Abstract
Transformation of Escherichia coli with purified plasmids containing DNA damage is frequently used as a tool to characterize repair pathways that operate on chromosomes. In this study, we used an assay that allowed us to quantify plasmid survival and to compare how efficiently various repair pathways operate on plasmid DNA introduced into cells relative to their efficiency on chromosomal DNA. We observed distinct differences between the mechanisms operating on the transforming plasmid DNA and the chromosome. An average of one UV-induced lesion was sufficient to inactivate ColE1-based plasmids introduced into nucleotide excision repair mutants, suggesting an essential role for repair on newly introduced plasmid DNA. By contrast, the absence of RecA, RecF, RecBC, RecG or RuvAB had a minimal effect on the survival of the transforming plasmid DNA containing UV-induced damage. Neither the presence of an endogenous homologous plasmid nor the induction of the SOS response enhanced the survival of transforming plasmids. Using two-dimensional agarose-gel analysis, both replication- and RecA-dependent structures that were observed on established, endogenous plasmids following UV-irradiation, failed to form on UV-irradiated plasmids introduced into E. coli. We interpret these observations to suggest that the lack of RecA-mediated survival is likely to be due to inefficient replication that occurs when plasmids are initially introduced into cells, rather than to the plasmid’s size, the absence of homologous sequences, or levels of recA expression.
Keywords: Transformation, E. coli, nucleotide excision repair, recA, recF, recBC
1. INTRODUCTION
Transformation of cells with plasmids containing DNA damage is one approach that has been frequently used to characterize cellular repair pathways in both prokaryotic and eukaryotic systems (Bichara et al., 2007; Bichara et al., 2009; Kucherlapati et al., 1984; Roberts and Strike, 1981; Saffran and Cantor, 1984; Schmid et al., 1982; Strike et al., 1979; Wang et al., 2001). UV irradiation of Escherichia coli causes the formation of cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts in DNA that can block polymerases and prevent replication of the genome (Setlow et al., 1963). A number of repair and tolerance pathways are known to contribute significantly to survival in the presence of UV-induced lesions on the chromosome. Nucleotide excision repair removes these lesions from the genome reducing the frequency that replication encounters this type of damage, and contributes significantly to cell survival (Courcelle et al., 1999; Courcelle et al., 2005; Courcelle et al., 2006). In the absence of repair, translesion polymerases can incorporate bases opposite to the damage that would otherwise prevent replication from being completed (Bagg et al., 1981; Courcelle et al., 1999; Courcelle et al., 2005; Courcelle et al., 2006). Under most conditions, these polymerases have only modest effects on survival, but can have large effects on the frequency of mutagenesis (Bagg et al., 1981; Courcelle et al., 1999; Courcelle et al., 2006; Radman, 1975). A large number of these processing events are dependent on RecA, and the absence of the protein renders cells hypersensitive to UV irradiation (Courcelle et al., 1999; Courcelle et al., 2003; Keller et al., 2001; West, 1996). RecA along with several RecF-pathway proteins are involved in processing replication forks that encounter DNA damage and are required for replication to resume (Courcelle et al., 1997; Courcelle et al., 1999; Courcelle et al., 2003). In addition, RecA along with RecBC- pathway proteins are needed to repair double-strand break intermediates that arise following several forms of DNA damage (Chow and Courcelle, 2007; Keller et al., 2001; Willetts and Clark, 1969). Strand exchange and Holliday junction intermediates that form during these RecA-mediated events are processed and resolved by the RuvABC and RecG resolvases (Donaldson et al., 2004; Lloyd and Sharples, 1993; Lloyd, 1991; West, 1996). Mutations inactivating recBC, recF, ruvABC, or recG reduce cellular resistance to UV-irradiation to varying degrees (Courcelle et al., 1997; Courcelle et al., 1999; McGlynn and Lloyd, 2001; McGlynn and Lloyd, 2002; West, 1996; Willetts and Clark, 1969).
Plasmids used to transform chemical- or electro-competent E. coli are taken up as double-strand circular DNA. Typically, in assays using plasmid transformation, the purified plasmid DNA is exogenously exposed to a damaging agent and then introduced into cells to monitor plasmid survival, DNA repair, or replication using various conditions and mutants. However, quantifying the effect that DNA damage has on plasmid survival can be challenging. Transformation frequencies can vary several-fold between experiments due to small variations in competent cell preparations, DNA-to-cell ratios, temperature, heat shock duration, or recovery times prior to selection (Froger and Hall, 2007; Norgard et al., 1978; Roychoudhury et al., 2009; Yoshida and Sato, 2009). In addition, several mutants, including recA and recBC, are known to affect the cell’s ability to take up or maintain plasmids, making it difficult to directly quantify and compare plasmid survival in different strains (Bassett and Kushner, 1984; Biek and Cohen, 1986; Summers and Sherratt, 1984).
A number of reports have suggested that differences exist between the repair pathways that operate on transforming plasmids and chromosomal DNA, but the underlying mechanism and overall conclusions have varied greatly between studies. A series of early studies by Roberts and Strike introduced UV-irradiated plasmids and found that plasmid survival depended on UvrA and RecBC, but not on RecA or RecF, irrespective of whether a homologous plasmid was present (Roberts and Strike, 1981; Roberts and Strike, 1986; Strike and Roberts, 1982; Strike et al., 1979). In contrast, a set of more recent studies reported that survival of transforming plasmids containing bulky adducts depended on both RecA and RecF function and required the presence of a homologous plasmid (Bichara et al., 2007; Bichara et al., 2009). However, an earlier study involving authors from this same group reported that RecA did not significantly contribute to survival of plasmids containing bulky lesions (Schmid et al., 1982). Another group using a similar approach to characterize repair of interstrand DNA crosslinks observed that a lack of RecA did not affect survival of transforming plasmids and concluded that recombinational processes may not be involved in repairing these lesions (Berardini et al., 1997; Berardini et al., 1999). In subsequent work, it was noted that recA mutants were hypersensitive to crosslinking agents and the authors speculated that a lack of homology to the plasmid might account for the observed differences between the plasmid and chromosome (Berardini et al., 1999). All of these studies utilized plasmids containing ColE1-type origins, making it unlikely that differences in replication origins could account for the observed variation in results. However, each of these studies relied on the introduction of a single, damaged plasmid into different preparations and strains of competent cells to draw conclusions about which proteins or pathways were important for survival. In some cases, damaged and undamaged plasmids were required to be prepared separately. Thus, some of the discrepancies between studies could be due to either variations in DNA concentrations, cell preparations, or inherent competency of cells from different genetic backgrounds.
In light of these issues, we developed an assay that utilizes a control plasmid to normalize for variations in transformation efficiencies between different experiments and strains. We then used this assay to quantify the relative contribution that various repair pathways have on the transforming plasmid’s survival. We found that only nucleotide excision repair, but not RecA, RecF, RecBC, RecG or RuvAB, contributes to the survival of transforming plasmids containing UV-induced lesions. Using two-dimensional agarose gel analysis, we observed that both replication-dependent and RecA-dependent processing intermediates are observed on established plasmids replicating endogenously, but not on plasmids that have been newly introduced into competent cells. Homology or SOS induction did not enhance the survival of transforming plasmids containing UV damage in the presence of RecA. Based on these observations, we infer that the inability of RecA-mediated pathways to operate is most likely due to inefficient replication of plasmids when they are initially introduced into cells, rather than to other aspects such as the plasmid’s size, the absence of homology, or levels of recA expression.
