window folding the range of 1–1400 nt of the target; Panel B: 250 nt window folding the range of 201–450 of the target; Panel C: 1283 nt window folding the range of 250–1532 of target; Panel D: 1283 nt window folding the range of 250–1532 of target). Sites targeted in this study in single stranded and folded regions are indicated.
Table 1.
Accessibility Parameters
| Region | Pa,DomSub | ssLoop | LFE | SFold | Convolution |
|---|---|---|---|---|---|
| 250 | 0.7682 | 0.8462 | 1.0 | 0.6547 | 0.4256 |
| 350 † | 0.1950 | 0.0513 | 0.0016 | 0.0397 | 6.35 × 10−7 |
| 485 † | 0.133 | 0.1026 | 0.4315 | 0.1167 | 6.87 × 10−4 |
| 780 | 0.9933 | 0.2051 | 0.5436 | 0.9954 | 0.1102 |
| 1190-1 | 0.3864 | 1.0 | 0.6902 | 0.4219 | 0.1125 |
| 1190-2 | 0.2163 | 0.3077 | 0.6902 | 0.0907 | 0.0042 |
| 1410 | 0.5955 | 0.3590 | 0.7050 | 0.8604 | 0.1297 |
Rank Order of Convolution: 250 > 1410 > 1190-1 > 780 > 1190-2 > 485 > 350
Pa,DomSub (MFold frequency of dominant substate) mean values were calculated from the ensembles containing the region of interest. ssLoop was obtained for the maximum single stranded platform of the dominant substate conformation and then normalized to the largest ss loop found in the entire dataset (Site 1190-1, 39 nt). LFE (target break energy) was calculated with a 15 nt window operated over all of the substates in a given ensemble. The value of the area under the peak of LFE around a region of interest was obtained after subtracting a linear regression line fit to the LFE map in that region. These values (positive and negative) were additively scaled so that all entries were positive and then normalized to the maximum value. SFold is the true probability of accessibility taken as an average over the primary sequence bounding the largest region of accessibility in the dominant substate as determined by MFold.
Sites 350 and 485: Pa was assigned on the basis of the local Mfold frequency value determined by sstrand output. The largest ssLoop identified in the most dominant substate of 350 was 2 nt and 4 nt for region 485.
Figure 3.
SFold analysis in regions of MFold predictors. The SFold true probability of access was determined in full length human rod opsin mRNA. SFold output was plotted in two regions containing targeted regions where MFold indicated accessible or inaccessible regions: (A) 200–850, (B) 882–1532. Target regions are indicated by overlying horizontal bars and numbers. Regions predicted to contain stable structures containing ss loops of varying size all have mean SFold probabilities greater than 0.5. The 350 and 485 regions, predicted to be inaccessible, have low SFold probabilities.
Figure 4.
OligoWalk analysis of LFE. OligoWalk was used to identify the local target break energy (LFE). A 15 nt window was used which is equivalent to the hhRz annealing platforms used in this study (7/7nt) including the single nt which does not form a Watson-Crick base pair at the site of cleavage. The raw output of OligoWalk provides the target break energy (LFE) for each nt. The horizontal bars and numbering indicate regions targeted in this study. Regions determined by MFold structural state analysis to contain stable ss annealing platforms, also confirmed by SFold analysis, are shown here to have more positive LFE levels, whereas the 350 region is a stable state with strong negative LFE.
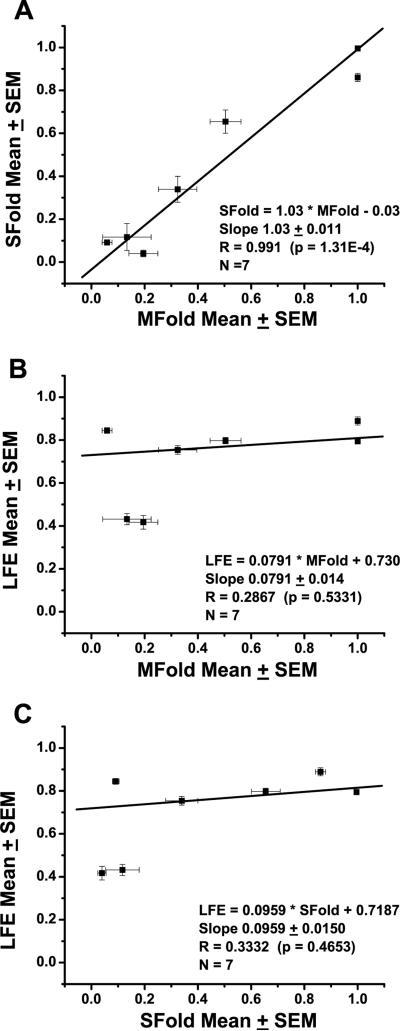
The energy minimization steric analysis (MFold) and the Boltzmann-weighted sampling of all states (SFold) lead to similar predictions at the sites examined in this study. A strong correlation was found between MFold and SFold measures at these sites (Fig. 5). However, weak correlation was found between the LFE measures and both MFold and SFold measures. These outcomes suggest that LFE is an independent parameter to estimate target accessibility, whereas MFold and SFold are sampling similar properties of steric accessibility. The 250 and 1410 regions were expected to be accessible for hhRz targeting because they satisfied four simultaneous conditions: 1) high frequency and probability of being sterically accessible by MFold and SFold respectively, 2) low (more positive) LFE, 3) a single dominant conformational substate with a large ss region, and 4) a number of hhRz NUH↓ sites. The 1190 region had intermediate characteristics with two predominant substates of moderate MFold access frequency, and moderate access probability by SFold. The 780 region had a singular highly accessible but small ss platform as shown by both MFold and SFold. This loop caps a highly stable stem, as determined by LFE analysis. This ss region contains a native GUC↓ cleavage site. We previously developed hhRzs to cleave successfully at GUC↓ 785 in vitro (Sullivan et al., 2002). These hhRzs proved ineffective when tested in live cells when both ribozyme and target RNAs were transcribed by a CMV promoter (unpublished results, Sullivan lab). We reasoned that while a small ss site may have high probability for being sterically accessible, its surrounding region may be so stable that full annealing of the hhRz is restricted and KD is impeded. LFE provides a measure of local and surrounding target energetic stability. Retesting this site with a much more powerful Pol-III promoter expressing the hhRz allowed a determination of the role of ss loop size in hhRz KD efficacy. The GUC↓ site at 356 appeared to be an inaccessible to hhRz annealing by both MFold and by SFold, and LFE analysis showed a broad negative energy peak in this region. On the basis of this computational analysis several NUH↓ cleavage sites were selected for targeting within the large predicted accessible regions (262, 266, 272, 1197, 1410, 1414), and optimal GUC↓ sites were chosen in the expected restricted and presumed inaccessible 785 and 356 sites, respectively. The GUC↓ site at 485 nt was evaluated late in the study by our HTS approach after the report of this agent (Gorbatyuk et al., 2007). The 485 target site has low estimates for accessibility by all three algorithms employed (Figs. 3–5, Table 1).
Figure 5.
Correlation of MFold, SFold, and LFE parameters. (A) We plotted the mean MFold probability (and SEM) for the dominant substate(s) and the mean SFold probability over the regions of interest initially established by MFold analysis. A linear regression line was fit to the data set without constraint to the origin. There is a high degree of correlation (R = 0.991, p = 1.31E-4) between these estimators. The fitted line passes unconstrained essentially through the origin (0.03) and the slope is 1.03 ± 0.011. (B) Comparison of a scaled positive LFE measure with the MFold frequency found no significant correlation (R = 0.2867, p = 0.5331). (C) Compared of a scaled positive LFE measure with the SFold probability also found no significant correlation (R = 0.3332, p = 0.4653). The MFold and SFold estimators of accessibility measure similar features, while the LFE measure appears to be an entirely independent estimator. Note the deep LFE values for the predicted inaccessible sites at 350 and 485.
The MFold access frequency of the dominant substate, the size of the ss loop in the dominant substate, the local integral of the area of the LFE map, and the mean SFold parameter over the region were treated as independent estimators of accessibility and multiplied together with equal weightings to achieve combined accessibility estimates which were rank ordered (Table 1). The stable secondary structures supporting the large ss loops at regions 250, 1410, and the largest loop substate at 1190 were ranked as the top three accessible sites, followed by the small loop at 780, the smaller loop at 1190, and then the regions around 385 and 350. The stable structures selected for hhRz targeting in this study are shown (Fig. 6).
Figure 6.
Ribozyme targeted regions. Stable high frequency recurrent stem loop structural motifs containing single stranded annealing platforms were selected as suitable candidate platforms for hhRz targeting. The 33 nt ss loop in region 250 extends from nts 249–281 in the human RHO mRNA sequence (NM000539). The 8 nt ss loop in region 780 extends from nts 780–787. The 39 nt ss loop in region 1190 extends from nts 1182–1220 and the 12 nt ss substate loop extends from nts 1178–1189. The 14 nt ss loop extends from nts 1406–1419. Available hhRz cleavage sites within these structural motifs are shown by arrows. The NUH↓ sites targeted in this study are shown by arrowheads.
3.2. Experimental Evaluation of Ribozymes
3.2.a. hhRz Chimeric Expression Construct
Previous studies demonstrated the catalytic effectiveness of chimeric hhRzs embedded in a modified adenoviral VA1 RNA against several intracellular RNA targets (Lieber and Strauss, 1995; Lieber and Kay, 1996; Lieber et al., 1996; Xie et al., 1997; Prislei et al., 1997) (Fig. 7A, 7B). VAI-hhRz chimera constructs are transcribed at very high levels by RNA polymerase-III, and traffic to the cytoplasm where they colocalize with most target mRNAs. The intracellular stability of the VA1 RNA protects the hhRz from degradation over periods of hours. The hhRz construct used in this study is designed with an extended Stem II (6 bp), and an ultrastable stem-capping loop (5' UUCG 3') in the preferred closure context for stability (Fig. 7C) (Tuerk et al., 1988; Dale et al., 2000). These changes energetically bias hhRz Stem II folding into the desired secondary structure. Two 7 nt AS arms surrounding the target cleavage site nucleotide were used for expected specificity (15 nt total antisense span). Based on prior studies the use of the Pol-III VAI promoter to express VAI-hhRz chimeras is expected to result in a substantial excess of hhRz enzyme over opsin mRNA substrate, and with critical colocalization of enzyme with substrate in the cytoplasm (Svensson and Akusjarvi, 1984; Thompson et al., 1995; Bertrand et al., 1997; Hormes et al., 1997; Koseki et al. 1999). Conditions of enzyme excess allow a direct test of target mRNA accessibility in vivo that is not conditionally dependent upon enzymatic turnover.
