1997 marked the 15th anniversary of the prion protein (PrP) and the hypothesis that this molecule is the major component of the proteinaceous infectious agents, “prions,” that cause transmissible neurodegenerative diseases in mammals. This anniversary represented a zenith for the hypothesis, at least in media coverage, as two events unfolded. First, experimental transmissions indicated a causal relationship between the bovine spongiform encephalopathy (BSE) epidemic and the appearance of new variant Creutzfeldt–Jakob disease in humans. Second, Stanley Prusiner was awarded the Nobel prize in Physiology or Medicine and a free garage space at the University of California for framing the prion hypothesis (1). While only time can tell this award’s impact, given the already grim parking situation in San Francisco, the “Frontiers” symposium provided an opportunity to assess the prion’s scientific progress.
PrP was discovered during studies of experimental scrapie disease in rodents; other candidate prion proteins are associated with heritable cytoplasmic traits in yeast and fungi (2–4). All prion proteins are host-encoded and come in at least two varieties. The benign “cellular” form is referred to as PrPC, a molecule that is most probably present in all mammals and expressed on the surface of neurons via a glycophosphatidylinositol anchor. NMR–derived structures have been established for recombinant mouse and hamster prion proteins that approximate PrPC. These reveal a globular C-terminal domain preceeded by a far larger, yet unstructured, N-terminal region (5, 6) (Fig. 1). The mischievious form of the prion protein is known as PrPSc (for scrapie) or alternatively as PrPres (for protease-resistant). PrPC serves as a necessary precursor to PrPSc in a posttranslational remodeling event that encompasses changes in secondary and tertiary structure (7, 8). Point mutations and deletions of rodent PrP genes profoundly affect the course of prion infections, and in humans missense mutations appear to cause “familial” prion diseases. Pathogenic PrPs engendered by D178N (Fatal familial insomnia) and E200K (Familial Creutzfeldt–Jakob disease) mutations have distinct conformations, as assessed by the protease sensitivity of their amino termini, and breed true in serial transmission. This result suggests that different conformations of PrP caused by point mutations are responsible for prion “strains” (9), as indicated by earlier analyses of transmissible mink encephalopathy “strains.”
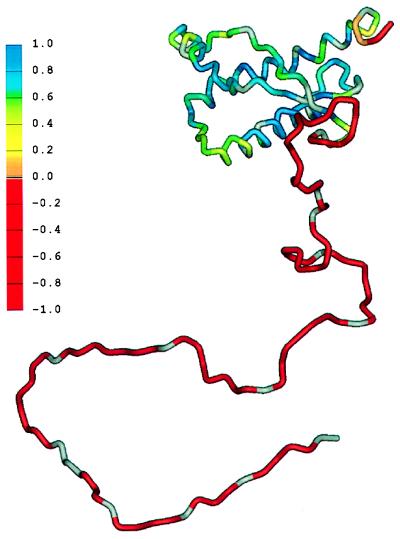
Figure 1.
Schematic diagram showing the flexibility of the polypeptide chain for PrP(29-231). The structure of the portion of the protein representing residues 90–231 was taken from the coordinates of PrP(90-231) (17). The remainder of the sequence was hand-built for illustration purposes only. The color scale shows the heteronuclear [1H]-15N NOE data, from red for the lowest (most negative) values, where the polypepide is most flexible, to blue for the highest (most positive) values in the most structured and rigid regions of the protein. (Courtesy of the University of California, San Francisco and reproduced with permission from ref. 6.)
The foremost stumbling block to acceptance of the prion hypothesis is the lack of pure preparations of infectious prions. Because the minimum dose of purified hamster scrapie necessary to establish an infection contains ≈100,000 PrPSc molecules, it is unclear whether subtypes of PrPSc are the true infectious agent or whether such preparations harbor cryptic agents. Such agents might be viruses or a hypothetical combination of a host-encoded macromolecule in conjunction with a small nucleic acid (an entity dubbed a “virino”; (ref. 10). The key role of PrP seen in genetic experiments can be rationalized in terms of a “viral” receptor. Proponents of these alternative hypotheses stress the ability of distinct scrapie strains to propagate in the same host, implying a nucleic acid “genome” that confers strain characteristics. Ultimately, the debate will only be resolved by experimentation. If, as some believe, familial “prion” diseases are really caused by a PRNP-linked proviruses, then it should be possible to retrieve these by molecular cloning and to transmit infectivity by DNA transfection. With regard to PrPC serving as a receptor, it may be possible to graft the “viral” binding site onto a different glycophosphatidylinositol-anchored protein (e.g., Thy-1) and rescue the “infectability” of PrP gene ablated mice, even though such animals would be incapable of expressing the protease-resistant core of PrP (“PrP27-30”) equated with the infectious agent.