2. MATERIALS AND METHODS
2.1 Strain Construction
The strains used in this work are presented in Table 1. All strains were derived from the parental strain ER1793 (New England Biolabs) using standard P1 transduction.
TABLE 1.
E. coli K-12 strains used
| Strain | Relevant Genotype | Source, Reference or Construction |
|---|---|---|
| ER1793 | F−
fhuA2 •(lacZ)r1 gln44e14−(McrA−) trp13-31 his-1 rpsL104 xyl-7 mtl-2 metB1 •(mcrC-mrr)114::IS10 |
New England Biolabs |
| KM21 | (recBCD)::kan | (Murphy, 1998) |
| TP539 | recG6201:: kan | (Murphy et al., 2000) |
| TP605 | sulA6209::tet | (Murphy et al., 2000) |
| TP638 | recQ6216::tet | (Murphy et al., 2000) |
| DY329 | (lacU169) nadA::Tn10, gal490, Lambda cI857, (cro-bioA) | (Yu et al., 2000) |
| CL1922* | sulA::frt-cam-frt | DY329 x linear PCR product Primers sulA-frtFOR and sulA-frtREV |
| DY351 | lacU169, gal490, cI857, (cro- bioA), D(kil-git::cam) | (Yu et al., 2000) |
| CL1016* | polB::tet | DY351 x linear PCR product Primers polBF-tetF and polBR-tetR |
| HL759 | uvrA6, zjd::Tn5 | Dr. Ann Ganesan |
| HL921 | recA::tet | (Courcelle et al., 1997) |
| HL924 | recJ284::Tn10 | (Courcelle and Hanawalt, 1999) |
| HL940 | lexA71::Tn5 | (Crowley and Hanawalt, 1998) |
| HL952 | uvrA::Tn10 | (Courcelle et al., 1999) |
| CL575 | umuC122::Tn5 | (Courcelle et al., 2005) |
| CL577 | ruvC53eda-51::Tn10 | (Donaldson et al., 2006) |
| CL578 | ruvAB6204::kan | (Donaldson et al., 2004) |
| CL579 | recF6206::tet | (Courcelle et al., 2003) |
| CL634 | dinB::kan | (Courcelle et al., 2005) |
| Strains isogenic to ER1793: | ||
| CL1312 | recJ284::Tn10 | ER1793 x P1 (HL924) |
| CL1313 | recA::tet | ER1793 x P1 (HL921) |
| CL1314 | ruvC53eda-51::Tn10 | ER1793 x P1 (CL577) |
| CL1315 | uvrA::Tn10 | ER1793 x P1 (HL952) |
| CL1316 | recQ6216::tet | ER1793 x P1 (TP638) |
| CL1317 | polB::tet | ER1793 x P1 (CL1016) |
| CL1318 | dinB::kan | ER1793 x P1 (CL634) |
| CL1319 | recF6206::tet | ER1793 x P1 (CL579) |
| CL1320 | ruvAB6204::kan | ER1793 x P1 (CL578) |
| CL1321 | recG6201:: kan | ER1793 x P1 (TP539) |
| CL1323 | umuC122::Tn5 | ER1793 x P1 (CL575) |
| CL1473 | (recBCD)::kan | ER1793 x P1 (KM21) |
| CL1605 | sulA6209::tet | ER1793 x P1 (TP605) |
| CL1623 | sulA6209::tet lexA71::Tn5 | CL1605 x P1 (HL940) |
| CL1919 | sulA::frt-cam-frt | ER1793 x P1 (CL1922) |
| CL1921 | sulA::frt-cam-frt lexA71::Tn5 | CL1919 x P1 (HL940) |
| CL1923* | sulA::frt lexA71::Tn5 | CL1921 x pCP20 (Cherepanov and Wackernagel, 1995) |
| CL 1507 | uvrA6, zjd::Tn5 | ER1793 x P1 (HL759) |
| CL 1566 | uvrA6, zjd::Tn5, recA::tet | CL1313 x P1 (HL759) |
| CL1736 | sulA::frt lexA71::Tn5 uvrA::Tn10 | CL1923 x P1 (HL952) |
see Materials and Methods for details; tet: tetracycline resistance; cam: chloramphenicol resistance; kan: kanamycin resistance
A gene replacement of polB was constructed using the recombineering strain DY351 (Yu et al., 2000). The tetracycline resistance cassette was inserted into polB using primers polBF-tetF and polBR-tetR, 5′CAAGCCTGGTTTTTTGATGGATTTTCAGCGTGGCGCAGGCACTCGACATCTT GGTTACCG and 5′TTGCTCAAAATAGCCCAAGTTGCCCGGTCATAAGTGTAGCCAAGAGGGTCATTATATTTCG, respectively, to amplify the tetR cassette from Tn10. The PCR product was used to transform DY351 to generate CL1016 (polB::tet,). CL1317 (ER1793 polB::tet) was constructed by P1 transduction of the polB::tet allele from CL1016 into ER1793.
A gene replacement of sulA was constructed using the recombineering strain DY329 (Yu et al., 2000). The chloramphenicol resistance cassette flanked by FRT sequences was inserted into sulA using primers sulA-frtFOR and sulA-frtREV, 5′CTGTACATCCATACAGTAACTCACAGGGGCTGGATTGATTGTGTAGGCTGGAGCTGCTTCG and 5′TGGGCGACAAAAAAAGTTCCAGGATTAATCCTAAATTTACCATATGAATATCCTCCTTA, respectively, to amplify the frt-cam-frt cassette from pKD3 (Datsenko and Wanner, 2000). The PCR product was then used to transform DY329 to generate CL1922 (sulA::frt-cam-frt). CL1919 (ER1793 sulA::frt-cam-frt) was constructed by P1 transduction of the sulA::frt-cam-frt allele from CL1922 into ER1793. CL1921 (ER1793 sulA::frt-cam-frt lexA71::Tn5) was constructed by P1 transduction of the lexA71::Tn5 allele from HL940 into CL1919. To generate a markerless sulA mutant, CL1923 (ER1793 sulA::frt lexA71::Tn5), the helper plasmid pCP20 (Cherepanov and Wackernagel, 1995) was introduced into CL1921 and FLP recombinase expression was induced to eliminate the chloramphenicol resistance cassette as described previously (Datsenko and Wanner, 2000).
Transductants were verified following selection with the appropriate antibiotic and by their sensitivity to UV irradiation when appropriate. Antibiotics were used at the following concentrations: Ampicillin 100 μg/mL, Chloramphenicol 20 μg/mL, Kanamycin 30 or 70 μg/mL, Tetracycline 16 μg/mL.
2.2 Plasmids
pBR322, pBluescriptII SK (pSK+) and the pBluescriptII KS (pKS+/−) vectors confer resistance to ampicillin and have been described previously(Alting-Mees et al., 1992; Watson, 1988). pBluescript BC SK+ (pBC, Fermentas) contains a chloramphenicol marker. pET30 (Novagen) contains a kanamycin resistance cassette.