3.2.b. Quantitative Tests of hhRz Efficacy in Cultured Human Cells
HhRzs were designed to target NUH↓ motifs in the large predicted 250 loop (CUU↓ 262, CUC↓ 266, CUC↓ 272), a folded stem region (GUC↓ 356), a small loop capping a long and stable stem (GUC↓ 785), the large predicted 1190 loop (UUC↓ 1197), and the moderately sized 1410 loop (CUC↓ 1411, AUC↓ 1414). pGVAL-ad constructs [without (control) or with cloned hhRzs] were co-transfected with pCDNA3-WT-RHO and pEGFP expression plasmids into equal numbers of naïve HEK293E cells. At 48 hours post transfection 10 cm plates were scanned for EGFP fluorescence and experiments continued only if EGFP fluorescent was uniform across the set of plates. Cells were harvested after 48 hrs and equivalent amounts of measured total cytoplasmic protein were electrophoresed into PAGE-SDS gels and nitrocellulose blots were probed for rod opsin protein and imaged on the Storm 860. Opsin on western blots was quantified by Cy5 fluorescence volume counting in ImageQuant. The dynamic range of measure was determined and found to extend linearly over several log orders with high sensitivity (6.52 volume counts/attomoleCy5) (Fig. 8A). Moreover, the mean raw Cy5 volume counts from opsin western blots obtained for each of the conditions evaluated (control, hhRz 262, 266, etc) across the entire data set used for analysis (see below) were commensurate with the linear dynamic range of Cy5 measure. Thus for a relative measure of opsin in experimental vs control samples we have strong evidence for linearity of measure. In these experiments we did not attempt to make absolute measures of opsin protein, which would have required a separate development of a standard curve as previously used to quantify opsin expression in HEK293 cell lines (Sullivan and Satchwell, 2000). Nevertheless, our measures are consistent with a direct linear proportionality between the numbers of opsins present and the Cy5 volume counts. The minimum number of Cy5-labeled antibody molecules detectable was 2.86 × 108, which corresponds to detection levels of RHO in approximately 50 cells of a stable expressing cell line (Sullivan and Shukla, 1999; Shukla and Sullivan, 1999). From the slope of the fitted line we determined that the sensitivity of detection is 1.08 × 10−5 fluorescent counts/Cy5 molecule. In the absence of a ribozyme KD agent the concentration of RHO expected in 1 ml of cell extraction buffer from 1×107 transfected cells is approximately 675 nM (2.7 μg/ml of RHO) (Sullivan and Shukla, 1999; Sullivan and Satchwell, 2000). With 20 μg of total protein (appx. 2.0 mg/ml) added per gel lane (0.135% RHO) there are 0.67 × 10−12 moles of RHO (4×1011 RHO molecules). Given the measured sensitivity of Cy5 detection (above), and assuming a 1:1:1 relationship of RHO: 1D4 antibody: Cy5-secondary we expect a maximum output signal of approximately 4×107 volume fluorescent counts, which is also within the high linear Cy5 dynamic range of measure determined on the Storm fluorimeter using the Cy5 secondary antibody alone. To normalize for varying amounts of total cell protein added in different experiments, and to address intrinsic blot-to-blot variability with Western analysis, we obtained a ratio of opsin volume counts for each pGVAL-hhRz sample divided by the opsin volume counts from the control sample (pGVAL without any hhRz) on the same blot. A representative western blot for quantifying opsin is shown (Fig. 8B). For each sample in a single experiment we tabulated the remaining fraction of opsin (REM) and hence the KD fraction (KD = 1-REM). Since the control is normalized to itself the Control REM is always 1.0 and its Control KD is always 0 in each experiment. The outcomes of a large set of experiments (n = 29) were subjected to statistical analysis (Fig. 8C). The mean REM level and standard error of mean are shown for each pGVAL-hhRz construct tested relative to pGVAL control. A critical outcome of these experiments is that there is KD from all pGVAL-hhRz constructs targeting NUH↓ sites in regions predicted to be accessible, whereas there is no KD at regions predicted to be inaccessible (GUC↓ 356, GUC↓ 785). It is interesting that the level of KD was not uniform across the three hhRzs used to target the 5' end (262), the middle (266) and the 3' end (272) of the 250 loop. This may indicate that even within a single structural region of predicted accessibility there are local constraints to annealing (see Discussion). Rz525/hhRz485 was note evaluated by western analysis approach.
A one way analysis of variance refuted the null hypothesis of no differences of REM between control and experimental samples for active hhRzs (p = 0.037). This indicated that at least one of the pGVAL-hhRz constructs exerted significant opsin KD. Variance was shown to be unequal (Levine's test, p < 1× 10−5), which was expected because control self-normalization in each experiment leads to zero variance for the control sample set. Several post-hoc nonparametric tests (Games-Howell, Tamhane's T2, Dunnett T3) were used to compare REM means given that sample variances that were known to be unequal. Discrete REM values in each pGVAL-hhRz sample were within 1.5 SD of the mean for all samples, which indicates the absence of strong outliers and a relatively homogeneous distribution. Relative to pGVAL control all tests showed statistically significant (p ≤ 0.05) KD by both hhRz CUC↓ 266 (p = 0.002, 0.003, 0.003) and hhRz CUC↓ 1411 (p = 0.018, 0.029, 0.025) (p values shown for Games Howell, Dunnett T3, and Tamhane's T2 testing, respectively). There was no difference in KD by any test between the CUC↓ 266 and the CUC↓ 1411 hhRzs. As a further test of statistical significance the pGVAL-hhRz constructs were compared in a fixed factor analysis (single population t-test) to a value of 1.0 REM (representing the control level normalized mean). There was statistically significant KD for CUC↓ 266 (p = 7.41 × 10−5), CUC↓ 1411 (p = 8.12 × 10−4), and also AUA↓ 1414 (p = 0.023). The mean levels of KD measured permit efficacy rank ordering of 1st generation anti-opsin pGVAL-hhRz constructs (Table 2).
Table 2.
Rank Ordering of hhRz by Experimental KD Efficacy
| hhRz | Mean REM ± SEM | Mean KD (%) | Rank Order |
|---|---|---|---|
| 1411 | 0.73579 ± 0.0658 | 26.42 | 1 |
| 266 | 0.76261 ± 0.04885 | 23.73 | 2 |
| 1414 | 0.78687 ± 0.08429 | 21.31 | 3 |
| 1197 | 0.80263 ± 0.10941 | 19.74 | 4 |
| 262 | 0.80278 ± 0.09761 | 19.74 | 5 |
| 272 | 0.947 ± 0.10776 | 5.30 | 6 |
| Control | 1.0 ± 0.0 | 0.00 | 7 |
| 785 | 1.05818 ± 0.13844 | −5.80 | 8 |
| 356 | 1.07545 ± 0.18671 | −7.55 | 9 |
pG-VAL-hhRz constructs targeting bolded sites showed statistically significant knockdown (p ≤ 0.05). “Control” is the same pGVAL vector that expresses the engineered VAI RNA but does not contain a hhRz sequence. Note that these values were obtained by Western analysis and do not contain evaluation of the hhRz 485 candidate.
For the CUC↓ 266 and CUC↓ 1411 active pGVAL-hhRz constructs the catalytic core of the enzyme was mutated (G5C, see Fig. 7C) to obviate enzymatic activity, and KD levels were compared between active and inactive expression constructs (Fig. 9). While the numbers of experiments with inactive hhRzs was substantially smaller, the levels of KD were not significantly different between active and inactive hhRzs. This outcome indicates that the KD effect results from one of at least two possible mechanisms: an antisense effect, or catalytic inhibition of the hhRz due to the 6 bp Stem II extension. An antisense effect involves annealing of the hhRz to the target but without enzymatic cleavage, or annealing and cleavage but without product release (catalytic antisense effect indistinguishable with this assay from a pure AS inhibition). The stem II extension to 6 bp could also have inhibited catalytic activity of the core. Regardless of the mechanism the KD effect indicates that accessibility to annealing is present in the regions around 250 and 1410 of the RHO mRNA.
Figure 9.
Comparison of active vs. inactive 266 and 1411 hhRzs to promote opsin knockdown. A single catalytic core mutation (G5C) was used to inactivate the catalytic function of the hhRz to test for reversal of significant KD effects in active 266 and 1411 pGVAL-hhRzs (Number of experiments for each condition: Active hhRz 266: 23; Inactive hhRz 266: 7; Active hhRz 1411: 19; Inactive hhRz 1411: 3). Pairwise comparisons show no significant differences in KD between the active and inactive hhRzs at site 266 (t = −0.1865, p = 0.8534) and site 1411 (t = −1.588, p = 0.1279).
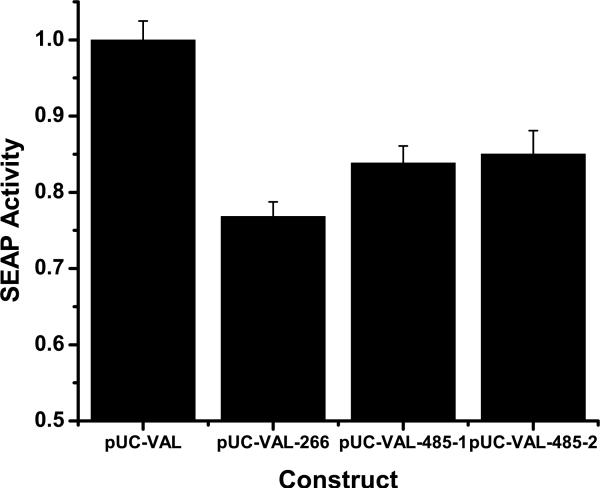
For RHO mRNA the best performing mutation independent (KD) hhRz agent previously realized (Rz525/hhRz485, Gorbatyuk et al., 2007) was able to KD 46% of mouse opsin P23H mRNA in the rat model of retinal degeneration. This hhRz partially rescued ERG a-wave and b-wave amplitudes and outer nuclear layer thickness as indices of efficacy to ameliorate the rate of photoreceptor degeneration (Gorbatyuk et al., 2007). “Rz525” was initially named after the position of the GUC↓ cleavage motif in the canine mRNA sequence (Gorbatyuk et al., 2005, 2007). This sequence corresponds precisely to the region around the equivalent GUC↓ 485 in human rod opsin mRNA. We call Rz525 by the name Rz525/hhRz 485 to locate the position of the site in the humanRHO mRNA sequence. We tested the efficacy of hhRz GUC↓ 485 in comparison to our lead hhRz CUC↓ 266 against human RHO mRNA with our novel HTS assay. Both hhRzs target NUH↓ sites in the coding region of RHO mRNA. To protect hhRzs from degradation inside cells they were expressed from the adenoviral VAI chimera, pUC-VAL, which is our next generation derivative of pGVAL with a reengineered hhRz insertion site (Yau and Sullivan, 2006, 2007; Yau et al., 2008; Yau and Sullivan, submitted). The six bp stem II form of hhRz 266 and two forms of hhRz 485 were evaluated [6 bp Stem II (hhRz 485-1) and 4 bp Stem II (hhRz 485-2); hhRz 485-2 is identical to the hhRz used in Gorbatyuk et al. (2007)]. We determined that the hhRz 266 design (with a 6 bp Stem II design) as identified in this study performs consistently and significantly better (23.17% KD) compared to both a six bp stem II Rz525/hhRz 485 (16.16% KD, p = 0.03) and a classical 4 bp stem II Rz525/hhRz485 (14.97% KD, p = 0.02) that target the same sequence in human RHO mRNA that Rz525 was designed to target in mouse or dog RHO mRNAs (Fig. 10). We attribute the better performance of hhRz 266 vs Rz525/hhRz 485 to a higher level of accessibility in the region around the CUC↓ 266 site in comparison to the GUC↓ 485 site in human RHO mRNA. This outcome is supported by bioinformatics analysis (see Figs. 3–5, Table 1). This identifies hhRz CUC↓ 266 as a higher efficacy agent for preclinical therapeutic testing with human RHO mRNA as the PTGS target.
Figure 10.
