Chromosomal rearrangements of the mixed lineage leukemia (MLL) gene with numerous partner genes are frequently found in acute myeloid and acute lymphoblastic leukemia.1, 2 Although the pathomechanism of t(4;11)-mediated leukemia is still being discussed, expression of the AF4•MLL fusion was found to enhance the repopulating potential of CD34+ cells and lead to the development of predominantly proB-acute lymphoblastic leukemia in a mouse model.1, 2 The AF4•MLL protein contains cleavage sites for threonine aspartase-1 (Taspase1).1, 2, 3, 4 Upon processing by Taspase1, the AF4•MLL cleavage products form a protein complex resistant to SIAH-mediated degradation and activate oncogenic programs.3, 5 Furthermore, Taspase1 is overexpressed in liquid and solid human cancers, suggesting that Taspase1 is co-opted to promote and sustain tumorigenesis.6 As genetic deletion of Taspase1 in the mouse produced no overt deficiencies,3 inhibition of Taspase1 may offer novel anticancer strategies, including the treatment of leukemias. Human Taspase1 encodes a protease of 420 amino acids cleaving substrates in trans by recognizing a conserved peptide motif (Q3[F,I,L,V]2D1↓G1′x2′D3′D4′).4 Unfortunately, Taspase1's activity is not affected by common protease inhibitors, therefore currently precluding the assessment of its clinical and therapeutic relevance.3, 4, 7
Here, we present our endeavors to target Taspase1's oncogenic potential by (i) overexpressing inactive Taspase1 variants, and (ii) testing a putative Taspase1 inhibitor (Figure 1a).
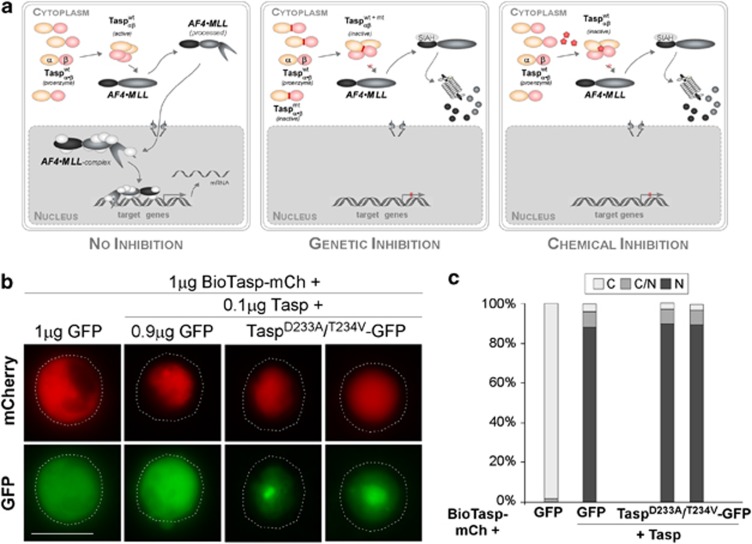
Figure 1.
(a) Strategies targeting Taspase1's oncogenic activity. Autoproteolysis of the Taspase1 proenzyme triggers formation of the active αββα heterodimer, hydrolyzing the AF4•MLL fusion protein and driving oncogenesis (left). Inhibition by overexpression of trans-dominant Taspase1 mutants results in the formation of inactive heterodimers, precluding AF4•MLL processing, and the activation of oncogenic programs (middle). Chemical Taspase1 inhibitors affect its proteolytic activity, preventing AF4•MLL processing and activation of pathological pathways (right). (b) Catalytically inactive Taspase1 mutants are not inhibitory. K562 cells were transfected with 1 μg of red fluorescent BioTaspR and 0.1 μg of Tasp-BFP, together with the indicated amounts of inactive Taspase1-green fluorescent protein (GFP) mutants or GFP expression plasmid. Even co-transfection of a ninefold excess of plasmids encoding the inactive Taspase1 variants did not affect BioTaspR processing. Localization was analyzed 48 h post transfection. GFP/mCherry (mCh) were visualized by fluorescence microscopy. Dashed lines mark cytoplasmic cell boundaries. (c) The number of cells showing cytoplasmic (C), cytoplasmic and nuclear (C/N), or nuclear (N) fluorescence was counted in at least 200 BioTaspR-expressing cells. Whereas the number of cells displaying cytoplasmic fluorescence significantly decreased by co-transfection of 0.1 μg Tasp-BFP expression plasmid, no significant trans-dominant-negative effect was evident for Taspase1 mutants.