2.3 Plasmid Purification and Irradiation
All plasmids used in this study were prepared and purified from JM109 cells using the Eppendorf Fastplasmid Mini kit and then gel purified (0.5X TAE, 0.5% Agarose, 0.3 μg/mL ethidium bromide ran at 1.5V/cm) to obtain supercoiled circular DNA. For survival experiments, 1 μg of purified plasmid in TE was UV irradiated at the indicated dose, using a Sylvania 15 W germicidal light bulb (254 nm) at an incidence rate of 0.9 J/m2/s. For all experiments, an equal molar amount of unirradiated, purified plasmid (pBC) was mixed with the UV-irradiated plasmids prior to the transformation analysis.
2.4 Preparation of Competent Cells and Transformation
Fresh overnight cultures of each strain were diluted 1:100 in 50 ml of Luria-Bertani medium (LB) and grown to an OD600 of 0.4. Cells were then pelleted, washed with 50 ml of 80 mM MgCl2, 20 mM CaCl2 solution, repelleted and then resuspended in 1 ml of 100 mM CaCl2 and 10% glycerol. Cells were stored at −80°C until needed.
Competent cells that were induced for the SOS response were prepared by irradiating the culture with 20 J/m2 UV light, and allowing the culture to recover at 37°C for 20 min before cells were pelleted and prepared using the protocol described above.
To measure transformation frequency of damaged plasmids, a single mixture of UV-lesion containing pKS or pBR322, was mixed with undamaged pBC in a 1:1 molar ratio. This same mixture was then used to transform wild-type cells and all mutants to be compared. For transformation, 200 μl competent cells were thawed on ice, mixed with 10 ng of the pKS/pBC plasmid mixture and heat shocked at 42°C for 90 sec. The mixture was then returned to ice for 1–2 min before 800 μl of fresh LB was added and the mixture was incubated at 37°C for 30 min. At this time, cultures were serially diluted and duplicate 100-μl aliquots were spread on LB plates containing ampicillin (100 μg/ml) or chloramphenicol (20 μg/ml). Plates were incubated overnight at 37°C before the number of surviving colonies on each plate was counted. The ratio of ampicillin-resistant colonies to chloramphenicol-resistant colonies was then determined for each mutant and compared to wild-type cultures.
2.5 Quantification of Induced DNA Damage
Lesion frequency on plasmids was measured by sensitivity to T4 endonuclease V (TEV, Epicentre), which cleaves DNA at sites containing cyclobutane pyrimidine dimers, generating a single strand nick. 20 U of TEV was added to 1 μg of purified, irradiated or unirradiated plasmid DNA in 25 μl TE buffer and incubated at 37°C for one hour. Reactions were stopped by the addition of an equal volume of chloroform and the DNA was separated on a 0.7% agarose gel containing ethidium bromide. Supercoiled plasmids containing lesions were thus converted to a nicked, open circle conformation that can be resolved on an agarose gel. For each dose, the number of lesions per plasmid molecule was then calculated from the fraction of plasmids that remained lesion free (the zero class), based on the Poisson expression, P(0 lesions) = −ln (n), where n represents the average number of lesions per plasmid.
2.6 Two-dimensional Agarose Gel Electrophoresis
For transformation experiments, electrocompentent cells were prepared by growing cultures in 50 ml LBthy media to an OD600 of 0.6 before they were washed 4X with ice-cold water and resuspended in 500 μl of ice-cold 10% glycerol. 40 μl of cells were then transformed via electroporation with 500 ng of pBR322, washed with 1 ml of LB media, pelleted, resuspended in 1 ml LB media, and allowed to recover at 37°C. During the recovery, 0.15-ml aliquots of the transformed cell culture were taken at the indicated times and transferred to 0.75-ml ice-cold NET buffer (100 mM NaCl, 20 mM EDTA, pH 8.0, 10 mM Tris, pH 8.0).
For experiments with cells containing an endogenous pBR322 plasmid, cultures were grown overnight in Davis medium supplemented with 0.4% glucose, 0.2% casamino acids (DGC), and 40 μg/ml each of histidine (his) and tryptophan (trp) in the presence of 100 μg/ml ampicillin. 0.2 ml of the overnight cultures were pelleted and resuspended in 20-ml DGC medium supplemented with his and trp in the absence of ampicillin and grown in a 37°C shaking water bath to an OD600 of 0.5. At this time, cultures were irradiated with 55 J/m2 of UV light, which produces an average of 1 lesion per strand of the plasmid, and allowed to recover at 37°C. At the times indicated, a 0.75-ml aliquot of culture was removed and transferred to an equal volume of NET buffer.
To purify genomic and plasmid DNA, cells were pelleted, resuspended in 0.15 ml of lysis buffer (1.5 mg/ml lysozyme, 0.5 mg/ml RNAseA in 10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0) and incubated at 37°C for 30 min. Then, 10 μl each of 10 mg/ml Proteinase K and 20% Sarkosyl was added to the samples and returned to 37°C for 30 min. Samples were then extracted with 4 volumes of phenol:chloroform (1:1), followed by an extraction with 4 volumes of chloroform, dialyzed against 200 ml of TE (2 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0) for one hour on a floating 47-mm Whatman 0.05-μm pore disk, and digested with PvuII (Fermentas) overnight at 37°C. Samples were extracted with an equal volume of chloroform and loaded directly onto a gel.
Genomic DNA was separated initially on a 0.4% agarose gel in 1X TBE at 1 V/cm for 15 h. For the second dimension, lanes were excised from the gel, rotated 90°, recast in a 1% agarose gel in 1X TBE and electrophoresed at 7 V/cm for 6 h. DNA in the gels was transferred to Hybond N+ (GE BioSciences) membrane using standard Southern blotting protocols. The plasmid DNA was detected by probing with 32P-labeled pBR322 that was prepared using nick translation (Roche) and visualized using a Storm 840 phosphorimager (GE Biosciences) and its associated ImageQuant analysis software.
3. RESULTS
3.1 Several RecA-mediated pathways that contribute to cell survival after UV irradiation do not contribute to survival of UV-irradiated plasmids introduced into competent cells
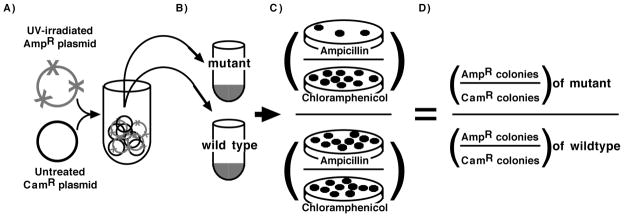
To characterize the repair pathways that operate on transforming plasmid DNA, we employed an assay in which the survival of plasmids containing DNA damage could be compared directly between different strains and transformation experiments (Fig. 1). In this assay, a mixture containing equal molar amounts of a UV-irradiated plasmid (pBR322 or pKS, as indicated) and an undamaged control plasmid (pBC) were used to transform wild-type and various mutant cells. Duplicate aliquots of each transformation were then plated on both ampicillin and chloramphenicol media. Survival of the damaged plasmid in each mutant was determined by calculating the ratio of surviving UV-damaged plasmids (AmpR colonies) to undamaged control plasmids (CamR colonies). The inclusion of the second, undamaged plasmid served to normalize for differences in the transformation frequency due to genetic mutations that affect DNA uptake and maintenance as well as to control for variations between experiments due to cell competence, DNA-cell concentrations, or heat shock durations (Berry and Kropinski, 1986; Dower et al., 1988).