Comparison of active hhRz 266 against two forms of Rz525/hhRz485. Two Stem II forms of hhRz 485 were evaluated [6 bp Stem II (hhRz 485-1) and 4 bp Stem II (hhRz 485-2)]. hhRz 485-2 is identical to the hhRz used in Gorbatyuk et al. (2007), which was not expressed in a VAI chimera in that study. All hhRz cDNAs were expressed in the adenoviral VAI chimera (pUC-VAL, a next generation derivative of pGVAL used above). Constructs were evaluated by transfection of the ribozyme or control plasmid into HEK293S cells stably expressing a dicistronic construct, Rho-IRES-SEAP. Secreted alkaline phosphatase (SEAP) levels were measured at 72 hours post transfection by a high throughput fluorescence enzyme assay. The maximum KD possible with this assay is no more than 50%. One-way ANOVA indicated a refutation of the null hypothesis that all the means were identical (F = 20.44, p = 4.33E-10). Within group t-tests were used to identify which pairs were significantly different (criterion p ≤ 0.05). pUC-VAL (control VAI RNA without hhRz) was compared to hhRz 266 (t = −7.40, p = 7.40E-10, significant), hhRz 485-1 (6 bp Stem II) (t = −4.34, p = 8.55E-5, significant), and hhRz 485-2 (4 bp Stem II) (t = −3.72, p = 5.66E-4, significant). All hhRzs promoted significant KD. HhRz 266 was compared with hhRz 485-1 (t = 2.27, p = 0.03, significant) and hhRz 485-2 (t = 2.38, p = 0.02, significant). VAI-HhRz CUC↓ 266 is more potent than VAI-Rz525/hhRz485 against the human RHO mRNA. Finally the two forms of hhRz 485 were compared and found not to be significantly different (t = 0.31, p = 0.75).
4. Discussion
4.1. Computational Approaches to mRNA Accessibility Prediction
Computational algorithms to predict secondary structures of mRNAs of average size are improving. It is now estimated that these algorithms, operating on available oligonucleotide annealing thermodynamic data, are able to predict the base pairs of major secondary conformational states of average sized mRNAs with accuracy as high as 73% (Mathews 2004, 2005; 2006; Mathews et al., 1999b, 2004). We used three in silico bioinformatics approaches to predict accessibility in the secondary structure of mRNAs. The first uses an energy minimization algorithm (MFold) that explores conformational space to identify the single structure with the MFE, and an ensemble of substructures that have lower stability than the MFE. The number of substructures increases exponentially with the size of the mRNA and the allowed differences between conformers with similar energy (Zuker, 1989, 2003; McCaskill, 1990; Zuker et al., 1999; Wuchty et al., 1999). Questions remain as to whether the set of structures close to the MFE contain the native structure, or even if there is a single native structure. RNA is known to undergo kinetic fluctuations in local and perhaps global secondary structure (Wu and Tinoco, 1998; Tinoco and Bustamante, 1999; Treiber and Williamson, 1999; Thirumalai et al., 2001; Onoa and Tinoco, 2004). Also, there is the general expectation that mRNA is folding during transcription into stable secondary structures that precede and support the tertiary fold (Tinoco and Bustamante, 1999). If this is correct then the nature of the structures that form will be exceedingly biased and conditionally dependent upon the local sequence context and perhaps the dynamics of formation of local secondary structure. Algorithms that minimize folding energy do not embrace these putative constraints because they seek the global energy minimized outcome over all possible base pairs. The second approach (SFold), which is not dependent upon energy minimization, involves a Boltzmann-weighted sampling of the statistical distribution of all structures (Ding and Lawrence, 2001, 2003; Ding et al., 2004). SFold assumes that the process of settling to a distribution of conformational substates is an equilibrium process. Probability varies for any secondary structure in the ensemble of all structures. For the regions targeted we found a strong significant correlation between MFold and SFold predictions (Fig. 5). This suggests that both algorithms can be used to identify stable secondary structures with substantial ss annealing platforms.
With the combined in silico computational approaches used here to assess target accessibility we were able to rationally design lead candidate VAI-hhRzs that significantly suppressed human RHO gene product expression in a cultured human cell line. In contrast, hhRzs targeted to regions predicted to be accessible proved to be ineffective. This is testament to use of combined computational approaches to assess RNA accessibility. This aggregate approach (mppRNA) has potential for predicting accessible sites, but will require confirmation by deeper bioinformatics analysis, validation on other mRNA targets, and correlative experimental measures.
4.2. Success and Limitations of 1st Generation VAI-hhRz Designs
We stress that both target accessibility and optimized hhRz performance are essential to achieve the maximized KD of target mRNA and protein. We identified three lead chimeric VAI-hhRz expression constructs, targeting sites 266, 1411, and 1414 in human RHO mRNA, which promote statistically significant RHO protein KD. The protein KD by these leads is not especially deep (at best 30%) at 48 hrs, but the intended identification of accessible target regions now creates further opportunity to optimize these lead hhRz candidates to improve efficacy. These existing lead candidate VAI-hhRzs already have potential for long term stable RHO KD in vivo. We evaluated the extent to which the 266 and 1411 VAI-hhRz constructs were functioning as catalytic agents in vivo by mutational inactivation of the hhRz enzyme. There was no significant reduction in the levels of KD achieved by catalytically inactivated hhRzs compared to active cognate VAI-hhRzs. Understanding the potential causes of this effect is essential to achieve optimized performance of the hhRz at the proven accessible sites. The lack of KD reversal by enzyme mutation may suggest that an antisense or catalytic antisense effect (cleavage with no release of products from the AS flanks-product inhibition) is the dominant form of current target suppression by these VAI-hhRzs. In a pure catalytic hhRz suppression of target the level of KD is expected to completely reverse by inactivating catalytic core mutation. Unfortunately, it is not possible to distinguish a pure antisense effect from a catalytic antisense (single turnover catalysis) by a mutation of the hhRz catalytic core in a cellular testing paradigm. There are several underlying mechanisms as to why the lead candidate VAI-hhRzs could be functioning in this manner. The nature of hhRz hybridization to an accessible region is dependent upon the size of the annealing platform as well as the energy of the hybrids formed, both before and after cleavage. The hhRz could anneal to the accessible region stably, with or without cleavage, and not dissociate from the target on a practical time scale. If the hhRz is capable of cleavage but the products are never released from the hhRz antisense flanks, this results in a form of enzyme product inhibition (catalytic antisense effect). Or, the products may be released slowly from the AS flanks of the hhRz, which constrains potential for enzymatic turnover of the target mRNA population. Preliminary results based on the hhRz kinetic model (Stage-Zimmermann and Uhlenbeck, 1998) suggest that the AS flanks of the lead candidate hhRzs could be optimized to enhance product leaving rates in order to improve catalytic turnover. Such hhRzs could potentially be optimized by decreasing the length and/or the composition of each antisense flank (Bertrand et al., 1994). It is also important to evaluate the extent to which the lead hhRzs fold correctly into active enzymatic structures within the context of the VAI chimeric RNA. MFold and SFold algorithms are highly reliable tools for investigating the folding behavior of small RNAs such as a hhRz, or a hhRz embedded within an small chimeric RNA. A hhRz within a VAI chimera with a 6 bp Stem II (extended 2 bp for stability) may not be enzymatically active. Classical 4 bp Stem II hhRzs embedded within the same chimera realized highly potent activity in cellulo and in vivo (see Lieber and Strauss, 1995). While long extensions of Stem II are known to inhibit hhRz activity, 6 bp appeared to be a reasonable choice to exert additional stabilizing energy over hhRz folding (Homann et al., 1994). Finally, the presence of the hhRz embedded in a chimeric RNA may change the nature of performance when compared to the same hhRz functioning in isolation. These issues are being addressed in an ongoing optimization study.
mRNA target properties clearly influence the level of KD achieved. First, we must consider the extent to which the regions targeted successfully are indeed fully accessible, or perhaps partially constrained by local mRNA structure(s) that impair rapid hhRz AS flank annealing. Experimental tests of small nucleic acid annealing over the regions of predicted accessibility are necessary to correlate with levels predicted by in silico analysis. We have developed novel experimental approaches that directly explore mRNA target accessibility to confirm computational predictions (Sullivan et al., 2007). Potentially interesting features of focal target accessibility arise in analysis of performance of hhRzs designed to target the 250 region loop. While hhRz CUC↓ 266 exerts statistically significant KD, and hhRz CUU↓ 262 hhRz exerts substantial but not significant KD, hhRz CUC↓ 272 exerts minimal KD. Yet, all three NUH↓ sites were predicted to reside within the large (33 nt) accessible ss loop. Even though the AS flanks of each of these hhRzs were designed to hybridize only to predicted ss sequences within this large predicted loop, we expect that focal accessibility within this loop may not be uniform. In contrast, hhRz CUC↓ 1411 and hhRz AUC↓ 1414 exerted significant KD, when the target cleavage sites reside within a single predicted 14 nt ss loop and are separated by only 3 nt. The single site test at UUC↓ 1197 promoted substantial but not statistically significant KD. AS inhibition studies have clearly shown that accessibility in expected ss loops is not necessarily uniform (Lima et al., 1992). Local stable internal hybridization sites or dynamic bp fluctuations within a proven accessible region would lead to nonuniform presentation of the expected ss platform for hhRz annealing. Or, local protein docking within an accessible mRNA region could influence hhRz annealing. With MFold we have observed that there are often several subconformational states around a given region of interest that have varying degrees of access probability. This outcome infers that dynamic fluctuations around a given accessible region could occur, especially since the substates have free energies that are nearly equivalent at physiological temperature. SFold also predicts that the probability of access within the large 33 nt loop predicted by MFold is not uniform but instead has fine structure (Fig. 3). It may also be important that the 266 hhRz targets the apex of the predicted 33 nt loop, whereas the 262 and 272 hhRzs target regions of the loop closer to the supporting stable stem structure. SFold predicts less fine structure in accessibility in the 1410 region. The region around 1190 was predicted by MFold analysis to exist in two well defined substates with ss platforms of 39 and 12 nt. SFold showed only some accessibility in the region. Fine structure mapping of accessibility within these regions could be obtained by experimental oligonucleotide annealing. Another target factor is that there is a known difference in catalytic efficacy of the hhRz when targeting different NUH↓ sites when there is no expected target RNA secondary structure (Zoumadakis and Tabler, 1995). The 266 and 272 hhRzs both target CUC↓ sites while the 262 hhRz targets a CUU↓ site. The 1411 hhRz targets a CUC↓ site while the 1414 hhRz targets an AUC↓ site. The site at 1197 is a UUC↓. Naturally occurring GUC↓ and GUA↓ sites have higher intrinsic cleavage rates with the standard hhRz than all of the other NUH↓ motifs. GUC↓ motifs were targeted in the control sites that were predicted to be poorly accessible and provided no KD. CUC↓ and CUU↓ have comparable rates. CUC↓ has a higher intrinsic rate than AUC↓ such that the slight efficacy difference between the 1411 (CUC↓) and 1414 (AUC↓) might be explained on the basis of the intrinsic cleavage rates.
4.3. Cell culture studies are appropriate for initial PTGS evaluation
PTGS by ribozymes or RNAi takes place in the context of cellular housekeeping functions and components. The PTGS silencing processes neither requires nor utilizes specialized properties of the differentiated cellular phenotype. The target mRNA is transcribed in the nucleus, is spliced, capped and polyadenylated before being trafficked to the cytoplasm. Within the cytoplasm the mature mRNA can reside along trafficking streams, dock at potential storage sites, or be used to translate target protein at the level of the ribosome. The mature mRNA is expected to have associated RNA binding proteins, the bulk of which are expressed in every human cell type. The ribozyme or RNAi will also associate with proteins which are again expressed in every human cell type. The physical chemical environment (ionic composition, ionic strength, pH, viscosity) around the target mRNA within the human cytoplasm is also a housekeeping function that is expected to be maintained across diverse human cellular lineages. The ribozyme or RNAi experiences the same housekeeping environment in a human cultured cell as it would otherwise experience in a disease target cell type in vivo. While in the cytoplasm the target mRNA or ribozyme may undergo structural conformational changes with a broad range of kinetics. These fluctuations are expected to be intrinsic to the local stability of the RNA at physiological temperature. The action of a ribozyme against any mRNA target is expected to take place by the same reaction mechanism regardless of what type of cell it operates within (e.g. HEK293S or rod photoreceptor cell). The cellular protein components of the RNAi-induced silencing complex also reside in housekeeping functions. Because of these facts, albeit not formally investigated, it is highly unlikely that some differentiated cell type specific factor (e.g. a unique RNA binding protein) will be proven to markedly influence PTGS reactions in different types of human or mammalian cells. PTGS operates at the level of cellular housekeeping, the set of functions and components that are shared among all cell types in an organism and are necessary and essential for survival, vitality and support of the overlying differentiated function. The reactions of a ribozyme or RNAi and its cognate mRNA target are only dependent upon the relative molar ratios of the two operative species (enzyme and target RNAs) that specify diffusion-limited collision frequency within the functional enzymatic environment. And, for a ribozyme, no additional cellular machinery is necessary to effect RNA based catalysis after the critical annealing event.