The simplest form of the “protein-only” prion hypothesis suggests that infectious molecules can be created by coercing PrPC to adopt PrPSc-like conformations Assuming controls to exclude contamination by preexisting prions, such experiments would constitute powerful evidence in favor of the prion hypothesis. Thus far, radiolabeled PrPC has been converted to a protease-resistant form by denaturation and prolonged incubation with preparations of PrPSc (11). Although these “conversion reactions” mimic strain and species-barrier phenomena associated with prion biogenesis in vivo, ambiguities remain. The radiolabeled PrPC derives from immunoprecipitations rather than a recombinant source, and because PrPSc cannot be purified to homogeneity, the identity of the active ingredient(s) is hard to pin down. More importantly, it has not been possible to prove that the newly synthesized protease-resistant PrPs are infectious. Nonetheless, these studies favor templated protein folding arising from physical interactions between PrPC and PrPSc, a concept close to the heart of the “protein-only” hypothesis.
What function does PrPC serve? Phenotypes under review in PrP ablated mice include lymphocyte activation, altered neurotransmission, perturbed circadian rhythms, loss of Purkinje cells, and alterations in superoxide dismutase activity (SOD-1). Deficits in the copper content of brain membranes have been described in two lines of ablated mice, an observation paralleled by studies revealing that copper binding to the N terminus of PrP is highly cooperative and more avid than previously thought (12); these data indicate that metal-bound forms of PrPC may exist in vivo. The issue of PrPC function may be important in providing insights into disease. For example, unique structural properties of PrPSc can be deduced only by using PrPC as a point of reference. Also, as PrPC is a precursor to PrPSc, a therapeutic approach is to reduce substrate availability. This approach might be effected by manipulating interactions with protein ligands (13); such a crucial ligand has been deduced from transgenetic studies of “species barriers” to prion transmission.
Matching between the primary sequence of PrPSc in inocula and host-encoded PrPC can affect transit of prions from one species to another, as exemplified in studies in which a barrier to infection of mice with hamster prions is alleviated by expression of hamster PrPC. Although initial efforts to extend this paradigm to human prions were not successful (14), the problem was solved by superimposing human PrP transgenes on a background lacking mouse PrPC or by transgenic expression of mosaic mouse-human-mouse PrPC molecules (15). These data imply a ligand, presumably a protein and denoted “protein X,” that interacts with PrPC in a species-specific manner; human PrPC interacts weakly with mouse protein X such that it cannot form functional complexes in the presence of competing mouse PrPC. Although the protein X binding site is mapped to the C terminus of PrPC, protein X itself has yet to be identified. Conversely, although no less than 11 putative PrP ligands have been identified by in vitro binding or the yeast two-hybrid system, there are questions here, too. Some ligands are located in incorrect cellular compartments (assuming that PrPC is displayed at the cell surface, internalized in endosomes, and degraded in lysosomes), and in no case have (i) independent methodologies identified the same ligand in an unequivocal fashion, (ii) binding sites been mapped on PrPC, (iii) binding affinities been estimated, or (iv) genetic strategies been exploited, e.g., ligand gene knockouts, to demonstrate binding in vivo. Clearly, an important task for the future is to define the true PrPC ligands.
In summary, although the prion hypothesis is not universally accepted, it provides a useful framework to approach the biology of these infectious diseases. At one end of the spectrum, even skeptics concede that the prion protein is a “gatekeeper” controlling disease susceptibility. At the other extreme, prion proteins comprise the prototype of a new class of infectious pathogen, establish protein misfolding as a novel mechanism of disease pathogenesis, and prompt the suggestion that simple organisms use prion-like mechanisms to switch physiological states and thereby adapt to new environments (16). As more detailed structural and biochemical analyses of prion proteins are completed, we can anticipate that fascinating surprises and puzzles will emerge as fast as current issues are laid to rest.
ABBREVIATION
- PrP
prion protein
References
- 1.Harris R F. Curr Biol. 1997;7:R668. doi: 10.1016/s0960-9822(06)00348-4. [DOI] [PubMed] [Google Scholar]
- 2.Wickner R B. Science. 1994;264:567–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 3.Chernoff Y O, Lindquist S L, Ono B-i, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 4.Coustou V, Deleu C, Saupe S, Begueret J. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 6.Donne D, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caughey B, Raymond G J. J Biol Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 8.Borchelt D R, Scott M, Taraboulos A, Stahl N, Prusiner S B. J Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Telling G C, Parchi P, DeArmond S J, Cortelli P, Montagna P, Gabizon R, Mastriani J, Lugaresi E, Gambetti P, Prusiner S B. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson A G, Outram G W. In: Genetic Aspects of Unconventional Virus Infections: The Basis of the Virino Hypothesis. Bock G, Marsh J, editors. Chichester, U.K.: Wiley; 1988. pp. 63–83. [DOI] [PubMed] [Google Scholar]
- 11.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Jr, Caughey B. Nature (London) 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 12.Brown D R, Qin K, Herms J, Madlung A, von Bohlen A, Manson J, Strome R, Fraser P E, Kruck T, Schulz-Schaeffer W, Giese A, Westaway D, Kretzschmar H A. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 13.Prusiner S B. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 14.Telling G C, Scott M, Foster D, Yang S-L, Torchia M, Sidle K C L, Collinge J, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1994;91:9936–9940. doi: 10.1073/pnas.91.21.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 16.Lindquist S. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 17.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]