FIGURE 1. Schematic of transformation assay.
A) Equal amounts of UV-damaged and untreated plasmids are mixed and used to B) transform a set of competent wild-type and mutant cells. C) The ratio of colonies formed on ampicillin plates (UV-damaged) to those formed on chloramphenicol plates (untreated) are determined for each mutant and D) normalized to the ratio observed in wild-type.
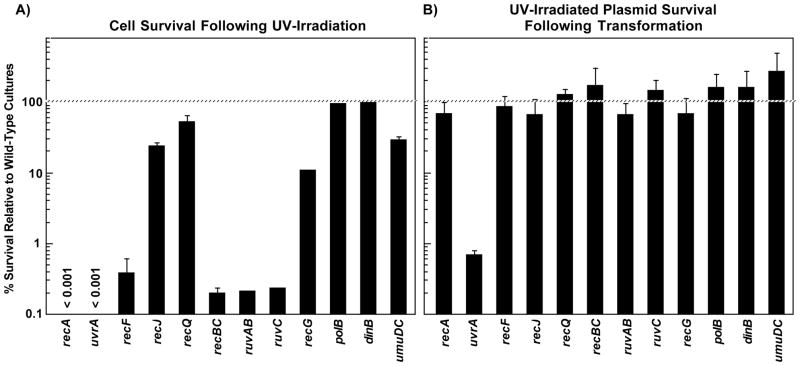
We used ER1793, which lacks the two restriction endonucleases encoded by the E. coli genome, as our parental strain. Isogenic mutants in genes known to be involved in processing UV-induced damage on the chromosome were then constructed in this background. For the purpose of comparison to the plasmid assay, the survival of each strain relative to the parental strain is shown following irradiation with 25 J/m2 (Fig. 2A). In strains lacking UvrA, RecA, RecF, RecBC, RuvAB, or RuvC, cell survival was reduced by greater than two orders of magnitude relative to wild-type cells after a dose of 25 J/m2. In recG mutants, survival was more than one order of magnitude lower relative to wild-type cells. At this dose, an average of 1 lesion per 10-kb DNA strand is induced ((Courcelle et al., 2003) and Fig. 3A) and approximately 70% of wild-type cells survive to form colonies.
FIGURE 2. Many RecA-mediated processes that contribute to cell survival do not contribute to the survival of transforming plasmids.
A) The survival of each mutant is plotted relative to the survival of wild-type cells following irradiation with 25 J/m2 of UV. B) The survival of UV-irradiated plasmids (160 J/m2) introduced into each mutant is plotted relative to the survival of plasmids introduced into wild-type cells. Data represents an average of at least 3 independent experiments. Error bars represent standard error of the mean.
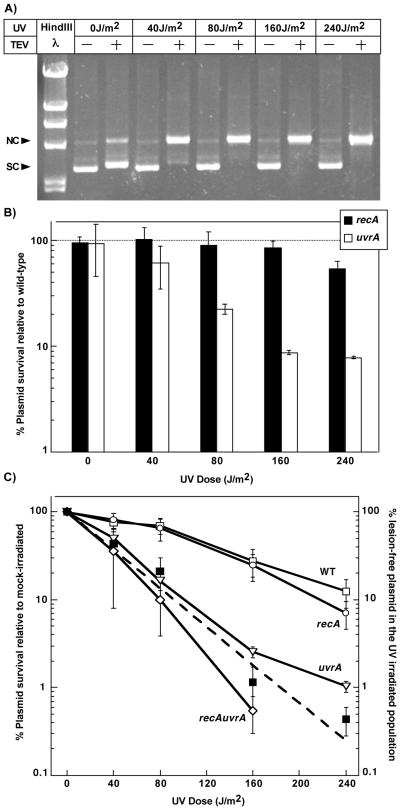
FIGURE 3. UvrA is nearly essential for the survival of transforming plasmids containing UV-induced damage.
A) The formation of the predominant UV-induced lesion, the cyclobutane pyrimidine dimer (CPD), increases linearly with UV dose. The presence of lesions was determined by sensitivity to T4 Endonuclease V (TEV), which cleaves DNA at sites containing CPDs converting supercoiled plasmid (SC) to a nicked open circular form (NC). These forms are differentiated by their migration pattern following agarose-gel electrophoresis (Spivak and Hanawalt, 1995). B) The survival of UV-irradiated plasmids following transformation in recA (filled bars) and uvrA (unfilled bars) mutants is plotted relative to the survival of plasmids introduced into wild-type cells at each dose. Survival was calculated as described in Figure 1. Plots represent an average of at least 2 independent experiments. Error bars represent standard error of the mean. C) The survival of UV-irradiated transforming plasmid is plotted relative to the survival of the mock-irradiated plasmid for wild-type (open squares), recA (open circles), uvrA (open triangles) and recA uvrA (open diamonds) at the indicated doses. The predicted (dashed line) and measured (filled squares) fraction of lesion-free plasmids in the UV-irradiated population is plotted. Measured lesion-free fractions were calculated from the amount of plasmid remaining resistant to TEV cleavage as shown in Figure 3A. Predicted lesion-free plasmids were calculated as the zero-class of plasmids using the Poisson expression. Graphs represent an average of at least 2 independent experiments. Error bars represent standard error of the mean.
To measure the survival of introduced plasmid DNA in these strains, a purified preparation of pBR322 was UV-irradiated with 110 J/m2. This dose produces an average of 2 lesions per plasmid strand ((Courcelle et al., 2003) and Fig. 3A). The irradiated plasmid was then mixed with an equal amount of a purified preparation of undamaged pBC plasmid and 20 ng of this mixture was then used to transform each strain, as described above. Based on the ratio of transformants obtained when selecting for single or double antibiotic resistances, less than 5% of the transformants received both plasmids. In addition, at the DNA concentrations used, the overall transformation frequency was typically in the range of <1 transformant per 1000 colony-forming units (cfu). These transformation frequencies suggest that cells generally received only a single plasmid and argue that plasmid incompatibility or homology-based repair processes would not affect plasmid survival rates.
When transformation efficiency was normalized to the undamaged control plasmid, we observed that the survival of plasmids containing UV-induced damage in recA mutants was similar to that in wild-type cells (Fig. 2B). By contrast, in uvrA mutants, which are deficient in nucleotide excision repair, plasmid survival was reduced by approximately 2 orders of magnitude relative to the parental strain.
Consistent with the results described above, the absence of other RecA-dependent repair processes also did not contribute to the survival of plasmids containing UV-induced damage. UV-irradiated plasmids introduced into recF, recJ, recQ, or recBC mutants survived as well as those introduced into the parental strain (Fig. 2B). Similarly, plasmid survival was also unaffected by mutations inactivating RecG, RuvAB or RuvC (Fig. 2B).
No effect on plasmid survival was observed in the absence of any single translesion DNA polymerase gene, umuC, polB or dinB. However, of the translesion DNA polymerase genes, only UmuC contributes to cell survival after high doses of UV (Courcelle et al., 2006). Similar results were obtained when survival of UV-irradiated Phi X 174 was examined following infection of wild-type, uvrA and recA cells (data not shown). Taken together, these results suggest that nucleotide excision repair, but not recA-mediated processes contribute to the survival of introduced DNA containing UV-induced damage.