As the Taspase1 proenzyme is autoproteolytically cleaved and assumed to assemble into an active αββα-heterodimer, we reasoned that overexpressing inactive Taspase1 mutants would inhibit the formation of active protease dimers. To analyze Taspase1's processing of AF4•MLL substrates in living cells, we employed our cell-based biosensor assay4 (Supplementary Figure S1a). Ectopic expression of Taspase1 promoted cleavage and complete nuclear accumulation of the autofluorescent BioTasp protein, containing the AF4•MLL cleavage site. Co-expression of catalytically inactive Taspase1 mutants, in which the catalytic nucleophile, Thr234, was changed into Val (TaspT234V) or Asp233 was mutated into Ala (TaspD233A), resulted in neither cleavage nor nuclear translocation (Figure 1b). Importantly, our assay as well as immunoblot analysis demonstrated that even co-transfecting a ninefold excess of the inactive Taspase1 mutants over the wild-type Taspase1 expression plasmid did not affect Taspase1's processing of the AF4•MLL biosensor (Figures 1b-d; Supplementary Figure S1b). Similar results were obtained using HA-tagged or untagged Taspase constructs, and these results were also confirmed for the Taspase1 targets TFIIA and USF2 (data not shown). Our results demonstrate that enforced expression of inactive Taspase1 mutants, aiming to inhibit formation of active protease dimers, was not inhibitory. One might speculate that Taspase1 is active already as an αβ-monomer, providing a mechanistic explanation why overexpression of inactive mutants was not trans-dominant.
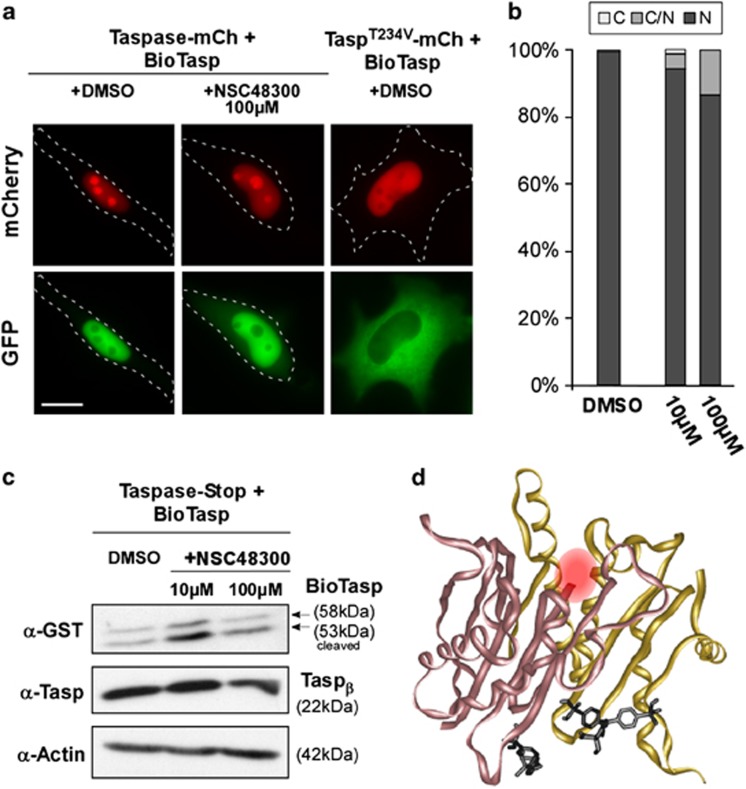
Besides genetic approaches, chemical decoys allowing the targeted inhibition/activation of proteins also allow to dissect and regulate molecular pathomechanisms. Consequently, we next tested (4-[(4-arsonophenyl)methyl]phenyl) arsonic acid (NSC48300), a recently described Taspase1 inhibitor.8 Prior to experimentation, the identity of the used batch of NSC48300 was confirmed by mass spectrometry (Supplementary Figure S2). NSC48300's potential to inhibit Taspase1's processing of the AF4•MLL substrate was examined in adherent and leukemic cell lines. Surprisingly, NSC48300 did not affect Taspase1's trans-cleavage activity, as indicated by the nuclear accumulation of the AF4•MLL biosensor at concentrations ranging from 10 to 500 μℳ (Figures 2a and b; Supplementary Figures S2c and d). The possibility that nuclear accumulation of the biosensor was indirectly mediated through the inhibition of nuclear export by NSC48300 was excluded by microinjection experiments (Supplementary Table S1). Albeit treatment with 500 μM NSC48300 impaired cell vitality, this effect was independent of endogenous Taspase1 levels (Supplementary Figures S2e and f). These results were confirmed by immunoblot analysis, revealing that NSC48300 did also not prevent Taspase1's autoprocessing (Figure 2c), and also further confirmed for the Taspase1 targets TFIIA, DPOLZ and USF2 (data not shown). To provide a molecular rationale for the observed lack of inhibition, we performed molecular docking. Albeit high-affinity NSC48300 binding sites in both the active and inactive Taspase1 structure7 were identified, no binding was detectable at or close to the catalytic nucleophile, Thr234 (Figure 2d, data not shown).