The critical variables underlying cellular hhRz functionality are: 1) the identification of a bonafide accessible site in the target mRNA that is stable over time within the cellular milieu, 2) a hhRz that folds into a structural state in which it can both interact with target mRNA by bp complementarity to perform catalysis, and 3) colocalization of the hhRz into the same compartment as the target mRNA. The structure of the target mRNA and the presence of an accessible site are dependent upon secondary and tertiary RNA structural folding that is entirely conditional upon the primary sequence of the RNA and the physicochemical conditions (ionic strength, temperature, pH) that are variables in this process. The information needed for RNA secondary and tertiary structure is specified in the primary sequence. The spontaneous process of target RNA folding is expected to follow the same kinetic landscape regardless of the cell type in which this folding occurs due to the commonality of the controlling physicochemical variables in all cells. Similarly, the primary sequence of the hhRz and its chimera specifies the RNA folding landscape that may or may not sponsor catalytic activity. HhRz and chimera folding is expected to occur with the same kinetic landscape regardless of cell type. Another critical variable is the specification by the hhRz/chimera RNA sequence of structure(s) that permit a long half-life within cells. At equivalent levels of expression a hhRz with a longer half life will establish a higher steady state enzyme level ([E]). A hhRz with a short half life decreases the steady state [E]/[S] ratio and predicts a lower efficacy measured as target KD (↓[S]). A hhRz that is degraded in the cell with a half life of 30 min will have little potential to effect a strong KD of a target mRNA with a relatively stable half life of 10 or more hours, and even less efficacy against a target with a shorter intrinsic half-life. Ideally, the hhRz primary sequence would be designed to engineer a long cellular half-life to provide the best conditions for high level efficacy. The only way that the hhRz can effect target mRNA KD is if it colocalizes with the target mRNA in the same cellular compartment or subcompartment. Colocalization of hhRz and target mRNA is essential to sponsor a high frequency of biophysical collisions at physiological temperature that are needed to promote effective molecular recognition (annealing) and formation of the enzyme: substrate complex. Clearly, the folded structure of the hhRz RNA modulates the cellular trafficking and eventual localization of the enzyme. Colocalization is dependent upon cellular trafficking signals and motifs that are found in the folded hhRz and target and specified in the primary RNA sequences. Finally, to effect a high frequency of productive collisional interactions in a diffusion-limited reaction the hhRz/chimera concentration must be in substantial molar excess over the target mRNA.
To insure the strong expression, stability, and appropriate trafficking and compartmental localization, the hhRz can be embedded in a chimeric RNA that has the desired prerequisite properties. It is for these reasons that we have chosen the adenoviral VAI RNA as a chimera in which to express hhRzs. VAI-type RNAs have a well established secondary and tertiary structure, are expressed by strong intragenic Pol-III promoter (106–108 copies per transfected cell), have long cellular half lives (~ 7–8 hours), and mostly traffic to the cellular cytoplasm (Svensson and Akusjarvi, 1984; Prislei et al., 1997). All mRNA targets currently relevant to ocular gene therapy spend the bulk of their intrinsic lifetimes in residence within the cytoplasm, where a VAI-hhRz chimera naturally colocalizes. With a VAI-hhRz at such high levels of expression and, at the extreme, an abundantly expressed target mRNA (104 – 105 copies per cell) transcribed by a Pol-II promoter, one expects that the [E]/[S] ratio will be at least on the order of 10-100:1. With moderate or low levels of mRNA target abundance the [E]/[S] ratio would be one to two orders of magnitude greater. Under these conditions of ([E] >(>) [S]) designed into our PTGS cellular testing paradigm, the measured KD efficacy is expected to be solely a property of the accessibility of the target mRNA at the intended annealing site in cellulo. In the challenge to identify lead hhRz candidates for therapeutic development the first step is clearly to determine if the site of potential attack is accessible in vivo. Optimization of such an agent can then occur after lead candidate identification. The investigative paradigm established here seeks to identify lead candidate hhRzs as a first essential step in a stratified RNA drug development platform that includes target mRNA accessibility determination, lead candidate identification, lead optimization, preclinical animal model testing, and human clinical trials.
4.4. On the Kinetics of Ribozyme Mediated Gene Therapy
Is the timescale of our measures of RHO protein and mRNA KD by VAI-hhRzs over 48–72 hours consistent with the expected properties of the target mRNA and protein in photoreceptors? That is, are these experiments representative outcomes of likely in vivo performance? To evaluate this question we need to know the half life of the target mRNA inside the cells in which it is expressed. A number of studies suggest that the half life for RHO mRNA in photoreceptors can be estimated to be approximately 12 hrs (Korenbrot and Fernald, 1989; McGinnis et al., 1992; Pierce et al., 1993). The function of a PTGS agent is to reduce the mean half-life of the target mRNA in the cell which results in a new lower steady state target mRNA level and hence reduced target protein translation. Intrinsic steady state target mRNA levels are the result of the ratio of transcriptional synthetic rates and natural mRNA degradation rates. For assumed constant transcriptional rates, we can simulate the impact of a ribozyme on mRNA decay, which is a well known problem from various engineering and biophysical disciplines. Assuming the natural half life of RHO mRNA at 12 hrs, the occurrence of complete (100%) hhRz mediated KD at time t=0 hrs would result in the decay of RHO mRNA as shown graphically (Fig. 11). The new steady state (0% mRNA remaining) is reached between 4–6 half lives. At one half life (12 hrs) there is approximately 37% (1/e) mRNA target reduction or 63% remaining. A single exponential will decay to 1% of initial value at 5 times the half-life. Obviously, if there is no impact of the PTGS agent then the steady state level of the target mRNA will not change over practical time of analysis (horizontal line). The graph shows the simulated single exponential decay of an mRNA species from an initial steady state concentration (y0 intercept) assuming that all new transcription of the target stopped at time t =0. This would be equivalent to a complete PTGS knockout of target mRNA instantaneously upon transfection. A PTGS agent adds a component to the intrinsic rate of decay of the mRNA, which in effect decreases the half life of the mRNA target. If the PTGS agent is able to KD only a fraction of the total RHO mRNA a new steady state (yf) will emerge where the exponential flattens at a level that represents the ratio of new mRNA synthesis rate to the decay rate (not shown). For simplicity here, we assume that at time t = 0 that all of the synthesis of the target mRNA was obviated by the PTGS agent (100% KD). How long does it take to approach the point of zero remaining mRNA? For the intrinsic 12 hr half life this clearly occurs between 48–72 hrs (4–6 times the half life). In this study opsin protein levels were measured at 48 hrs post transfection so we expect that we are sampling at the new steady state. As a hhRz acts to enhance the decay rate, we have also modeled decay with total decay half times of 9 hrs, 6 hrs, and 3 hrs. In these cases the decay to the final steady state occurs more rapidly than the time scale of our experimental measures. Hence, we expect that our measures at 48 and 72 hrs fully embrace a new steady state that has emerged for the target RHO mRNA in the cellular expression system.
Figure 11.
Model outcomes for the kinetics of RHO mRNA suppression by a hhRz. Mammalian RHO mRNA has a half life (τ) of approximately 12 hrs. In a quantitative model of a steady state expression cellular system the numbers of target mRNAs remains constant. This means that the rate of synthesis is equal to the intrinsic rate of natural decay of the mRNA, or the ratio of synthesis to decay is equal to 1.0. If the rate of synthesis suddenly decreased to zero, then the decay of the numbers of target mRNAs would follow the natural decay time course with a τ equal to 12 hrs. Note that at the 48 or 72 hr readout points, or between 4–6 times the intrinsic expected half life of the RHO mRNA target (τ = 12 hrs), that the steady state has clearly been reached. With a hhRz contributing an increasingly stronger additional component to the decay process (time constant of decay now 9hrs, 6 hrs, or 3 hrs), again without any new transcription, the percent of target would decrease progressively even faster and the 48 or 72 hr assay readout points would again reliably sample the new steady states that emerge. Even if we underestimated the target half life and it was 2 fold greater (τ = 24 hrs) then the 48 and 72 hr time points would still give reasonable estimates of the relative KD between effective and ineffective agents.
We next asked the question as to how we could expect our lead VAI-hhRzs to perform in photoreceptor cells. To do so we need a reliable estimate of the copy number of the RHO mRNA in rod photoreceptors. There are approximately 2 × 108 copies of RHO protein in the normal mammalian rod photoreceptor outer segment. The RHO mRNA traffics selectively to the outer segment side of the nucleus and is translated on polyribosomes there. Approximately 10% of the total amount of RHO protein is lost each day as shed old discs, and to maintain a steady-state outer segment length and visual pigment concentration, there must be synthesis of an equivalent number of new RHO protein molecules (2.0 × 107 opsins/day = 1.33 × 104 opsins/min). Logically, we assume that there is a steady state concentration of RHO mRNA that is maintained in rod photoreceptors to support the steady state level of daily RHO protein translated. This steady state concentration of RHO mRNA must reflect the relative rates of synthesis (transcription) and intrinsic first order decay. From general molecular and cellular biology we assume that each mRNA can be loaded with ribosomes (polyribosome) up to a maximum value of one ribosome each 100 nt (coding region), and that the ribosomes move along the mRNA at a speed of 100 nt/sec. With a coding region of 1044 nt there are approximately 10 ribosomes/opsin mRNA. At the expected translation speed it will take 1 min to make the full RHO polypeptide. With 10 ribosomes translating on the RHO mRNA coding region at steady state then the intrinsic RHO protein translation or synthetic rate (ks,P) is 10 opsins/min-mRNA or 1.44 × 104 opsins/mRNA-day. The number of RHO mRNAs at steady state (NR,ss) in the mammalian photoreceptor is then estimated at:
With an intrinsic RHO mRNA half life of 12 hrs we can calculate an intrinsic degradation rate (kd,R) of RHO mRNA which is 9.63/min With the expected steady state number of RHO mRNAs/cell and the intrinsic degradation rate we can estimate the transcriptional synthetic rate (ks,R) (ks,R = NR,ss *kR,d) (ks,R = 1.35 mRNAs/min). With the daily loss of 10% of the rod outer segment the intrinsic half life (τd,P) of RHO protein loss (to reach 1/e or 63.2% of maximum; e = 2.718) is 3.7 days. The intrinsic decay rate of RHO protein is then 1.30 × 10−4/min. The steady state numbers of RHO protein (NP,ss) can then be estimated (Almagor and Paigen, 1988):
| (1) |
where kd,hhRz is the rate of RHO mRNA degradation contributed by the hhRz. Assuming the rates calculated above and initially that kd,hhRz is zero, then Np,ss is found to be 1.08 × 108 RHO protein molecules per photoreceptor outer segment, which is clearly on the correct order of known biology.