3.2 UvrA is nearly essential for survival of plasmids containing UV-induced damage following transformation
To further characterize the role of nucleotide excision repair and recA-mediated pathways in processing introduced DNA, we examined the survival of plasmids over a range of UV doses. To this end, aliquots of a purified plasmid preparation of pKS+ were either mock irradiated or irradiated with 40, 80, 160 or 240 J/m2 of UV to produce an average of 0, 1, 2, 4 and 6 CPDs on the 3-kb plasmid, respectively. The lesion frequency was confirmed by treating the irradiated samples with T4 endonuclease V (TEV), which incises DNA at sites containing CPDs. Following incision by TEV, supercoiled plasmids containing UV lesions are converted to a relaxed open circular conformation that can be monitored in an agarose gel (Fig. 3A). The UV-irradiated plasmid preparations were then mixed with an unirradiated control pBC plasmid and the mixture was used to transform a set of wild-type, recA, or uvrA cultures as described above. As shown in Fig. 3B, the survival of UV-irradiated plasmids in recA mutants (filled bar) was similar to wild-type cells up to doses of 160 J/m2. By comparison, plasmid survival in uvrA mutants (unfilled bar) decreased relative to survival in wild-type cultures as the lesion frequency increased.
To assess plasmid survival within each strain as a function of UV dose, we also plotted plasmid survival relative to the survival of a mock-irradiated plasmid in each strain. Because we are not comparing survival to wild-type cultures, this type of analysis does not control for differences in transformation efficiencies that may exist between strains. However, the use of the undamaged control plasmid still normalizes for differences in transformation efficiencies that occur between experiments and allows one to determine how survival is reduced within each strain as a function of UV dose. By this type of analysis, plasmid survival in wild-type cells decreased linearly as the number of lesions on the plasmid increased over the doses we examined (Fig. 3C). In recA mutants, plasmid survival decreased at the same rate as that observed in wild-type cells. The lack of any RecA dependence on plasmid survival across the range of doses examined argues that the observed effect is not a result of the lesion frequency being below or beyond the capacity of RecA pathways to operate.
In contrast to recA cultures, plasmid survival in uvrA and recA uvrA mutants decreased more rapidly than wild-type cultures as the UV dose increased. At each dose examined, the percent plasmid survival was approximately equal to the fraction of plasmids that, by chance, remained lesion-free within the UV-irradiated samples (compare Figs. 3A and 3C). The result indicates that when lesions are present on the newly introduced DNA, nucleotide excision repair is nearly essential for survival. Other mutants that were examined, including recBC, recF and umuC, did not exhibit dose-dependent hypersensitivity to UV-induced damage on plasmid DNA (data not shown), consistent with the idea that other RecA-dependent processes are not contributing to plasmid survival.
3.3 RecA-dependent processing intermediates are observed on endogenously replicating plasmids, but not when plasmids are first introduced into cells
The inability of RecA-mediated pathways to contribute to survival of transforming plasmid DNA may be due either to the plasmid’s small size, its structure, or some aspect associated with the transformation process. To differentiate between these possibilities, we compared the processing intermediates observed on newly introduced plasmids following transformation to those observed on established, endogenously replicating plasmids using two-dimensional (2D) agarose-gel electrophoresis. This technique is able to differentiate and identify the structural properties of plasmids during repair and replication. To examine the structures induced on established, endogenous plasmids, cultures of E. coli containing the plasmid pBR322 were UV irradiated with 55 J/m2 and then allowed to recover. To examine structures on newly introduced plasmids, purified pBR322 was first UV-irradiated with 110 J/m2 and then used to transform competent preparations of E. coli. In both cases, samples were then taken at various times during the recovery period and total genomic DNA (plasmid and chromosomal) was purified. The genomic DNA was then digested with PvuII to linearize the plasmid at its unidirectional origin of replication and analyzed by 2D agarose-gel electrophoresis.
On endogenous plasmids in the absence of DNA damage, nonreplicating plasmid molecules migrate as a linear 4.4-kb fragment and form the prominent spot observed in this analysis. Replicating, Y-shaped molecules migrate more slowly due to their larger size and nonlinear structure and appear as an arc that extends out from the prominent nonreplicating spot (Fig. 4A and D). In the presence of UV-induced DNA damage, a unique class of intermediates is transiently observed that migrates more slowly than the replicating Y-arc, forming a cone region. These intermediates represent molecules having four arms or two branch points and have been shown to be resolved at a time that correlates with repair of the lesion and resumption of replication (Courcelle et al., 2003).
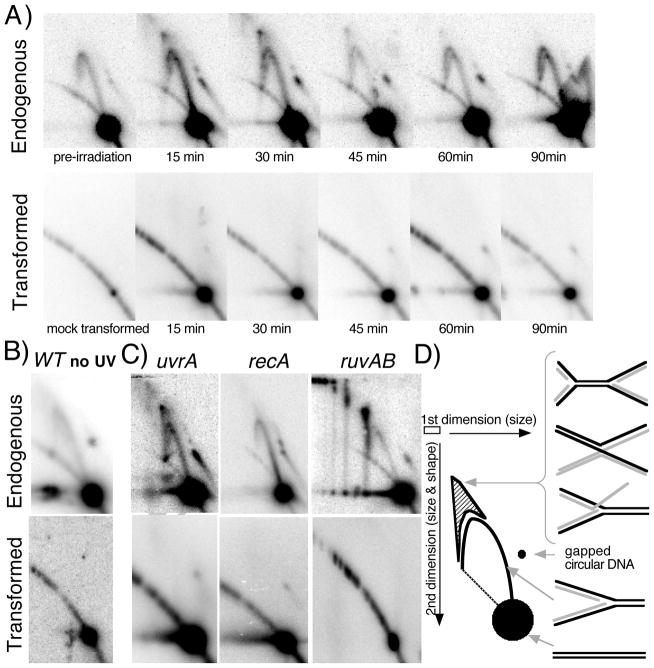
FIGURE 4. Replication- and RecA-dependent processing intermediates are not observed on newly introduced plasmids.
A) Replication-associated structural intermediates observed on endogenous plasmids containing UV-induced DNA damage are not observed on plasmids following transformation. Wild-type cells containing an endogenous plasmid pBR322 were UV-irradiated with 55 J/m2 (top) or wild-type cells were transformed with a plasmid that was UV-irradiated with 110 J/m2 (bottom). Genomic DNA was then purified, digested with PvuII, and analyzed by two-dimensional agarose-gel analysis at the times indicated. B) Non-damaged plasmids also fail to generate replication intermediates when first introduced into cells. Wild-type cells containing an endogenous plasmid pBR322 were mock UV-irradiated (top) or wild-type cells were transformed with a plasmid that was mock UV-irradiated, and then analyzed as in (A) following a 60-min recovery period. C) On endogenous plasmids containing DNA damage, RecA-dependent structural intermediates persist and accumulate in uvrA and ruvAB mutants (top), but are not observed when UV-irradiated plasmids are introduced into these mutants (bottom). uvrA, recA, and ruvAB mutants were analyzed as described in (A) following UV irradiation of cultures containing an endogenous plasmid or following introduction with UV-irradiated plasmid, pBR322. uvrA and recA mutants were analyzed at 30 min following treatment, ruvAB mutants were analyzed 60 min following treatment. D) Migration pattern of replication- and UV-induced structural intermediates following PvuII digestion of pBR322 during two-dimensional agarose-gel analysis. Nonreplicating plasmids run as a linear 4.4-kb fragment. Normal replicating fragments form Y-shaped structures and migrate more slowly because of their larger size and nonlinear shape, forming an arc that extends out from the linear fragment. Double Y- or X-shaped molecules migrate in the cone region and are observed after UV-induced damage on endogenous plasmids.
