Abstract
Radiology reports are permanent legal documents that serve as official interpretation of imaging tests. Manual analysis of textual information contained in these reports requires significant time and effort. This study describes the development and initial evaluation of a toolkit that enables automated identification of relevant information from within these largely unstructured text reports. We developed and made publicly available a natural language processing toolkit, Information from Searching Content with an Ontology-Utilizing Toolkit (iSCOUT). Core functions are included in the following modules: the Data Loader, Header Extractor, Terminology Interface, Reviewer, and Analyzer. The toolkit enables search for specific terms and retrieval of (radiology) reports containing exact term matches as well as similar or synonymous term matches within the text of the report. The Terminology Interface is the main component of the toolkit. It allows query expansion based on synonyms from a controlled terminology (e.g., RadLex or National Cancer Institute Thesaurus [NCIT]). We evaluated iSCOUT document retrieval of radiology reports that contained liver cysts, and compared precision and recall with and without using NCIT synonyms for query expansion. iSCOUT retrieved radiology reports with documented liver cysts with a precision of 0.92 and recall of 0.96, utilizing NCIT. This recall (i.e., utilizing the Terminology Interface) is significantly better than using each of two search terms alone (0.72, p = 0.03 for liver cyst and 0.52, p = 0.0002 for hepatic cyst). iSCOUT reliably assembled relevant radiology reports for a cohort of patients with liver cysts with significant improvement in document retrieval when utilizing controlled lexicons.
Keywords: Controlled vocabulary, Natural language processing, Information storage and retrieval
Introduction
Radiology reports are permanent legal documents that serve as official interpretation of radiology tests. The clinical indication(s) for each examination, relevant clinical history, and pertinent findings are often recorded in the form of narrative text. Several studies have evaluated various methods including natural language processing (NLP) for extracting information from these unstructured text reports for different purposes—from differentiating significant versus normal radiology results, to finding patients who have specific findings (e.g., lung mass or pulmonary embolus) [1–6]. In addition, multiple studies have looked at semantic structures that were specifically constructed in order to represent findings within radiology reports [1, 7].
Enormous volumes of radiology reports resulting from tests indicated for screening, follow-up, evaluation, and treatment planning provide an excellent data resource for research and for clinical quality improvement activities [8, 9]. Retrieving cohorts of patients based on findings described in the textual reports provides a step towards analyzing patient outcomes as well as provider behavior. A major drawback is that wading through hundreds of thousands of reports to retrieve relevant patients is cost and time prohibitive. Nevertheless, retrieval of reports with pertinent findings is a critically important task. This study aims to implement and demonstrate the use of a toolkit for retrieving radiology reports that describe clinically relevant findings, using the specific clinical case of liver cysts.
Several NLP and information retrieval algorithms have been developed to enable information extraction from narrative reports and document retrieval based on content [7, 10–14]. However, these implementations require technical and programming skills to use [15]. We developed an ontology augmented NLP toolkit that automatically retrieves radiology reports based on relevant clinical findings described in the text. This toolkit provides several components that can be utilized alone or in combination, without the need for further customization or programming. In addition to searching based on ontologically defined synonyms, our algorithm exploits a valuable feature of radiology reports—headings that provide some structure to the text (e.g., indication, technique, findings) to further enhance search and retrieval.
This study describes the development and initial evaluation of our toolkit, Information from Searching Content with an Ontology-Utilizing Toolkit (iSCOUT), and compared precision and recall with and without using synonyms for query expansion from a controlled terminology.
Materials and Methods
iSCOUT is comprised of a core set of modules, including the Data Loader, Header Extractor, Terminology Interface, Analyzer, and Reviewer described below. In addition, ancillary components include Stop Word Remover and Negator functions. The toolkit enables users to search for specific findings, identifying radiology reports containing exact and/or similar term matches. To improve retrieval, we enable two complementary methods for including lexical and semantic variants of the query terms—an interface to a list of terms provided by a domain expert, and an interface to controlled terminologies. In this implementation, we utilize the Radiology Lexicon (RadLex) as well as the National Cancer Institute Thesaurus (NCIT) [16, 17].