To understand how a hhRz might impact RHO protein expression the photoreceptor we need to estimate kd,hhRz. Fortunately, we can obtain an estimate of this value from HEK293 expression experiments. We can use available data to estimate the number of opsin mRNAs in HEK293S cells under stable transfection conditions. We are able to purify by immunoaffinity approximately 20 μg of human rod opsin from 6.6 × 107 cells (two 15 cm dishes of confluent cells) of WT-HEK293S cells (line 12), which is equivalent to 0.5 × 10−9 moles of protein or 3 × 1014 molecules (Sullivan and Satchwell, 2000; Sullivan and Shukla, 1999). In this stable cell line the opsin cDNA is expressed from a CMV promoter/enhancer. The steady state value of 5 × 106 opsin molecules/cell is consistent with our prior measures by spectrophotometry and electrophysiology in this same exact cell line (Sullivan and Shukla, 1999; Shukla and Sullivan, 1999; Sullivan and Satchwell, 2000). In a transient transfection experiment, as used in the current set of experiments, there is at least 20 μg of opsin synthesized in a 48 hr period post transfection. Initially assuming equivalence of the rate of RHO protein translation per mRNA between the photoreceptor and HEK293 cell (ks,P = 1.44 × 104 opsins/mRNA-day) then for 5 × 106 opsins made in a 48 hr period per cell, or a minimum of 2.5 × 106 opsins/day (1 × 105 opsins/hr) then we can estimate NR,ss equal to 174 RHO mRNAs/cell. When naïve HEK293S cells are transfected transiently with an equivalent CMV expression plasmid, as conducted in this experimental design, there are at least 2–3 fold greater amounts of opsin which are synthesized in the same number of cells over 48 hrs. By proportionality this would raise the average number of RHO mRNAs to approximately 500 mRNAs/cell or about 40% of the level in a mammalian rod photoreceptor at steady state. It appears that the HEK293S cell system under transient CMV promoter expression of RHO cDNAs is fairly efficient and generates approximately 40% of the amount of RHO mRNAs at steady state and within one log order of the number of RHO protein molecules per day. This is not entirely surprising because the CMV promoter, like the rod opsin promoter, is quite strong. With 500 RHO mRNA molecules at steady state and an estimate of the intrinsic kd,R (9.63 × 10−4/min) we can calculate the expected transcriptional rate of RHO in HEK293 cells (ks,R,293 = 0.48 mRNAs/min). With 500 RHO mRNAs/cell and the photoreceptor value of ks,p we can estimate that 1.44 × 107 opsin proteins are made per cell over the 48 hr period of the experimental paradigm used here. A 30% maximum KD of RHO protein leads to an estimated decrease of 4.32 × 106 opsins/cell. From this estimate we can use ks,P to calculate the number loss of RHO mRNA molecules from each cell (a decrease of 150 RHO mRNAs) which leaves approximately 350 RHO mRNAs remaining per cell. Assuming that this is a new steady state value we can use the HEK293-specific RHO mRNA synthetic rate (ks,R,293 = 0.48 mRNA/min) to calculate the total RHO mRNA degradation rate (kd,T,R = kd,R + kd,R,hhRz) finding that kd,T,R is estimated at 1.38 × 10−3/min. This leads us to a new half life of the RHO mRNA of 504 min (8.4 hrs). We use our estimates of kd,T,R and kd,R to calculate kd,R,hhRz(4.12 × 10−4/min). It is relevant that this value is within the measured range of hhRz cleavage rates for long target mRNAs that occur in vivo (Birikh et al., 1997; Hendrix et al., 1996; Bertrand et al. 1994). This rate is the product of the cellular hhRz concentration and the second order reaction rate (both unknowns). We will assume that the hhRz (e.g. hhRz 266) would behave according to the same second order reaction rate in a photoreceptor and that the hhRz concentration (e.g. expressed by delivered AAV genomes) is likewise the same. Using equation (1) and the estimates obtained we find that NP,ss is approximately 70% of normal or a 30% KD of RHO protein. Hence, with a combination of a few critical assumptions and measured values our model suggests that an equivalent level of RHO protein KD could occur in photoreceptors with the current lead hhRz candidates. The impact over many half lives of the protein would be a shorter rod outer segment.
4.5. Preclinical and potential clinical applications of 266 and 1411 hhRzs
The target used in this study is a human mRNA and all hhRzs were designed to precisely recognize human RHO mRNA target by Watson-Crick base pairs. All of the lead candidate hhRzs identified in this study could be used in all or nearly all currently known human RHO mutations (N > 125) (Gal et al., 1997). Since these hhRzs were designed as mutation-independent “KD” agents it might be necessary to design allelic variant genes that encode WT or WT-like proteins that are translated from mRNAs that are resistant to cleavage by the cognate lead hhRz. We have examined the capacity to form allelic variant WT opsins for the hhRz cleavage sites used in this study (Table 3). All of the sites targeted by the hhRzs could be simply and silently altered to form allelic variant WT constructs with no protein coding changes, except for an allelic variant KD-RECON target for hhRz 262, but this did not become a lead candidate. On a preclinical level it is ideal to test ribozymes in small or large scale animal models that are humanized for the target mRNA of concern. Mouse models of human RP have been generated on the mouse rod opsin knockout background with human mutant RHO genomic constructs (Li et al., 1996, 1998). However, animal models not humanized for the desired target have also been used in preclinical testing for ribozymes or RNAi (Lewin et al., 1998; LaVail et al., 2000; Gorbatyuk et al., 2005, 2007). We examined the extent of primary sequence conservation among human, pig, dog, rat, and mouse RHO mRNAs for the annealing of successful 266, 1411, and 1414 lead human hhRzs (Table 4). The site targeted by Rz525/hhRz485 is also shown for comparison. For the CUC↓ 266 hhRz annealing site there is high complementarity among all organisms compared. Mouse and rat rod opsin mRNA primary sequences are identical to a region of human RHO mRNA targeted, and dog and pig mRNAs have only one nt difference. This is not surprising given that the 266 hhRz is designed to cut human RHO mRNA in the coding region, and there is an exceptionally high degree of conservation of rhodopsins at both the protein and mRNA levels. There is minimal primary sequence complementarity of the mouse and rat RHO mRNAs to human mRNA target sites in the 1410 loop (e.g. 5 of 14 nts are different for hhRz 1411). The canine RHO mRNA 3' UTR shows surprising primary sequence conservation with the human RHO mRNA in the 1410 region. Pig 3' UTR sequences were not available for comparison. Even though regions of high primary sequence complementarity occur around the CUC↓ 266 hhRz cleavage site in the coding region of rod opsin mRNA, we emphasize that this does not imply that there will be equivalent accessibility at this local sequence region within the secondary and/or tertiary structures among the human, monkey, mouse, rat, pig, or dog mRNAs. This factor is often unconsidered in preclinical studies, yet could play a substantial role in eventual clinical efficacy outcomes (Sullivan et al., 2007). A successful PTGS agent designed, for example, against a mouse, dog, or pig mRNA target and tested in small or larger scale mammalian models that express animal target mRNAs could fail when transferred into a human clinical trial, even if an identical region in primary RNA sequence is targeted. This could result because the region is inaccessible to hhRz annealing at the secondary and tertiary structural levels in the human target. Similarly, a PTGS agent designed against the human mRNA could fail in a preclinical trial where the targets are animal mRNAs. To establish the best conditions for equivalent clinical translation of efficacy the best target for preclinical studies seeking proof-of-principle support for human therapeutic trials is a human mRNA expressed in a mammalian model with knockouts in native target genes (Humphries et al., 1997; Lem et al., 1999). Currently, such models are exceedingly rare, but much needed.
Table 3.
Mutational alleles susceptible to hhRzs and potential to generate allelic variant WT opsin proteins.
| Ribozyme | WT Sequence | Susceptible Mutations | aWT Sequence | Coding Mutation in aWT | ||||
|---|---|---|---|---|---|---|---|---|
| hhRz 262 | AAC | UUC↓ | CUC | all but T58R | AAC | UUG | CUC | yes F56C |
| N55 | F56 | L57 | N55 | C56 | L57 | |||
| hhRz 266 | UUC | CUC↓ | ACG | all but T58R | UUC | CUG | ACG | no |
| F56 | L57 | T58 | F56 | L57 | T58 | |||
| hhRz 272 | ACG | CUC↓ | UAC | all but T58R | ACG | CUG | UAC | no |
| T58 | L59 | Y60 | T58 | L59 | Y60 | |||
| hhRz 356 | AUG | GUC↓ | CUA | all but V87D and G89D | AUG | GUG | CUA | no |
| M86 | V87 | L88 | M86 | V87 | L88 | |||
| Rz525/hhRz 485* | GUG | GUC↓ | CUG | all but L131P | GUG | GUG | CUG | no |
| V129 | V130 | L131 | V129 | V130 | L131 | |||
| hhRz 785 | ACC | GUC↓ | AAG | all | ACC | GUG | AAG | no |
| T229 | V230 | K231 | T229 | V230 | 1 K231 | |||
| hhRz 1197 | ACC | UUC↓ | CCC | all | ACC | UUG | CCC | no |
| ϕ | ϕ | ϕ | ϕ | ϕ | ϕ | |||
| hhRz 1411 | CAA | CUC↓ | AUC | all | CAA | CUG | AUC | no |
| ϕ | ϕ | ϕ | ϕ | ϕ | ϕ | |||
| hhRz 1414 | CUC | AUC↓ | UUU | all | CUC | AUG | UUU | no |
| ϕ | ϕ | ϕ | ϕ | ϕ | ϕ | |||
Cleavage site is shown in the WT sequence. ϕ indicates that the triplet targeted is in a noncoding mRNA region. The same cleavage site is attacked in most if not all known human rod opsin mutants (Susceptible Mutations). Conversion of NUH↓ to NUG creates a noncleavable site in an allelic variant cDNA and mRNA (aWT). Allelic variation to create a NUG site may promote a new mutation which could be deleterious to local or global protein structure or function (e.g. F56C in aWT for hhRz 262 would alter the primary sequence underlying the first transmembrane helix of rhodopsin). While cleavage potential is considered here there is also the potential for pure antisense effects. hhRzs in bold are lead candidate therapeutics.
Rz525/hhRz485is the lead candidate hhRz of Gorbatyuk et al (2007).
Table 4.