When we analyzed the structure of plasmids following transformation, we observed a distinct absence of both replicating structures and processing intermediates. We did observe a prominent 4.4 kb linear spot that corresponded to nonreplicating molecules (Fig. 4B). To estimate the amount of plasmid DNA that was not taken up by cells but detected in these assays, non-transformed samples were also analyzed after plasmid DNA was simply mixed with competent cell preparations. Based on the amount of plasmid detected in these mock-transformed samples, more than 96% of the signal detected in the Southern analysis represents plasmids taken up by cells, suggesting that plasmids not taken up by cells were effectively removed from the media by the washing step before DNA was prepared from the culture (Fig. 4A). To determine whether the lack of any observed intermediates was due to the presence of DNA damage on the transforming plasmid DNA, we also examined undamaged plasmids using the same approach and observed a similar lack of replication intermediates on plasmids newly introduced to competent cells (Fig. 4B). These observations suggest that newly introduced plasmids do not replicate efficiently and therefore fail to generate replication intermediates associated with processing lesions at replication forks.
On established, endogenous plasmids, the formation of the transient UV-induced structural intermediates requires ongoing replication and depends on RecA. By contrast, the UV-induced intermediates fail to resolve in uvrA mutants when the lesions cannot be removed. Thus, in these mutants, the intermediates are not transient but are seen to persist and accumulate. Similarly, in ruvAB mutants, which are unable to resolve these replication intermediates, the structures have been shown to accumulate, and go on to form higher order structures when other RecA-dependent processes are absent ((Courcelle et al., 2003; Donaldson et al., 2006) and Fig. 4C). Thus, if the transient nature of these intermediates prohibited their detection following transformation in wild-type cells, we reasoned that we should observe them in uvrA or ruvAB mutants where they are not resolved and continue to accumulate. However, as shown in Fig. 4C, when we examined the structure of newly introduced plasmids in each of these mutants, only nonreplicating linear fragments were detected in transformed uvrA, recA or ruvAB mutants. The lack of intermediate structures seen in these mutants argues that these replication-dependent processing events are not occurring at high frequencies following transformation.
We interpret these observations to be consistent with the idea that the inability of RecA-mediated processes to contribute to survival is due to an aspect of the transformation process. The presence of RecA-dependent intermediates on endogenous plasmids argues against the idea that the plasmid is either too small or is otherwise inappropriate as a substrate for RecA-mediated processes to operate. The absence of any replication-associated structural intermediates on the newly introduced plasmids suggests that the plasmid replicates inefficiently when it is initially introduced into host cells and that this is likely to account for the lack of processing by RecA-mediated pathways.
3.4 SOS induction or plasmid homology does not enhance survival of the transforming plasmid DNA
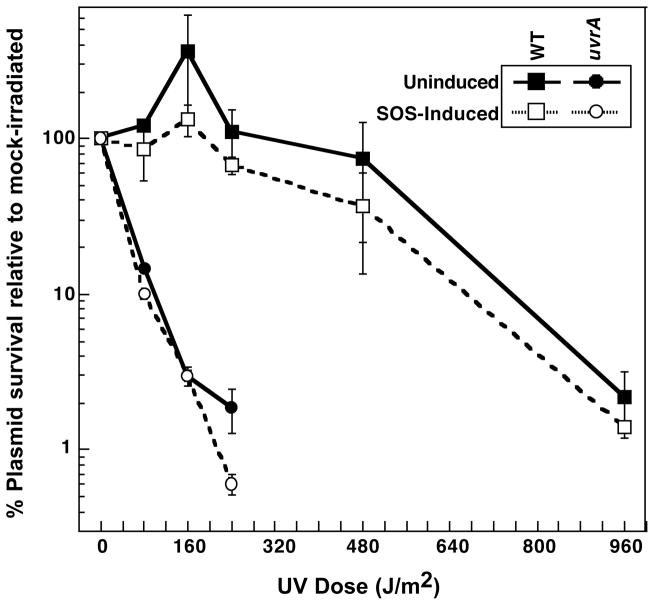
Following DNA damage on the chromosome, RecA is upregulated as part of the SOS response (Radman, 1975; Sassanfar and Roberts, 1990). Thus, it is possible that higher levels of RecA expression are required to observe RecA-mediated survival or processing on the newly introduced UV-irradiated plasmids. To address this possibility, we constructed wild-type and uvrA mutants that lacked the LexA repressor, allowing constitutive expression of RecA and all other SOS-regulated gene products. We then examined pKS plasmid survival following transformation as a function of UV dose in each of these strains as described before. By this type of analysis, constitutive expression of recA and other SOS-regulated genes had no effect on plasmid survival at any of the doses examined compared to their LexA+ counterparts (Fig. 5), indicating that the inability of RecA-mediated processes to operate in transforming plasmid DNA is not due to the expression level of recA or other SOS-regulated genes.
FIGURE 5. Constitutive recA expression does not enhance the survival of transforming UV-damaged plasmids.
The survival of UV-irradiated plasmids following introduction into wild-type (filled squares), lexA (open squares, dotted lines), uvrA (filled circles), and uvrA lexA (open circles, dotted lines) cells is plotted relative to the survival of mock-irradiated plasmids. Plots represent an average of at least 2 experiments. Error bars represent standard error of the mean.
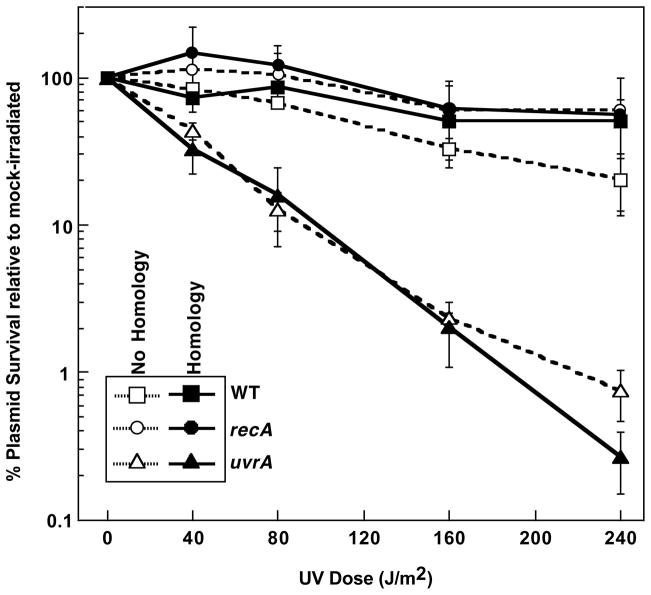
A second possible reason that RecA-dependent survival is not observed on newly introduced plasmids is the lack of established homologous sequences in the cell at the time of transformation. In the case of endogenously replicating plasmids, the plasmid is present in multiple copies inside the cell. Thus, it is possible that RecA-dependent survival would increase significantly if homologous sequences were present inside the cell at the time of transformation. To test this possibility, we used a mixture of a UV-irradiated plasmid, pSK+, and control plasmid, pET30, to transform competent preparations of wild-type, recA, and uvrA cells that either contained no homologous plasmid, or contained an endogenous, homologous plasmid, pBC. The pBC plasmid is entirely homologous to the UV-irradiated pSK+ plasmid except for its antibiotic resistance cassette but is not homologous to pET30. As shown in Fig. 6, the survival of UV-irradiated plasmids in uvrA cultures was reduced to the same extent regardless of whether or not cells contained the homologous plasmid and no RecA-dependent survival was observed. We interpret these results to indicate that the presence of homology does not limit the ability of RecA-mediated processes from operating on plasmids following transformation.