The tools were tested in two use cases in order to demonstrate how individual components interact with each other for retrieving radiology reports. NCIT synonyms for the search term “liver cyst” were utilized in the use cases because there were no synonyms available in RadLex.
Materials
All iSCOUT components were originally developed for this study, written in Java programming language and distributed as jar files. Several guidelines were followed in the design of the individual tools, including enhanced usability (e.g., “ease of use”), efficient performance, and portability. These guidelines enable widespread usability of the toolkit for researchers in an informatics research setting who have technical as well as nontechnical backgrounds. One of the authors (RL) wrote the initial programs and utilized iterative refinement from input given by clinicians and investigators at our institution to make the tools more robust. Currently, we utilize the toolkit to generate cohorts for research as well as for quality improvement activities by automating retrieval of relevant radiology reports.
With Institutional Review Board (IRB) approval, radiology reports for 338 recently completed abdominal computed tomography (CT) scans were obtained from our institution’s Emergency Department in June 2010. All radiology reports were processed as regular text files, without further formatting requirements. The requirement for obtaining informed consent was waived by the IRB for this Health Insurance Portability and Accountability Act-compliant study.
Components
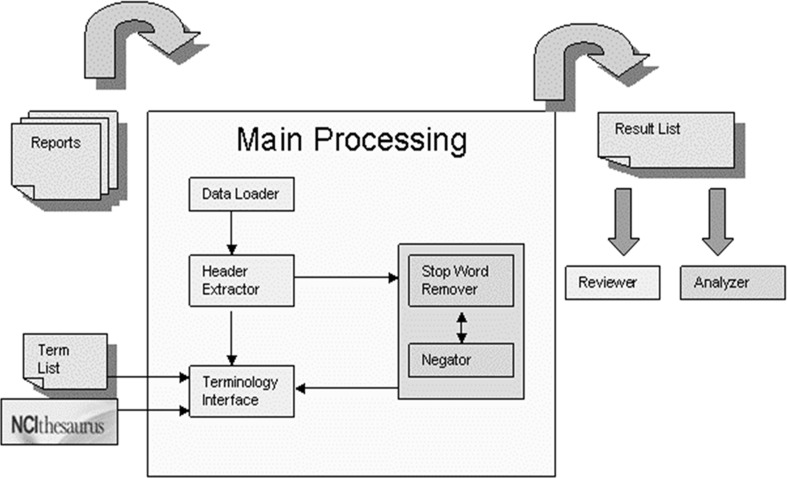
Figure 1 illustrates an overview of iSCOUT. The main processing component uses a string matcher to look for each of the search terms in a regular sentence within the data. A sentence is defined as a unit of one or more words, concluding with appropriate end punctuation (e.g., period). In the simplest case wherein there is a single search term (e.g., “liver cyst”), the search begins by assessing whether all words in the search term are contained in any one sentence within the report. If so, then this report is retrieved. Otherwise, the search is terminated. If more search terms are provided, the process iterates through all possible search terms, one search term at a time, until a match is obtained or the list of terms is exhausted. Various mechanisms for providing more search terms, also known as query expansion, will be discussed further in the Controlled Terminology section. After automated processing, a results list is generated for review and analysis.
Fig. 1.
iSCOUT toolkit architecture
Preprocessing
The first step in any textual analysis involves processing an input file into an analytical file. For document retrieval involving radiology reports, a report with a unique identifier (e.g., accession number) is typically considered the unit of analysis. In iSCOUT, the Data Loader accepts as input a single file containing concatenated radiology reports in regular text format. Each radiology report is stored in the picture archiving and communication system, uniquely identified by an accession number [18]. A database query enables extraction of reports, typically within a specified time frame, in a single file. This is easily accessed as a text file. The only requirements are that each individual report starts with an identifying number as the first token separated from the rest of the report with the character “~”, and that each report is separated by a line of white space from the next one, as illustrated in Fig. 2. The central engine of the Data Loader is a file separator. The file separator parses the text file into tokens (e.g., words) and sentences. A sentence is defined as a sequence of tokens, separated from the next sentence by any of the following punctuation marks—“.”, “?”, “!”. It then delineates individual reports, each identified by the report accession number. In addition, the Data Loader removes extraneous characters (e.g., +, ~), white spaces, and extra lines between text reports. The output of the Data Loader is a new file that is suitable for further processing by other components of the iSCOUT system (e.g., Header Extractor).