Comparison of mammalian RHO primary sequences around lead hhRz cleavage sites and the 485 site.
| hhRz CUC↓ 266 Target Sites | |||
| 266 ↓ | |||
| Homo sapiens | 249 | 5' ---- UUCCCCAUCAACUUCCUC↓ACGCUCUACGUCACC ---- 3' | 281 |
| Macaca fascicularis | 249 | 5' ---- UUCCCCAUCAACUUCCUC↓ACGCUCUACGUCACU ---- 3' | 281 |
| Sus scrofa | 154 | 5' ---- UUCCCCAUCAACUUCCUC↓ACGCUCUACGUCACG ---- 3' | 186 |
| Canis familiaris | 290 | 5' ---- UUCCCCAUCAACUUCCUC↓ACGCTCUACGUCACA ---- 3' | 322 |
| Mus musculus | 232 | 5' ---- UUCCCCAUCAACUUCCUC↓ACGCUCUACGUCACC ---- 3' | 264 |
| Rattus norvegicus | 237 | 5' ---- UUCCCCAUCAACUUCCUC↓ACGCUCUACGUCACC ---- 3' | 269 |
| hhRz CUC↓1411 and 1414 AUC↓ Target Sites | |||
| 1411↓ 1414↓ | |||
| Homo sapiens | 1398 | 5' ---- CCUCCUCCCAACUC ↓AUC↓ UUUCAGGAA ---- 3' | 1423 |
| Macaca fascicularis | 1409 | 5' ---- CCUCCUCCCAGCUC ↓AUC↓ UUUCAGGAA ---- 3' | 1434 |
| Sus scrofa | n.a. | ||
| Canis familiaris | 1422 | 5' ---- CCUCCCCCCAACUC ↓AUC↓ UCUCAGGAA ---- 3' | 1447 |
| Mus musculus | 1383 | 5' ---- CCCUCCCUCAGCUU ↓GUC↓ UAUCAGGAA ---- 3' | 1408 |
| Rattus norvegicus | 1385 | 5' ---- CCUCCCCUCAGCUU ↓GUG UCUCAGGAA ---- 3' | 1410 |
| Rz525/hhRz485 | |||
| 485 ↓ | |||
| Homo sapiens | 474 | 5' ---- UCCUUGGUGGUC↓CUGGCCAUCGAGCGG ---- 3' | 500 |
| Macaca fascicularis | 474 | 5' ---- UCCUUGGUGGUC↓CUGGCCAUCGAACGG ---- 3' | 500 |
| Sus scrofa | 379 | 5' ----UCCUUGGUGGUC↓CUGGCCAUUGAACGG---- 3' | 405 |
| Canis familiaris | 515 | 5' ----UCUUUGGUGGUC↓CUGGCCAUUGAGCGG---- 3' | 541 |
| Mus musculus | 457 | 5' ---- UCCCUGGUGGUC↓CUGGCCAUCGAGCGC---- 3' | 483 |
| Rattus norvegicus | 462 | 5' ---- UCCCUGGUAGUC↓CUGGCCAUUGAGCGC---- 3' | 488 |
hhRz NUH↓ cleavage sites are written in bold. Nucleotides different from human are written in bold underlined font. For the hhRz 266 target site the entire single stranded region of the predicted loop in RHO mRNA is shown. For the hhRz 1411 and 1414 target sites the entire single stranded region of the predicted loop is shown including additional nucleotides on each side. The regions underlined in the human RHO mRNA segments are the total primary sequence span to which hhRz 266, 485, or 1411 is expected to anneal. The 1414 target site is shifted three nt to the right of the one for site 1411.
The hhRz 266 and hhRz 485 target sites are located in the coding region while the hhRz 1411 and 1414 target sites are located in the 3' UTR. Note the high complementarity across mammalian targets for hhRz 266 and hhRz 485 and the lower degree of complementarity (especially for mouse and rat) for hhRz 1411 and 1414 sites. The GenBank accession numbers for the RHO mRNAs and their sizes are: human (NM000539, 2767 nt), monkey (S76579, 1848 nt), mouse (NM145383, 3249 nt), rat (NM033441, 1493 nt), dog (X71380, 2589 nt), and pig (AF008947, 1150 nt). n.a: the published pig rod opsin mRNA is incomplete has no 5'UT and an incomplete 3'UT.
4.6. How much knockdown is enough for long term therapeutic rescue?
The issue of the minimum or maximum amount of RHO suppression needed to generate and maintain therapeutic rescue remains unsolved and is a difficult problem indeed. Previous studies showed that surprisingly low levels of mutant mRNA KD (~ 15%) were sufficient to rescue photoreceptor degeneration over short and long term periods (1 year) in a rat model expressing a mutant mouse rod opsin transgene (P23H) (Lewin et al., 1998; LaVail et al., 2000). In this line of rats there are approximately equal levels of normal rat opsin mRNA and mutant mouse P23H mRNA. The ribozymes used were mutation-specific agents targeting an engineered cleavage site in the mouse mRNA. These elegant proof-of-principle experimental studies indicated that low to moderate levels of KD may be sufficient in the long term to rescue rod photoreceptors from mutant opsin expression, at least in rodent models. Unfortunately, these hhRz agents cannot translate to large scale mammalian preclinical models or to human clinical trials, which is the goal of RNA drug discovery. Two mutation-independent hhRzs (Rz397 and Rz525) have been designed and tested in vitro, in cultured cells, and in the rat P23H mouse RHO model of retinal degeneration (Gorbatyuk et al., 2005, 2007). Both hhRzs were designed against conserved primary mRNA sequences present in mouse, dog, and human RHO mRNAs, with the obvious intent of stepping successful agents in mouse up to a canine retinal degeneration model due to a natural RHO mutation (Kijas et al., 2002). In particular, the full AS span of Rz525/hhRz485 can precisely anneal to human RHO if the region was accessible. After AAV-mediated transduction Rz397 was able to KD remaining mouse RHO mRNA and protein by 60% and 80%, respectively, in Rho+/− mice which express approximately 50% of normal levels of mouse RHO (Gorbatyuk et al., 2005). A better performing Rz525/hhRz 485, that targets a different region of the mouse RHO mRNA, was able to KD 46% of mouse RHO P23H mRNA in the same rat degeneration model and partially rescued ERG measures and retinal histology as additional indices of efficacy (Gorbatyuk et al., 2007). Knockdown of mutant mouse RHO at the protein level was, however, not detected. AAV vectors delivered Rz525/hhRz485 expression constructs subretinally on postnatal day 16. Expressed from a Pol-II promoter, Rz525/hhRz485 reduced ERG a-wave and b-wave amplitude loss by about 50% compared to control eyes (AAV GFP) when evaluated at 4, 8 and 12 weeks of age. Rz525/hhRz485 also rescued outer nuclear layer thickness in the superior retina by approximately 50% relative to control eyes. The Rz525/hhRz485 has proven preclinical efficacy against a mouse mRNA target.
In a HTS assay recently developed in this lab we compared the efficacy of Rz525/hhRz485 and hhRz 266 against the human rod opsin mRNA with both hhRzs expressed in the same VAI RNA chimera (Yau and Sullivan, 2006, 2007; Kolniak et al., 2007; Yau et al., 2008). The strong Pol-III based transcription from the VAI intragenic promoter is expected to substantially increase expression of Rz525/hhRz485 relative to any Pol-II promoter, and the VAI structure is expected to substantially increase the half-life of the Rz525/hhRz485 compared the poorly stabilized construct previously employed (Gorbatyuk et al., 2007). In the Pol-II expression constructs used thus far for Rz525/hhRz485 the RNA is processed 3' to the actual hhRz element by a downstream in cis hairpin ribozyme, which removes the stabilizing polyadenylation sequence. The mammalian cellular lifetime of such trimmed hhRzs that are capped but not polyadenylated has never been reported. The capacity of a 2'–3' cyclic phosphate at the 3' end of the trimmed hhRz RNA to prevent 3' to 5' RNA exonucleases is also unclear. In our HTS assay hhRz 266 (with a 6 bp Stem II design that generates a predominant AS effect) performs consistently and significantly better (23.2% KD) than both a six base pair stem II Rz525/hhRz485 (16.2% KD) and a classical 4 bp stem II Rz525/hhRz485 (15.0% KD) against human RHO mRNA when expressed in a RHO-IRES-SEAP dicistronic mRNA (Fig. 9). When tested under identical conditions, with all other contributing variables being controlled and uniform, the improved performance of hhRz CUC↓ 266 relative to Rz525/hhRz485 (GUC↓) is expected to result solely from a higher level of accessibility in the region around the CUC↓ 266 cleavage site in humanRHO mRNA relative to the region around GUC↓ 485. Additional bioinformatics and experimental measures of accessibility of the full length human RHO mRNA indicate that the region around 485 is not substantially detectable as an accessible site, when compared to the region around the hhRz 266 cleavage site (Sullivan and Taggart, 2007; Sullivan et al., in preparation; Taggart and Sullivan, in preparation).
This study indicates that with hhRz CUC↓ 266 we already have a PTGS agent for human RHO that is improved in comparison to prior best-performing allele-independent hhRz (Rz525/hhRz485) that exerted some rescue against retinal degeneration in the P23H rat model (Gorbatyuk et al., 2007). Tacitly, we would expect the existing hhRz CUC↓ 266 to have improved performance in vivo, especially when expressed within the VAI-hhRz chimera. Further work is necessary to determine the extent to which optimized versions of hhRz CUC↓ 266 can exert even greater target KD, or if hhRzs targeting other sites might prove more efficacious. Subsequent lead optimization studies are appropriately performed in cellulo prior to more costly animal experimentation. Animal experimentation can occur in an appropriate humanized model in which outcome results are indeed directly translatable to human clinical studies. The agents we have designed and tested are already proven-fit for targeting toxic gain-of-function mutations in human RHO mRNA. For a KD-RECON strategy one must also design and test an allelic variant cDNA and expression construct for a hhRz-resistant RHO mRNA that will reconstitute normal levels of wild type RHO expression in the rod photoreceptors. We demonstrated here how such allelic variant RHO targets can be generated. Long term therapy will likely require regulation of expression or activity of the PTGS agent, real time evaluation of efficacy through appropriate clinical testing measures, and certainly a much deeper knowledge of global photoreceptor gene expression over the time span of the retinal degeneration in appropriate animal models.
4.7. Conclusions
The problem to identify accessible regions for annealing in target RNAs is a particularly severe one that has limited the development of efficacious PTGS agents for the pharmaceutical market over the last three decades. A robust in silico RNA computational approach predicted accessible regions in a model human disease target mRNA and these regions supported successful rational hhRz development. The three lead hhRzs recognize substantial ss annealing platforms predicted in stable secondary structures. Regions predicted to be substantially hybridized or small ss annealing platforms on stable hybridized regions did not support successful hhRz development. These outcomes suggest that a bioinformatics approach has validity to detect accessible regions at least in RHO mRNA and suggests the types of regions that are susceptible to hhRz attack. This bioinformatics driven study contributes to validation of our hypothetical model that large stable ss regions are the best regions for hhRz targeting. The VAI-hhRz constructs (hhRz CUC↓ 266, hhRz CUC↓ 1411, hhRz AUA↓ 1414) that demonstrated significant target protein KD could be useful in preclinical gene therapy or functional genomics studies. These KD hhRzs target all known mutations in the human RHO gene that underlie autosomal dominant RP. The hhRz CUC↓ 266 performs better against human RHO mRNA than the best-performing established allele-independent hhRz (Rz525/hhRz 485) that was designed to target mouse, dog, and human RHO mRNAs. Optimization of the 266 CUC↓ hhRz, the 1411/1414 hhRzs, or identification of hhRzs that target other more accessible regions could ultimately lead to deeper target RHO protein suppression. The hhRzs tested under this rigorous experimental efficacy paradigm to determine regions of target mRNA accessibility in cellulo are proven against human disease target mRNAs expressed in cultured human cells. HhRzs and PTGS agents in general function at the level of cellular housekeeping properties, which reside below differentiated functions. Human cell testing strategies simulate most of the relevant operational biophysical variables for PTGS testing that otherwise exist in retinal cells that express mutant or overexpressed genes. The biocomplexity of RNA biology and PTGS is well represented within the housekeeping functions of all mammalian cell types and requires no functional specialization. While cellular model systems certainly do not preclude the need for animal model systems to test efficacy and toxicity, they are useful tools for development and optimization of allied PTGS agents prior to more costly and much slower testing in animal models of disease. Cellular model systems can also be used for robust screens of combinatorial PTGS expression libraries in search of the best agent against a given target mRNA. A cell based approach, developed in part in this report, is modular, versatile, and cost effective and can be applied to RNA drug discovery for any full length cloned human disease gene.