FIGURE 6. The presence of a homologous plasmid does not enhance the survival of transforming UV-damaged plasmids.
The survival of UV-irradiated plasmids following introduction into wild-type (squares), recA (circles) and uvrA (triangles) cells is plotted relative to the survival of mock-irradiated plasmids at each dose. The cells either contained an endogenous homologous plasmid (filled symbols) or no homologous plasmid (open symbols, dotted lines). Plots represent an average of at least 2 experiments. Error bars represent standard error of the mean.
4. DISCUSSION
We demonstrate that distinct differences exist between the battery of repair processes that operate on newly introduced plasmids as compared to those that operate on established, endogenous plasmids or the chromosome. On newly introduced plasmid DNA containing UV-induced damage, nucleotide excision repair is nearly essential for survival, whereas RecA-mediated processes do not significantly contribute to survival. The presence of replication and processing intermediates on endogenous plasmids, but not on newly introduced plasmids suggests that the lack of RecA-mediated processing is due to inefficient replication when plasmids are initially introduced into cells, rather than to other aspects such as the plasmid’s size, the presence of homology, or levels of RecA expression. Delayed or inefficient replication of the plasmid would allow more time for lesion repair to occur, obviating the need for RecA-mediated processing of arrested replication forks, as originally postulated by Strike and Roberts (Strike and Roberts, 1982).
The inefficient replication and processing by RecA-mediated pathways following transformation could reflect the need to establish additional protein factors on the purified DNA before replication or RecA-mediated processing can occur. Endogenous plasmids contain a number of associated structural proteins, including histone-like factors such as Hu, Ihf, or topoisomerases that are known to impair replication of plasmids when absent (Baker et al., 1986; Biek and Cohen, 1992; Gamas et al., 1986; Hwang and Kornberg, 1992; van der Ende et al., 1985). It is reasonable to consider that the newly introduced DNA may not replicate or be recognized efficiently by some enzymatic processes until these topology-altering factors are established.
A key difference between this study and previous studies involving the transformation of plasmids containing DNA damage involves the inclusion of an undamaged reference plasmid to normalize for transformation frequencies between different experiments and strains. This may help clarify a number of previous studies that have reached differing conclusions when characterizing repair mechanisms on transforming DNA. Strike and Roberts initially found that plasmid survival was dependent on RecBC, but curiously, it did not depend on RecA or RecF (Roberts and Strike, 1986). When normalized to an undamaged reference plasmid, we found that the survival of introduced plasmids containing UV-induced damage in recBC mutants was similar to wild-type cells and did not depend on RecA. A number of studies subsequent to Strike and Roberts work have shown that mutants defective in the RecBCD pathway have an impaired ability to maintain plasmids, which may account for the reduced survival that was observed in these early studies (Biek and Cohen, 1986; Niki et al., 1988; Summers and Sherratt, 1984).
Two previous studies also reached different conclusions as to whether a resident homologous plasmid enhances survival. Strike and Roberts concluded that RecA did not enhance survival of introduced plasmids even in the presence of homology (Strike and Roberts, 1982). Curiously, they did observe a modest increase in plasmid survival that was dependent on UvrA and UvrB, but not UvrC. In contrast, Bichara et al. using plasmids containing an AAF adduct, reported that survival depended on both RecA and RecF, required the presence of homology and involved a pathway that could be coupled to lesion removal by nucleotide excision repair (Bichara et al., 2007; Bichara et al., 2009). The RecA- and RecF-dependent survival was initially observed when using single-strand plasmids (Bichara et al., 2007). In a subsequent study, the authors suggested that the RecA-mediated effect also occurred when using double-strand plasmids, although survival of the transforming plasmid DNA was not specifically examined in this study (Bichara et al., 2009). An earlier study from this group reported that RecA did not contribute to survival following introduction of double-stranded plasmids containing either UV or AAF adducts (Schmid et al., 1982). When normalized to the reference plasmid, we did not observe any enhanced survival that was dependent on RecA or homology, consistent with the early results reported by Strike and Roberts, and Schmid et al. (Schmid et al., 1982; Strike and Roberts, 1982). Considering the essential role for nucleotide excision repair on transforming plasmids, it is likely that both RecA and the presence of a second, homologous template plays a more important role in the survival of single-stranded plasmids than on double-stranded plasmids following introduction into the cell, which may in part explain the discrepancies between these studies (Bichara et al., 2007; Bichara et al., 2009; Roberts and Strike, 1981; Schmid et al., 1982; Strike and Roberts, 1982). On double-stranded plasmids, a range of genetic and cellular observations suggest that arrested replication forks are processed by RecA to restore the region to a double-stranded form that can be repaired by nucleotide excision repair (Courcelle and Hanawalt, 2001; Courcelle et al., 1997; Courcelle et al., 1999; Courcelle et al., 2001; Courcelle et al., 2003). However, on single-stranded plasmids, repair of the lesion following the arrest of replication would require a second homologous plasmid to provide a template that can restore the lesion-containing region to a double-stranded form, as Bichara et al. observed (Bichara et al., 2007).
Highlights.
We used an internally normalized transformation assay to quantify plasmid survival.
Nucleotide excision repair is required for UV-damaged transforming plasmid survival.
RecA-mediated pathways do not enhance survival of UV-damaged transforming plasmids.
2D gel analysis shows transforming plasmids initially replicate inefficiently.