Fig. 2.
Sample of radiology reports for iSCOUT input. Each report begins with a unique identifier and is separated from the next one by a line of white space
Utilizing Document Structure
The Header Extractor application enables searching for terms contained in specific areas of the report (e.g., Findings), as opposed to searching from within the entire text of the report. Falsely retrieving reports commonly occurs when the Indications section is included in the text being queried. The following text demonstrates how a query for a lung nodule in the Indications section might be misleading, “INDICATION: Suspected lung nodule on prior chest x-ray.” iSCOUT would falsely retrieve the report that contains the aforementioned text, if the Header Extractor is not utilized. Without utilizing the Header Extractor, the search proceeds to include all headings in the report. With its use, only portions of the report which include current findings are included in the search fields. For example, Clinical History or Indications fields are excluded from search. Conversely, we have shown that for findings of a lung nodule, for example, the documentation of the presence of a lung nodule in the Findings section may not be consistent with its being documented in the Impression section of the reports, so errors could occur if only the Impression section were searched [19]. Using the Header Extractor component, we can exclude from search the history, clinical history, comparison(s), technique(s), and indication(s) sections, searching only the finding(s), impression(s), and conclusion(s) sections. Currently, the headers are identified using existing header terms, which occur in capitalized forms in our reports (e.g., HISTORY). Lexical variants (e.g., plural forms) are included in finding headers, as well as some semantic variants obtained from RadLex (e.g., conclusion and summary as synonyms for impression).
Term Matching
The Terminology Interface component performs the search procedure and takes a search term as input. The algorithm proceeds by utilizing each sentence within the report. A match is determined for a radiology report when a sentence contains all of the tokens within the search term, even when the tokens are not adjacent to each other. This retrieves the report, identified by its unique identifier. Otherwise, when all sentences in a report are exhausted and the search terms are not found in any single sentence, the report is not retrieved.
Controlled Terminology
The Terminology Interface allows query expansion based on synonymous or other related terms. Query expansion enables retrieval of more reports than are generally obtainable using a single query term by allowing more search terms (e.g., synonyms) to be utilized in the search. The Terminology Interface enables two approaches to accessing similar terms—by utilizing a controlled terminology (“terminology approach”) or by enabling an interface with a list of similar terms provided by a domain expert (“expert approach”). In the terminology approach, the module employs a Java Database Connectivity/Open Database Connectivity (ODBC) driver and utilizes the ODBC driver to connect to local copies of the NCIT and Radlex databases. Recent versions of the terminologies from the NCIT and RadLex websites were downloaded to a local database server [17, 20]. A user has the option of selecting which terminology to use. Synonyms of the term found in the lexicons are used to expand the query. Table 1 shows synonyms of several terms from NCIT and from RadLex. NCIT identifies a preferred term for a given concept (e.g., lung carcinoma—NCIT code C4878), as well as synonyms for the preferred term. RadLex, on the other hand, was utilized by finding synonyms of the search term or by postcoordinating words in the search term to find similar terms. For instance, the search term “lung carcinoma” does not have a unique RadLex concept and is therefore postcoordinated by appending the two words “lung” (Radlex ID RID1301) and “carcinoma” (RadLex ID RID4247) together. Synonyms of both words, if available, were also included in the list of similar terms.
Table 1.