Figure 2.
(a) NSC48300 does not inhibit Taspase1. HeLa transfectants coexpressing green fluorescent BioTaspG and red fluorescent (mCherry, mCh) wild type or inactive TaspT234V were treated with dimethylsulfoxide (DMSO)/NSC48300 and analyzed 48 h later. Cleavage-induced nuclear translocation of BioTaspG by Taspase1 was not affected by NSC48300. Inactive TaspT234V-mCh did not result in cleavage and nuclear accumulation of BioTaspG. Scale bar, 10μm. (b) Quantitation of BioTaspG processing. No significant inhibition of cleavage was observed upon treatment with NSC48300. Localization was analyzed 48 h post transfection. (c) Immunoblot analysis demonstrates that NSC48300 did not inhibit Taspase1's trans-cleavage, nor cis-cleavage. HeLa cells were transfected with 1 μg of BioTaspG together with 1 μg of untagged Taspase1 and treated for 48 h. Proteins were visualized using α-glutathione S-transferase (GST) and α-Taspase1 Abs. GapDH served as loading control. (d) Stereo diagram showing the molecular docking of NSC48300 (black) to activated Taspase1. The α subunit is shown in yellow, and the β subunit in rose. The catalytic Thr234 is marked by a red sphere.
Collectively, though NSC48300 interfered with cell migration and invasion,9 was patented as an anti-angiogenic compound, and inhibited the growth of breast and brain tumors in murine models,8 our results show that these effects are not primarily based on the inhibition of Taspase1. The reason why NSC48300 was reported to affect Taspase1 in an in vitro assay8 but not in vivo (this study) remains to be elucidated.
As it will be unlikely to inhibit Taspase1 by using strategies attempting to interfere with its heterodimer formation, experimental and in silico strategies should focus on the identification of specific chemical Taspase1 inhibitors by screening of compound libraries.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Bursen A, Schwabe K, Ruster B, Henschler R, Ruthardt M, Dingermann T, et al. The AF4bulletMLL fusion protein is capable of inducing ALL in mice without requirement of MLLbulletAF4. Blood. 2010;115:3570–3579. doi: 10.1182/blood-2009-06-229542. [DOI] [PubMed] [Google Scholar]
- Montes R, Ayllon V, Gutierrez-Aranda I, Prat I, Hernandez-Lamas MC, Ponce L, et al. Enforced expression of MLL-AF4 fusion in cord blood CD34+ cells enhances the hematopoietic repopulating cell function and clonogenic potential but is not sufficient to initiate leukemia. Blood. 2011;117:4746–4758. doi: 10.1182/blood-2010-12-322230. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Knauer SK, Fetz V, Rabenstein J, Friedl S, Hofmann B, Sabiani S, et al. Bioassays to monitor Taspase1 function for the identification of pharmacogenetic inhibitors. Plos One. 2011;6:e18253, 1–14. doi: 10.1371/journal.pone.0018253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless B, Oehm C, Knauer S, Stauber RH, Dingermann T, Marschalek R. The heterodimerization domains of MLL-FYRN and FYRC-are potential target structures in t(4;11) leukemia. Leukemia. 2011;25:663–670. doi: 10.1038/leu.2010.308. [DOI] [PubMed] [Google Scholar]
- Chen DY, Liu H, Takeda S, Tu HC, Sasagawa S, Van Tine BA, et al. Taspase1 functions as a non-oncogene addiction protease that coordinates cancer cell proliferation and apoptosis. Cancer Res. 2011;70:5358–5367. doi: 10.1158/0008-5472.CAN-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan JA, Dunn BM, Tong L. Crystal structure of human Taspase1, a crucial protease regulating the function of MLL. Structure. 2005;13:1443–1452. doi: 10.1016/j.str.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Chen DY, Lee Y, Van Tine BA, Searleman AC, Westergard TD, Liu H, et al. A Pharmacologic inhibitor of the protease Taspase1 effectively inhibits breast and brain tumor growth. Cancer Res. 2011;72:736–746. doi: 10.1158/0008-5472.CAN-11-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LP, Ouellette A, Bandle R, Chang WC, Zhou H, Misra RN, et al. Identification of small-molecule inhibitors of autotaxin that inhibit melanoma cell migration and invasion. Mol Cancer Ther. 2008;7:3352–3362. doi: 10.1158/1535-7163.MCT-08-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.