Acknowledgments
We thank Dr. Iwata-Reuyl and Dr. Fred Heffron for providing plasmids used in this study. This work was supported by the National Science Foundation (CAREER award MCB0551798); and the National Institute of General Medical Sciences, National Institutes of Health (AREA grant R15GM86839).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alting-Mees MA, Sorge JA, Short JM. pBluescriptII: multifunctional cloning and mapping vectors. Methods Enzymol. 1992;216:483–495. doi: 10.1016/0076-6879(92)16044-k. [DOI] [PubMed] [Google Scholar]
- Bagg A, Kenyon CJ, Walker GC. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 1981;78:5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Sekimizu K, Funnell BE, Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986;45:53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- Bassett CL, Kushner SR. Exonucleases I, III, and V are required for stability of ColE1-related plasmids in Escherichia coli. J Bacteriol. 1984;157:661–664. doi: 10.1128/jb.157.2.661-664.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J Bacteriol. 1999;181:2878–2882. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini M, Mackay W, Loechler EL. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry. 1997;36:3506–3513. doi: 10.1021/bi962778w. [DOI] [PubMed] [Google Scholar]
- Berry D, Kropinski AM. Effect of lipopolysaccharide mutations and temperature on plasmid transformation efficiency in Pseudomonas aeruginosa. Can J Microbiol. 1986;32:436–438. doi: 10.1139/m86-082. [DOI] [PubMed] [Google Scholar]
- Bichara M, Fuchs RP, Cordonnier A, Lambert IB. Preferential post-replication repair of DNA lesions situated on the leading strand of plasmids in Escherichia coli. Mol Microbiol. 2009;71:305–314. doi: 10.1111/j.1365-2958.2008.06527.x. [DOI] [PubMed] [Google Scholar]
- Bichara M, Pinet I, Lambert IB, Fuchs RP. RecA-mediated excision repair: a novel mechanism for repairing DNA lesions at sites of arrested DNA synthesis. Mol Microbiol. 2007;65:218–229. doi: 10.1111/j.1365-2958.2007.05790.x. [DOI] [PubMed] [Google Scholar]
- Biek DP, Cohen SN. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek DP, Cohen SN. Propagation of pSC101 plasmids defective in binding of integration host factor. J Bacteriol. 1992;174:785–792. doi: 10.1128/jb.174.3.785-792.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Chow KH, Courcelle J. RecBCD and RecJ/RecQ initiate DNA degradation on distinct substrates in UV-irradiated Escherichia coli. Radiat Res. 2007;168:499–506. doi: 10.1667/RR1033.1. [DOI] [PubMed] [Google Scholar]
- Courcelle CT, Belle JJ, Courcelle J. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J Bacteriol. 2005;187:6953–6961. doi: 10.1128/JB.187.20.6953-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:9154–9159. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Crowley DJ, Hanawalt PC. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J Bacteriol. 1999;181:916–922. doi: 10.1128/jb.181.3.916-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Donaldson JR, Chow KH, Courcelle CT. DNA Damage-Induced Replication Fork Regression and Processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Ganesan AK, Hanawalt PC. Therefore, what are recombination proteins there for? Bioessays. 2001;23:463–470. doi: 10.1002/bies.1065. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc Natl Acad Sci U S A. 2001;98:8196–8202. doi: 10.1073/pnas.121008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hanawalt PC. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci U S A. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley DJ, Hanawalt PC. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6–4 photoproducts, in UV-irradiated Escherichia coli. J Bacteriol. 1998;180:3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JR, Courcelle CT, Courcelle J. RuvAB and RecG Are Not Essential for the Recovery of DNA Synthesis Following UV-Induced DNA Damage in Escherichia coli. Genetics. 2004;166:1631–1640. doi: 10.1534/genetics.166.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JR, Courcelle CT, Courcelle J. RuvABC is required to resolve holliday junctions that accumulate following replication on damaged templates in Escherichia coli. J Biol Chem. 2006;281:28811–28821. doi: 10.1074/jbc.M603933200. [DOI] [PubMed] [Google Scholar]
- Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp. 2007:253. doi: 10.3791/253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, Burger AC, Churchward G, Caro L, Galas D, Chandler M. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Mol Gen Genet. 1986;204:85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- Keller KL, Overbeck-Carrick TL, Beck DJ. Survival and induction of SOS in Escherichia coli treated with cisplatin, UV-irradiation, or mitomycin C are dependent on the function of the RecBC and RecFOR pathways of homologous recombination. Mutat Res. 2001;486:21–29. doi: 10.1016/s0921-8777(01)00077-5. [DOI] [PubMed] [Google Scholar]
- Kucherlapati RS, Eves EM, Song KY, Morse BS, Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984;81:3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG, Sharples GJ. Dissociation of synthetic Holliday junctions by E. coli RecG protein. Embo J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Action of RuvAB at replication fork structures. J Biol Chem. 2001;276:41938–41944. doi: 10.1074/jbc.M107945200. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 2002;18:413–419. doi: 10.1016/s0168-9525(02)02720-8. [DOI] [PubMed] [Google Scholar]
- Murphy KC, Campellone KG, Poteete AR. PCR-mediated gene replacement in Escherichia coli. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Ichinose C, Ogura T, Mori H, Morita M, Hasegawa M, Kusukawa N, Hiraga S. Chromosomal genes essential for stable maintenance of the mini-F plasmid in Escherichia coli. J Bacteriol. 1988;170:5272–5278. doi: 10.1128/jb.170.11.5272-5278.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgard MV, Keem K, Monahan JJ. Factors affecting the transformation of Escherichia coli strain chi1776 by pBR322 plasmid DNA. Gene. 1978;3:279–292. doi: 10.1016/0378-1119(78)90038-0. [DOI] [PubMed] [Google Scholar]
- Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Strike P. Efficiency of Escherichia coli repair processes on uv-damaged transforming plasmid DNA. Plasmid. 1981;5:213–220. doi: 10.1016/0147-619x(81)90022-6. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Strike P. Repair in E. coli of transforming plasmid DNA damaged by psoralen plus near-ultraviolet irradiation. Mutat Res. 1986;165:81–88. doi: 10.1016/0167-8817(86)90063-5. [DOI] [PubMed] [Google Scholar]
- Roychoudhury A, Basu S, Sengupta DN. Analysis of comparative efficiencies of different transformation methods of E. coli using two common plasmid vectors. Indian J Biochem Biophys. 2009;46:395–400. [PubMed] [Google Scholar]
- Saffran WA, Cantor CR. Mutagenic SOS repair of site-specific psoralen damage in plasmid pBR322. J Mol Biol. 1984;178:595–609. doi: 10.1016/0022-2836(84)90240-7. [DOI] [PubMed] [Google Scholar]
- Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Schmid SE, Daune MP, Fuchs RP. Repair and mutagenesis of plasmid DNA modified by ultraviolet irradiation or N-acetoxy-N-2-acetylaminofluorene. Proc Natl Acad Sci U S A. 1982;79:4133–4137. doi: 10.1073/pnas.79.13.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Swenson PA, Carrier WL. Thymine dimers and Inhibition of DNA synthesis by ultraviolet irradiation of cells. Science. 1963;142:1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- Spivak G, Hanawalt PC. Determination of damage and repair in specific DNA sequences. METHODS: A Companion to Methods in Enzymology. 1995;7:147–161. [Google Scholar]
- Strike P, Humphreys GO, Roberts RJ. Nature of transforming deoxyribonucleic acid in calcium-treated Escherichia coli. J Bacteriol. 1979;138:1033–1035. doi: 10.1128/jb.138.3.1033-1035.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strike P, Roberts RJ. Resident enhanced repair: novel repair process action on plasmid DNA transformed into Escherichia coli K-12. J Bacteriol. 1982;150:385–388. doi: 10.1128/jb.150.1.385-388.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DK, Sherratt DJ. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984;36:1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- van der Ende A, Baker TA, Ogawa T, Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc Natl Acad Sci U S A. 1985;82:3954–3958. doi: 10.1073/pnas.82.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988;70:399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]
- West SC. The RuvABC proteins and Holliday junction processing in Escherichia coli. J Bacteriol. 1996;178:1237–1241. doi: 10.1128/jb.178.5.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts NS, Clark AJ. Characteristics of some multiply recombination-deficient strains of Escherichia coli. J Bacteriol. 1969;100:231–239. doi: 10.1128/jb.100.1.231-239.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Sato M. Plasmid uptake by bacteria: a comparison of methods and efficiencies. Appl Microbiol Biotechnol. 2009;83:791–798. doi: 10.1007/s00253-009-2042-4. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]