Synonyms of terms from NCIT and RadLex
| Preferred term | NCIT | RadLex |
|---|---|---|
| Lung carcinoma | Cancer of lung, cancer of the lung, carcinoma of lung, carcinoma of the lung, lung cancer, lung carcinoma | Lung + carcinoma |
| Kidney cyst | Renal cyst, cyst of kidney, cyst of the kidney, kidney cyst | Kidney + cyst |
The expert approach utilizes an interface with a text editor, wherein a domain expert can specify terms deemed similar to the search term. All that is required of the expert is that they enter similar terms separated by line spaces within a text file. Training on or familiarity with the toolkit is not required. The expert terms are then accessed by the Terminology Interface and used to expand the query. When the text file is left blank, the default approach is to search without the use of expert terms.
Review and Analysis
After a list of radiology reports are retrieved utilizing a search term with or without query expansion, the Reviewer module allows for a manual review of retrieved results for the purposes of validation. The Reviewer finds the entire report for each of the accession numbers returned and saves them all in a single text file for review offline using any text editor. The radiology reports are obtained from the original files, before preprocessing, which are easily readable by human reviewers. The process of generating a list of relevant accession numbers, as well as a file with corresponding radiology reports is automated.
An Analyzer was constructed for the purposes of evaluating the performance of iSCOUT. The Analyzer program accepts as input two lists—the accession numbers of all reports retrieved using iSCOUT, and all reports that should have been retrieved based on manual review by domain experts or curators. The latter represents the gold standard for comparison. These lists allow the Analyzer to calculate the precision and recall for the particular query, two frequently utilized performance metrics for information retrieval [21]. Precision is defined as the proportion of true positive reports to the total number of reports retrieved (see Table 2). Recall is defined as the proportion of true positives that were actually retrieved to all reports that should have been retrieved. Precision is similar to the positive predictive value, whereas recall is similar to test sensitivity.
Table 2.
Definition of precision and recall (TP = true positive, FP = false positive, TN = true negative, FN = false negative)
| Also known as | Formula | |
|---|---|---|
| Precision | Positive predictive value | 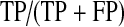 |
| Recall | Sensitivity | 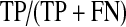 |
Ancillary Tools
Two ancillary tools commonly used for information retrieval include a Stop Word Remover and a Negator. Stop words are common words that frequently occur in the text and contain very little additional information [22]. We identified stop words by finding the 10 most common words in the entire dataset. These include “the,” “is,” “of,” “and,” “no,” “there,” “are,” “in,” “with,” and “or.” Stop Word Remover is an optional tool that removes these words from the data. The stop words selected are very similar to those published in the literature [19].
A simplified Negator identifies pertinent negatives in the radiology reports. More precisely, the Negator looks for search terms that are explicitly negated in the text (e.g., “no lung cancer”), and ensures that reports are not retrieved. Currently, the Negator consists of a set of rules for identifying negations, similar to commonly used negation detection algorithms [23, 24]. For instance, the following negation terms, “no,” “not,” and “unlikely,” when found in a sentence with the corresponding search term, considers the term negated. Thus, a report is not retrieved.
Demonstration
Two independent annotators manually searched for reported cases of liver cysts (the “search term”) within abdominal CT scan reports and identified 25 such cases by consensus. This is used as truth. To demonstrate iSCOUT capabilities for query and retrieval of radiology reports, we describe two use cases: one utilizing the default settings for searching using iSCOUT and another query utilizing the Terminology Interface to expand the query term. Report retrieval was evaluated against a set of reports retrieved by human annotators, comparing precision and recall when using a terminology for query expansion to that using a single search term.
Case 1: Query Using the Search Term “Liver Cyst”
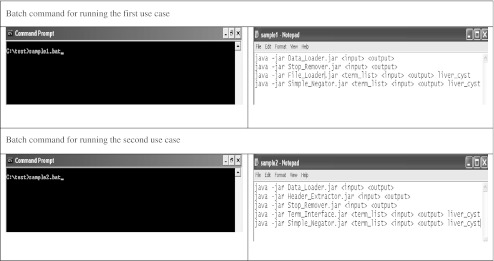
In the first query, the search term was used to look for abdominal CT scan reports that reported liver cysts in any portion of the radiology report. The Header Extractor was not used to refine the search to only certain areas of the reports because it was anticipated that utilizing the entire text of the reports would not substantially influence the query. The rationale: following up hepatic cysts are usually not the primary indication for an abdominal CT scan. Thus, the search term did not frequently appear in the Indication or Clinical History sections of the report. Rather, hepatic cysts are often mentioned in the Findings or Conclusion sections, both of which would result in appropriate report retrieval whether or not the Header Extractor was utilized. Thus, this query utilized only the following components of iSCOUT: Data Loader, Terminology Interface, Stop Word Remover, and Simple Negator. The top panel of Fig. 3 illustrates the batch file, utilizing these components in series. This query yielded 20 total reports. The Analyzer was then used to calculate precision and recall.