Acknowledgements
We acknowledge and thank the National Eye Institute (R01 EY13433) (PI: Sullivan) as the primary source of funding for this work. We acknowledge Research to Prevent Blindness Challenge Grants (to the Departments of Ophthalmology at Upstate Medical University and the University at Buffalo), and a grant from the Oishei Foundation (Buffalo, NY) to the Dept of Ophthalmology at University at Buffalo. Dr. Abdelmaksoud acknowledges the support of the Cultural Embassy of the Arab Republic of Egypt during her Ph.D. training in Neuroscience at SUNY- Upstate Medical University. We acknowledge the gift of pGVAL from Andre Lieber, M.D., Ph.D. (Univ. Washington). We acknowledge the assistance of Sean Higgins, Ph.D. (Upstate Medical University) who wrote the macro code to convert RNAFold outcome matrix into individual LFE measures for mapping in early analysis. We appreciate the input of Mae Gordon, Ph.D. (Dept. of Ophthalmology, Washington University School of Medicine) and Greg Wilding, Ph.D. (Dept. of Epidemiology, University at Buffalo) who advised on statistical data analysis. We appreciate the critical read of the manuscript by Lowell G. Sheflin, Ph.D. (Medical Research, VA Western New York Healthcare System), and Shahrokh C. Khani, M.D., Ph.D. (Dept. Ophthalmology, University at Buffalo) prior to its submission.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ABBREVIATIONS AS: antisense; ΔG: free energy (kcal/mol); hhRz: hammerhead ribozyme; HTS: high throughput screening; KD: knockdown; KD-RECON: knockdown-reconstitute approach to gene therapy; LFE: local folding energy; MFE: minimum folding energy; mppRNA: multiparameter prediction of RNA accessibility; Pa,DomSub: mean probability of accessibility for the most frequently occurring substate; PTGS: post transcriptional gene silencing; REM: remaining fraction of opsin; RHO: rod opsin; RP: retinitis pigmentosa; ss: single-stranded; ss loop: single-stranded loop; UTR: untranslated region; WT: wild type
References
- Almagor H, Paigen K. Chemical kinetics of induced gene expression: activation of transcription by noncooperative binding of multiple regulatory molecules. Biochemistry. 1988;27:2094–2102. doi: 10.1021/bi00406a042. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M, Brede G, Babale E, Grotli M, Sproat B, Prydz H. Secondary structure prediction and in vitro accessibility of mRNA as tools in the selection of target sites for ribozymes. Nucleic Acids Res. 2000;28:4113–4124. doi: 10.1093/nar/28.21.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. RNAi in moderation. Nat. Biotechnol. 2006;24:796–797. doi: 10.1038/nbt0706-796. [DOI] [PubMed] [Google Scholar]
- Beck J, Nassal M. Efficient hammerhead ribozyme-mediated cleavage of the structured hepatitis B virus encapsidation signal in vitro and in cell extracts, but not in intact cells. Nucleic Acids Res. 1995;23:4954–4962. doi: 10.1093/nar/23.24.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Pictet R, Grange T. Can hammerhead ribozymes be efficient tools to inactivate gene function? Nucleic Acids Res. 1994;22:293–300. doi: 10.1093/nar/22.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Castanotto D, Zhou C, Carbonnelle C, Lee NS, Good P, Chatterjee S, Grange T, Pictet R, Kohn D, Engelke D, Rossi JJ. The expression cassette determines the functional activities of ribozymes in mammalian cells by controlling their intracellular localization. RNA. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- Birikh KR, Berlin YA, Soreq H, Eckstein F. Probing accessible sites for ribozymes on human acetylcholinesterase RNA. RNA. 1997;3:429–437. [PMC free article] [PubMed] [Google Scholar]
- Brann MR, Young WS. Localization and quantitation of opsin and transducin mRNAs in bovine retina by in situ hybridization histochemistry. FEBS Lett. 1986;200:275–278. doi: 10.1016/0014-5793(86)81151-6. [DOI] [PubMed] [Google Scholar]
- Brauns EB, Dyer RB. Time-resolved infrared spectroscopy of RNA folding. Biophys. J. 2005;89:3523–3530. doi: 10.1529/biophysj.105.061531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Chu C-Y, Rana TM. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- Dale T, Smith R, Serra MJ. A test of the model to predict unusually stable RNA hairpin loop stability. RNA. 2000;6:608–615. doi: 10.1017/s1355838200992495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Lawrence CE. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res. 2001;29:1034–1046. doi: 10.1093/nar/29.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32(supp):W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty EA, Doudna JA. Ribozyme structures and mechanisms. Annu. Rev. Biophys. Biomol. Struct. 2001;30:457–475. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- Drenser KA, Timmers AM, Hauswirth WW, Lewin AS. Ribozyme-targeted destruction of RNA associated with autosomal-dominant retinitis pigmentosa. Invest Ophthalmol. Vis. Sci. 1998;39:681–689. [PubMed] [Google Scholar]
- Fradelizi J, Friederich E, Beckerle MC, Golsteyn RM. Quantitative measurement of proteins by Western blotting with Cy™-coupled secondary antibodies. BioTechniques. 1999;26:484–494. [PubMed] [Google Scholar]
- Gal A, Apfelstedt-Sylla, Janecke AR, Zrenner E. Rhodopsin mutations in inherited retinal dystrophies and dysfunctions. Prog. Ret. Eye Res. 1997;16:51–79. [Google Scholar]
- Giddings MC, Matveeva OV, Atkins JF, Gesteland RF. ODNBase- a web database for antisense oligonucleotide effectiveness studies. Oligodeoxynucleotides. Bioinformatics. 2000;16:843–844. doi: 10.1093/bioinformatics/16.9.843. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk M, Pang J-J, Thomas J, Jr., Hauswirth WW, Lewin AS. Knockdown of wild-type mouse rhodopsin using an AAV vectored ribozyme as part of an RNA replacement approach. Molecular Vision. 2005;11:648–656. [PubMed] [Google Scholar]
- Gorbatyuk M, Justilien V, Liu J, Hauswirth WW, Lewin AS. Preservation of photoreceptor morphology and function in P23H rats using an allele independent ribozyme. Exp. Eye Res. 2007;84:44–52. doi: 10.1016/j.exer.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marior P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Hendrix C, Anne J, Joris B, Van Aerschot A, Herdewijn P. Selection of hammerhead ribozymes for optimum cleavage of interleukin 6 mRNA. Biochem. J. 1996;314:655–661. doi: 10.1042/bj3140655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. Implications of ribozyme kinetics for targeting the cleavage of specific RNA molecules in vivo: more isn't always better. Proc. Natl. Acad. Sci. USA. 1991;88:6921–6925. doi: 10.1073/pnas.88.16.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ, Pardi A, Uhlenbeck OC, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach WL, Hodgson R, Symons RH. Numbering system for the hammerhead. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PG. RNA secondary structure: physical and computational aspects. Quart. Rev. Biophys. 2000;33:199–253. doi: 10.1017/s0033583500003620. [DOI] [PubMed] [Google Scholar]
- Homann M, Tabler M, Tzortzakaki S, Sczakiel G. Extension of helix II and an HIV-1-directed hammerhead ribozyme with long antisense flanks does not alter kinetic parameters in vitro but causes loss of the inhibitory potential in living cells. Nucleic Acids Res. 1994;22:3951–3957. doi: 10.1093/nar/22.19.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormes R, Homann M, Oelze I, Marschall P, Tabler M, Eckstein F, Sczakiel G. The subcellular localization and length of hammerhead ribozymes determine efficacy in human cells. Nucleic Acids Res. 1997;25:769–775. doi: 10.1093/nar/25.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MM, Rancourt D, Farrar GJ, Kenna P, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 1997;15:216–219. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Nelson Chau B, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis TC, Wincott FE, Alby LJ, McSwiggen JA, Beigelman L, Gustofson J, DiRenzo A, Levy K, Matulic-Adamic J, Karpeisky A, Gonzalez C, Woolf TM, Usman N, Stinchcomb DT. Optimizing the cell efficacy of synthetic ribozymes: site selection and chemical modifications of ribozymes targeting the proto-oncogene c-myb. J. Biol. Chem. 1996;271:29107–29112. doi: 10.1074/jbc.271.46.29107. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Suyama E, Iyo M, Taira K. siRNAs generated by recombinant human Dicer induce specific and significant but target site-independent gene silencing in human cells. Nucleic Acids Res. 2003;31:981–987. doi: 10.1093/nar/gkg184. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kijas JW, Cideciyan AV, Aleman TS, Pianta MJ, Pearce-Kelling SE, Miller BJ, Jacobson SG, Aguirre GD, Acland GM. Naturally occurring rhodopsin mutation in the dog causes retinal dysfunction and degeneration mimicking human dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. USA. 2002;99:6328–6333. doi: 10.1073/pnas.082714499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller A, Watzig H. Precision and variance components in quantitative gel electrophoresis. Electrophoresis. 2005;26:2470–2475. doi: 10.1002/elps.200500024. [DOI] [PubMed] [Google Scholar]
- Kolniak TA, Yau EH, Taggart RT, Sullivan JM. Identification of lead candidate ribozymes for human rod opsin therapeutics. Invest. Ophthalmol. Vis. Sci. 2007;48:3248, B498. [Google Scholar]
- Korenbrot JI, Fernald RD. Circadian rhythm and light regulate opsin mRNA in rod photoreceptors. Nature. 1989;337:454–457. doi: 10.1038/337454a0. [DOI] [PubMed] [Google Scholar]
- Koseki S, Tanabe T, Tani K, Asano S, Shioda T, Nagai Y, Shimada T, Ohkawa J, Tiara K. Factors governing the activity in vivo of ribozymes transcribed by RNA polymerase III. J. Virol. 1999;73:1868–1877. doi: 10.1128/jvi.73.3.1868-1877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham MP, Brown DJ, McCallum SA, Pardi A. NMR methods for studying the structure and dynamics of RNA. ChemBioChem. 2005;6:1492–1505. doi: 10.1002/cbic.200500123. [DOI] [PubMed] [Google Scholar]
- LaVail MM, Yasumura D, Matthes MT, Drenser KA, Flannery JG, Lewin AS, Hauswirth WW. Ribozymes rescue of photoreceptor cells in P23H transgenic rats: long-term survival and late-stage therapy. Proc. Natl. Acad. Sci. USA. 2000;97:11488–11493. doi: 10.1073/pnas.210319397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc. Natl. Acad. Sci. USA. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin AS, Drenser KA, Hauswirth WW, Nishikawa S, Yasamura D, Flannery JG, LaVail MM. Ribozyme rescue of photoreceptor cells in transgenic rat model of autosomal dominant retinitis pigmentosa. Nat. Med. 1998;4:967–971. doi: 10.1038/nm0898-967. [DOI] [PubMed] [Google Scholar]
- Li T, Snyder WK, Olsson JE, Dryja TP. Transgenic mice carrying the dominant rhodopsin mutation P347S: evidence for defective vectorial transport of rhodopsin to the outer segments. Proc. Natl. Acad. Sci. USA. 1996;93:14176–14181. doi: 10.1073/pnas.93.24.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Sandberg MA, Pawlyk BS, Rosner B, Hayes KC, Dryja TP, Berson EL. Effect of vitamin A supplementation on rhodopsin mutants threonine-17→ methionine and proline-347 → serine in transgenic mice and in cell cultures. Proc. Natl. Acad. Sci. USA. 1998;95:11933–11938. doi: 10.1073/pnas.95.20.11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber A, Strauss M. Selection of efficient cleavage sites in target RNAs by using a ribozyme expression library. Mol. Cell. Biol. 1995;15:540–551. doi: 10.1128/mcb.15.1.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber A, Kay MA. Adenovirus-mediated expression of ribozymes in mice. J. Virol. 1996;70:3153–3158. doi: 10.1128/jvi.70.5.3153-3158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber A, He C-Y, Polyak SJ, Gretch DR, Barr D, Kay MA. Elimination of Hepatitis C virus RNA in infected human hepatocytes by adenovirus-mediated expression of ribozymes. J. Virol. 1996;70:8782–8791. doi: 10.1128/jvi.70.12.8782-8791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WF, Monia BP, Ecker DJ, Freier SM. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry. 1992;31:12055–12061. doi: 10.1021/bi00163a013. [DOI] [PubMed] [Google Scholar]
- Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DM, Uhlenbeck OC. Kinetic characterization of intramolecular and intermolecular hammerhead RNAs with Stem II deletions. Proc. Natl. Acad. Sci. USA. 1994;91:6977–6981. doi: 10.1073/pnas.91.15.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH. Predicting a set of minimal free energy RNA secondary structures common to two sequences. Bioinformatics. 2005;21:2246–2253. doi: 10.1093/bioinformatics/bti349. [DOI] [PubMed] [Google Scholar]
- Mathews DH. Revolutions in RNA secondary structure prediction. J. Mol. Biol. 2006;359:526–532. doi: 10.1016/j.jmb.2006.01.067. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Burkard ME, Freier SM, Wyatt JR, Turner DH. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999a;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, et al. Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structures. J. Mol. Biol. 1999b;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- McGinnis JF, Whelan JP, Donoso LA. Transient, cyclic changes in mouse visual cell gene products during the light-dark cycle. J. Neurosci. Res. 1992;31:584–590. doi: 10.1002/jnr.490310325. [DOI] [PubMed] [Google Scholar]
- Millington-Ward S, O'Neill B, Tuohy G, Al-Jandal N, Kiang A-S, Kenna PF, Palfi A, Hayden P, Mansergh F, Kennan A, Humphries P, Farrar GJ. Strategems in vitro for gene therapies directed to dominant mutations. Hum. Mol. Genet. 1997;6:1415–1426. doi: 10.1093/hmg/6.9.1415. [DOI] [PubMed] [Google Scholar]
- Mohler WA, Charlton CA, Blau HM. Spectrophotometric quantitation of tissue culture cell number in any medium. BioTechniques. 1996;21:260–266. doi: 10.2144/96212st03. [DOI] [PubMed] [Google Scholar]
- Nathans J, Weitz CJ, Agarwal N, Nir I, Papermaster DS. Production of bovine rhodopsin by mammalian cell lines expressing cloned cDNA: spectrophotometry and subcellular localization. Vision Res. 1989;29:907–914. doi: 10.1016/0042-6989(89)90105-3. [DOI] [PubMed] [Google Scholar]
- O'Neill B, Millington-Ward S, O'Reilly M, Kiang AS, Kenna PF, Humpheries P, Farrar GJ. Ribozyme-based therapeutic approaches for autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 2000;41:2863–2869. [PubMed] [Google Scholar]
- Nakamaye KL, Eckstein F. AUA-cleaving hammerhead ribozymes: attempted selection for improved cleavage. Biochemistry. 1994;33:1271–1277. doi: 10.1021/bi00171a030. [DOI] [PubMed] [Google Scholar]
- Nathans J, Hogness DS. Isolation and nucleotide sequence of the gene encoding human rhodopsin. Proc. Natl. Acad. Sci. USA. 1984;81:4851–4855. doi: 10.1073/pnas.81.15.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoa B, Tinoco I. RNA folding and unfolding. Curr. Opin. Struct. Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Patzel V, Sczakiel G. Theoretical design of antisense RNA structures substantially improves annealing kinetics and efficacy in human cells. Nat. Biotechnol. 1998;16:64–68. doi: 10.1038/nbt0198-64. [DOI] [PubMed] [Google Scholar]
- Patzel V, Steidl U, Kronenwett R, Haas R, Sczakiel G. A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucleic Acids Res. 1999;27:4328–4334. doi: 10.1093/nar/27.22.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Green MR. Nonspecific concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce ME, Sheshberadaran H, Zhang Z, Fox LE, Applebury ML, Takahashi JS. Circadian regulation of iodopsin gene expression in embryonic photoreceptors in retinal cell culture. Neuron. 1993;10:579–584. doi: 10.1016/0896-6273(93)90161-j. [DOI] [PubMed] [Google Scholar]
- Prislei S, Buonomo SBC, Michienzi A, Bozzoni I. Use of adenoviral VAI small RNA as a carrier for cytoplasmic delivery of ribozymes. RNA. 1997;3:677–687. [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Szostak JW. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature. 2001;414:82–84. doi: 10.1038/35102081. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee J, C, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, Collins FS. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherr M, Rossi JJ, Sczakiel G, Patzel V. RNA accessibility prediction: a theoretical approach is consistent with experimental studies in cell extracts. Nucleic Acids Res. 2000;28:2455–2461. doi: 10.1093/nar/28.13.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherr M, Rossi JJ. Rapid determination and quantitation of the accessibility to native RNAs by antisense oligodeoxynucleotides in murine cell extracts. Nucleic Acids Res. 1998;26:5079–5085. doi: 10.1093/nar/26.22.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczakiel G, Tabler M. Computer-aided calculation of the local folding potential of target RNA and its use for ribozyme design. In: Turner PC, editor. Methods in Molecular Biology. Vol 74. Humana Press Inc; Totowa NJ: 1997. pp. 11–15. Ribozyme Protocols. [DOI] [PubMed] [Google Scholar]
- Sczakiel G, Homann M, Rittner K. Computer-aided search for effective antisense RNA target sequences of the human immunodeficiency virus type 1. Antisense Res. Dev. 1993;3:45–52. doi: 10.1089/ard.1993.3.45. [DOI] [PubMed] [Google Scholar]
- Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. USA. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Symons RH. Mutational analysis of a self-cleaving RNA. Nucleic Acids. Res. 1989;17:5679–5685. doi: 10.1093/nar/17.14.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibin Q, Coen M, Terran L. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005;33:1834–47. doi: 10.1093/nar/gki324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P, Sullivan JM. Normal and mutant rhodopsin activation measured with the early receptor current in a unicellular expression system. J. Gen. Physiol. 1999;114:609–636. doi: 10.1085/jgp.114.5.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann TK, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–879. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman BW, Gluzman Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol. Cell. Biol. 1985;5:2051–2060. doi: 10.1128/mcb.5.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JM, Shukla P. Time-resolved rhodopsin activation currents in a unicellular expression system. Biophys. J. 1999;77(3):1333–1357. doi: 10.1016/S0006-3495(99)76983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JM, Satchwell MF. Development of stable cell lines expressing high levels of point mutants of human opsin for biochemical and biophysical studies. Methods Enzymol. 2000;315:30–58. doi: 10.1016/s0076-6879(00)15833-1. [DOI] [PubMed] [Google Scholar]
- Sullivan JM, Pietras KM, Shin BJ, Misasi JN. Hammerhead ribozymes designed to cleave all human rod opsin mRNAs which cause autosomal dominant retinitis pigmentosa. Mol. Vis. 2002;8:102–113. [PubMed] [Google Scholar]
- Sullivan JM, Yau EH, Taggart RT, Butler MC, Kolniak TA. Bottlenecks in development of therapeutic post-transcriptional gene silencing agents. Vision Res. 2008;48:453–469. doi: 10.1016/j.visres.2007.09.011. doi:10.1016/j.visres. 2007.09.011 (published on line in 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JM, Taggart RT. Novel and enhanced approaches to determine local mRNA accessibility. Invest. Ophthalmol. Vis. Sci. 2007;48:4605, B442. [Google Scholar]
- Sullivan JM, Yau EH, Taggart RT, Butler MC, Kolniak TA. High throughput approaches to identify and optimize candidate post transcriptional gene silencing agents. Elsevier Meeting; May 4–5, 2007; 2007. Retinal Degenerations. [Google Scholar]
- Sung C-H, Schneider B, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. USA. 1991;88:8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson C, Akusjarvi G. Adenovirus VA RNAI: a positive regulator of mRNA translation. Mol. Cell. Biol. 1984;4:736–742. doi: 10.1128/mcb.4.4.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Breaker RR. Examination of the catalytic fitness of the hammerhead ribozyme by in vitro selection. RNA. 1997;3:914–925. [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Ayers DF, Malmstrom TA, McKenzie TL, Ganousis L, Chowrira BM, Couture L, Stinchcomb DT. Improved accumulation and activity of ribozymes expressed from a tRNA-based RNA polymerase III promoter. Nucleic Acids Res. 1995;23:2259–2268. doi: 10.1093/nar/23.12.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D, Lee N, Woodson SA, Klimov DK. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- Tinoco I, Jr., Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gauss P, Thermes C, Groebe DR, Gayle M, Guild N, Stormo G, D'Aubenton-Carafa Y, Uhlenbeck OC, Tinoco I, Jr., Brody EN, Gold L. CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc. Natl. Acad. Sci. USA. 1988;85:1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T, Eckstein F. Hammerhead ribozymes: importance of stem-loop II for activity. Proc. Natl. Acad. Sci. USA. 1993;90:6991–6994. doi: 10.1073/pnas.90.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish NK, Heaton PA, Eckstein F. Isolation of hammerhead ribozymes with altered core sequences by in vitro selection. Biochemistry. 1997;36:6495–6501. doi: 10.1021/bi963134r. [DOI] [PubMed] [Google Scholar]
- Watzig H. Quantitation and validation in electrophoresis: definitions and fundamentals. 2005 This is available at the following website: http://www.pharmchem.tu-bs.de/waetzig-supportive.html.
- Wu M, Tinoco I., Jr. RNA folding causes secondary structure rearrangement. Proc. Natl. Acad. Sci. USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S, Fontana W, Hofacker IL, Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49:145–165. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Xie Y, Chen X, Wagner TE. A ribozyme-mediated, gene “knockdown” strategy for the identification of gene function in zebrafish. Proc. Natl. Acad. Sci. USA. 1997;94:13777–13781. doi: 10.1073/pnas.94.25.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau E, Sullivan JM. Cell-based, high-throughput screening of rationally designed ribozymes. Invest. Ophthalmol. Vis. Sci. 2006;47:848. [Google Scholar]
- Yau EH, Sullivan JM. High throughput cellular screening for ribozyme development against arbitrary mRNA targets. Invest. Ophthalmol. Vis. Sci. 2007;48:1681. [Google Scholar]
- Yau EH, Kolniak TA, Taggart RT, Sheflin LG, Sullivan JM. Optimization of lead ribozyme agents for human rod opsin therapeutics. Invest. Ophthalmol. Vis. Sci. 2008;49:5344. [Google Scholar]
- Young RW, Droz B. The renewal of protein in retinal rods and cones. J. Cell Biol. 1968;39:169–184. doi: 10.1083/jcb.39.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. The renewal of photoreceptor cell outer segments. J. Cell. Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH. Biostatistical Analysis. Second Edition Prentice-Hall; Englewood Cliffs, NJ: 1984. p. 718. [Google Scholar]
- Zoumadakis M, Tabler M. Comparative analysis of cleavage rates after systematic permutation of the NUX↓ consensus target motif for hammerhead ribozymes. Nucleic Acids Res. 1995;23:1192–1196. doi: 10.1093/nar/23.7.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Sankoff D. RNA secondary structures and their prediction. Bull. Math. Biol. 1984;46:591–621. [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark BFC, editors. RNA Biochemistry and Biotechnology. Kluwer Academic Publishers; 1999. (NATO ASI Series). [Google Scholar]
- Zuker M. Calculating nucleic acid secondary structure. Curr. Opin Struct. Biol. 2000;10:303–310. doi: 10.1016/s0959-440x(00)00088-9. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]