Fig. 3.
Batch file for running two use cases
In order to ascertain that certain portions of the reports do not lead to falsely identifying liver cysts by allowing search in some fields of the report (i.e., searching through the Indication section), the Header Extractor was utilized in an additional query. As expected, the resulting retrieved reports were identical to the ones from the first query (20 reports).
A second query was performed using the search term “hepatic cyst.” This query yielded 13 reports. The Header Extractor was utilized in an additional query and, as expected, retrieved the same 13 reports. Again, the Analyzer was used to calculate precision and recall. The automated process took less than 2 s in total. No further customizing of the tools was performed and no additional terms were provided by the user or any expert in the domain.
Case 2: Query Using the Search Term “Liver Cyst” with the Terminology Interface
A third query was performed, utilizing the Terminology Interface component and not utilizing the Header Extractor. As expected, more search terms were utilized in the query. Instead of only searching for “liver cyst,” Table 3 shows several other search terms that were included in the query. The NCIT code or unique identifier for these terms was C3960, all corresponding to the preferred label “hepatic cyst”.
Table 3.
Query terms using the National Cancer Institute Thesaurus (NCIT)
| Search terms |
|---|
| Liver cyst |
| Hepatic cyst |
| Cyst of liver |
| Cyst of the liver |
This query utilized the following AART components: Data Loader, Header Extractor, Stop Word Remover, Terminology Interface, and Simple Negator and yielded 26 total reports. The bottom panel of Fig. 3 illustrates the batch file, utilizing these components. The Analyzer was then used to calculate precision and recall.
Results
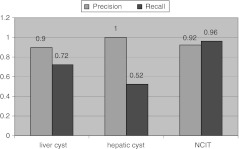
Figure 4 illustrates the performance measures using iSCOUT for the two cases described. As previously noted, 20 records were retrieved in case 1 (using the search term “liver cyst”), 18 of which were true positives, for a precision of 0.90 (18/20). Using the search term “hepatic cyst”, 13 records were retrieved, all of which were true positives, for a precision of 1.0 (13/13). A total of 26 records were retrieved in case 2, of which 24 were reports that contained liver cysts or the ontology synonyms, for a precision of 0.92 (24/26). Using Header Extractor did not change the results for either case.
Fig. 4.
Precision and recall of iSCOUT toolkit
Expert review identified a total of 25 radiology reports with liver cysts. The recall measures in case 1 were 0.72 (18/25) and 0.52 (13/25) for the search terms “liver cyst” and “hepatic cyst,” respectively. In case 2, the recall measure was 0.96 (24/25). The recall for the search utilizing the Terminology Interface is significantly better than using the either search term alone (p = 0.03 and p = 0.0002, McNemar’s exact test). Table 4 illustrates several example sentences from reports retrieved utilizing “liver cyst” and “hepatic cyst.”
Table 4.
Example report sentences
| Search term | Sentences |
|---|---|
| Liver cyst | There are multiple hypodensities in the liver which are unchanged, the largest which measures 1.5 cm in the right lobe and is consistent with a cyst |
| Several low attenuation liver lesions likely represent cysts | |
| Hepatic cyst | Impression: Hepatic cysts/biliary hamartomas |
| There is a 2.3 cm hepatic cyst within segment 4A of the liver |
Discussion
This study describes a publicly available toolkit we developed for retrieval of relevant radiology reports documenting a specific finding. The toolkit is implemented in a modular fashion, allowing various components to be used in combination, and does not require extensive end-user training. In addition, the components are designed to function without requiring an expert in the domain of discourse.
Case 1 illustrates how the toolkit can be utilized with a single search term. As demonstrated in this example, more radiology reports were retrieved using the search term “liver cyst” as compared to using the term “hepatic cyst”. Expanding the query by utilizing the Terminology Interface component to include search by synonymous terms significantly improved recall. In the second case example, query expansion was automatically done by utilizing the NCIT. The NCIT was developed by the National Cancer Institute primarily to support translational cancer research and not necessarily to search radiology reports [16]. Thus, although greatly expanded to support various clinical findings, further improvement of recall may be attained by utilizing other controlled terminologies [25–28].
The performance of iSCOUT compares favorably to several information retrieval and classification algorithms and tools currently in use [6, 12, 29–31]. The precision of a computer algorithm utilizing an entropy reduction approach, which was utilized to classify radiology reports into those with clinically important findings, yielded a precision and recall of 97.5% and 98.9% [6]. A machine learning algorithm utilized for tumor status classification of MRI reports yielded mean precision and recall of 82.4% and 80.6%, respectively [29].
A comparison of four terminologies for retrieving critical results using iSCOUT did not identify a terminology that consistently correlated with improved report retrieval [32]. Precision was most consistent with RadLex, with at least 93% precision for retrieving three distinct critical imaging findings [32]. Further evaluation will utilize the Terminology Interface module, focusing on expert approach in which experts will provide additional search terms. The expert approach in combination with a controlled terminology is expected to greatly improve recall. In addition, a more focused evaluation of iSCOUT for retrieving reports with critical imaging findings in a larger set of radiology reports is underway.
iSCOUT, and report retrieval in general, can be used in many ways, such as to search for and generate patient cohorts for research purposes or for routine monitoring for report quality assurance and process improvement, potentially impacting the quality of patient care. Ease of use, coupled with an end-user’s ability to combine various components as necessary, make this a valuable toolkit for radiology report retrieval.
Conclusion
iSCOUT reliably identifies and retrieves relevant radiology reports when the findings are described in the final report. The toolkit has acceptable precision and recall even without requiring further customization and training. Utilizing a terminology interface (that includes similar terms for retrieval from a controlled ontology) to expand the query, significantly improves recall compared to utilizing a single search term alone.
Acknowledgments
This work was partly funded by AHRQ grant 1R18HS019635.
References
- 1.Taira RK, Soderland SG, Jakobovits RM. Automatic structuring of radiology free-text reports. Radiographics. 2001;21(1):237–245. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 2.Mamlin BW, Heinze DT, McDonald CJ. Automated extraction and normalization of findings from cancer-related free-text radiology reports. AMIA Annu Symp Proc 420–424, 2003 [PMC free article] [PubMed]
- 3.Zingmond D, Lenert LA. Monitoring free-text data using medical language processing. Comput Biomed Res. 1993;26(5):467–481. doi: 10.1006/cbmr.1993.1033. [DOI] [PubMed] [Google Scholar]
- 4.Fiszman M, Haug PJ, Frederick PR. Automatic extraction of PIOPED interpretations from ventilation/perfusion lung scan reports. Proc AMIA Symp 860–864, 1998 [PMC free article] [PubMed]
- 5.Thomas BJ, Ouellette H, Halpern EF, Rosenthal DI. Automated computer-assisted categorization of radiology reports. AJR Am J Roentgenol. 2005;184(2):687–690. doi: 10.2214/ajr.184.2.01840687. [DOI] [PubMed] [Google Scholar]
- 6.Dreyer KJ, Kalra MK, Maher MM, Hurier AM, Asfaw BA, Schultz T, et al. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study. Radiology. 2005;234(2):323–329. doi: 10.1148/radiol.2341040049. [DOI] [PubMed] [Google Scholar]
- 7.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pines JM. Trends in the rates of radiography use and important diagnoses in emergency department patients with abdominal pain. Med Care. 2009;47(7):782–786. doi: 10.1097/MLR.0b013e31819748e9. [DOI] [PubMed] [Google Scholar]
- 9.Korley FK, Pham JC, Kirsch TD. Use of advanced radiology during visits to US emergency departments for injury-related conditions, 1998–2007. JAMA. 2010;304(13):1465–1471. doi: 10.1001/jama.2010.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meystre SM, Haug PJ. Comparing natural language processing tools to extract medical problems from narrative text. AMIA Annu Symp Proc 525–529, 2005 [PMC free article] [PubMed]
- 11.Xu H, Fu Z, Shah A, Chen Y, Peterson NB, Chen Q, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. AMIA Annu Symp Proc. 2011;2011:1564–1572. [PMC free article] [PubMed] [Google Scholar]
- 12.Uzuner O, South BR, Shen S, Duvall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meystre S, Haug PJ. Natural language processing to extract medical problems from electronic clinical documents: performance evaluation. J Biomed Inform. 2006;39(6):589–599. doi: 10.1016/j.jbi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham H, D Maynard, K Bontcheva, V Tablan. GATE: A framework and graphical development environment for robust NLP tools and applications. Proc 40th Assoc for Computational Linguistics, 2002
- 16.Coronado S, Haber MW, Sioutos N, Tuttle MS, Wright LW. NCI Thesaurus: using science-based terminology to integrate cancer research results. Stud Health Technol Inform. 2004;107(Pt 1):33–37. [PubMed] [Google Scholar]
- 17.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26(6):1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 18.Andriole KP, Khorasani R. Implementing a replacement PACS: issues to consider. J Am Coll Radiol. 2007;4(6):416–418. doi: 10.1016/j.jacr.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Gershanik EF, Lacson R, Khorasani R. Critical finding capture in the impression section of radiology reports. AMIA Annu Symp Proc. 2011;2011:465–469. [PMC free article] [PubMed] [Google Scholar]
- 20.National Cancer Institute. http://ncit.nci.nih.gov. 26 July 2010.
- 21.Hersh W. Evaluation of biomedical text-mining systems: lessons learned from information retrieval. Brief Bioinform. 2005;6(4):344–356. doi: 10.1093/bib/6.4.344. [DOI] [PubMed] [Google Scholar]
- 22.Su K, Ries JE, Peterson GM, Cullinan Sievert ME, Patrick TB, Moxley DE et al. Comparing frequency of word occurrences in abstracts and texts using two stop word lists. Proc AMIA Symp 682–686, 2001 [PMC free article] [PubMed]
- 23.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18(5):544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 25.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med. 1993;32(4):281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loy P. International classification of diseases—9th revision. Med Rec Health Care Inf J. 1978;19(2):390–396. [PubMed] [Google Scholar]
- 27.Cote RA, Robboy S. Progress in medical information management. Systematized nomenclature of medicine (SNOMED) JAMA. 1980;243(8):756–762. doi: 10.1001/jama.1980.03300340032015. [DOI] [PubMed] [Google Scholar]
- 28.Rogers FB. Medical subject headings. Bull Med Libr Assoc. 1963;51:114–116. [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng LT, Zheng J, Savova GK, Erickson BJ. Discerning tumor status from unstructured MRI reports—completeness of information in existing reports and utility of automated natural language processing. J Digit Imaging. 2010;23(2):119–132. doi: 10.1007/s10278-009-9215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng B, Titterington D. Neural networks: a review from a statistical perspective. Stat Sci. 1994;9(1):2–54. doi: 10.1214/ss/1177010638. [DOI] [Google Scholar]
- 31.Savova GK, Fan J, Ye Z, Murphy SP, Zheng J, Chute CG, et al. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA Annu Symp Proc. 2010;2010:722–726. [PMC free article] [PubMed] [Google Scholar]
- 32.Warden GI, Lacson R, Khorasani R. Leveraging terminologies for retrieval of radiology reports with critical imaging findings. AMIA Annu Symp Proc. 2011;2011:1481–1488. [PMC free article] [PubMed] [Google Scholar]