Abstract
The heart is a complex organ that is composed of numerous cell types, which must integrate their programs for proper specification, differentiation and cardiac morphogenesis. During cardiogenesis members of the Twist-family of basic helix-loop-helix (bHLH) transcription factors play distinct roles within cardiac lineages such as the endocardium and extra-cardiac lineages such as the cardiac neural crest (cNCC) and epicardium. While the study of these cell populations is often eclipsed by that of cardiomyocytes, the contributions of non-cardiomyocytes to development and disease are increasingly being appreciated as both dynamic and essential. This review summarizes what is known regarding Twist-family bHLH function in extra-cardiac cell populations and the endocardium, with a focus on regulatory mechanisms, downstream targets, and expression profiles. Improving our understanding of the molecular pathways that Twist-family bHLH factors mediate in these lineages will be necessary to ascertain how their dysfunction leads to congenital disease and adult pathologies such as myocardial infarctions and cardiac fibroblast induced fibrosis. Indeed, this knowledge will prove to be critical to clinicians seeking to improve current treatments.
Twist Family Transcription Factors and Heart Development
The complex process of cardiac development begins with the specification of the anterior lateral mesoderm, termed the cardiac crescent, which contributes to the first heart field (FHF) myocardium and a portion of the endocardium. (Lyons 1996; Olson and Srivastava 1996; Sucov 1998; Brand 2003; Prall, Menon et al. 2007). These cells first undergo migration to the embryo midline to form a linear heart tube. This heart tube is composed of two cell layers, an inner layer composed of endocardial (endothelial) cells and the outer being cardiomyocytes. The space between the layers is filled with extracellular matrix (ECM) commonly referred to as cardiac jelly (DeLaughter, Saint-Jean et al. 2011). After initial heart tube formation, pharyngeal mesoderm that lies anterior to the cardiac crescent, which is termed the second heart field (SHF), migrates into the heart tube from both the rostral and caudal ends, adding endocardium and myocardium to the now expanding heart (for review see Kelly 2005; Nakajima 2010; Vincent and Buckingham 2010; Zaffran and Kelly. 2012). As a consequence of the SHF cell contribution, the tube loops to the right and balloons to form common atrial and ventricular chambers, adjoined by a single common atrioventricular canal (AVC). Further expansion and remodeling (accompanied by atrial, ventricular, and AVC septation) results in a four chambered heart complete with two distinct AVCs - the right AVC being guarded by the tricuspid valve and the left by the mitral valve. During this remodeling process, cells originating from a developmental structure termed the proepicardial organ (PEO) envelop the heart to form the epicardium, while migratory cardiac neural crest cells colonize the outflow tract (OFT) and facilitate its septation (Snider, Olaopa et al. 2007; Gittenberger-de Groot, Winter et al. 2010). Twist family bHLH factors are dynamically expressed within both the FHF and SHF with numerous gain-of-function and loss-of-function studies showing that these factors are crucial for successful completion of cardiogenesis (Riley, Anson-Cartwright et al. 1998; McFadden, Barbosa et al. 2005; Chakraborty, Wirrig et al. 2010; Barnes, Firulli et al. 2011; Tsuchihashi, Maeda et al. 2011; Vincentz, Barnes et al. 2011).
bHLH proteins make up a class of transcription factors which share two common domains. The first is a motif consisting of basic residues that facilitate DNA binding to a canonical consensus sequence CANNTG, which is termed an E-box. The second is the helix-loop-helix domain, which contains two amphipathic α-helices separated by a loop of variable length. The HLH domains facilitate dimerization via juxtaposition of the hydrophobic faces of the α-helices from two different bHLH proteins. Dimer formation allows for correct positioning of the two basic domains that directly bind DNA (for extensive bHLH review see (Massari and Murre 2000)). Of the hundreds of proteins which contain a bHLH motif (Atchley and Fitch 1997) the Twist family is a small group of transcription factors that contain similar amino acid compositions, with particularly high conservation in the bHLH domain (Castanon and Baylies 2002). In mammals, the Twist family of bHLH transcription factors is composed of six members: Twist1, Twist2, Hand1, Hand2, Paraxis, and Scleraxis; although homologs of these factors can also be found in phylogenetically distant species such as flies, flatworms, and jellyfish (Gitelman 2007; Barnes and Firulli 2009). While Twist proteins show some diversity in their expression profiles, and thus in the developmental processes they regulate, Twist family members have several key characteristics in common. It is has been well established that Twist bHLH proteins functionally operate either as homodimers or heterodimers with other bHLH factors (Castanon, Von Stetina et al. 2001; Castanon and Baylies 2002; Tapanes-Castillo and Baylies 2004; Barnes and Firulli 2009). Twist family members can heterodimerize with non-Twist bHLH proteins, such as the ubiquitously expressed E-proteins. Dimer partner choice has been shown to dictate factor function (Castanon, Von Stetina et al. 2001; Firulli, Redick et al. 2007) and is affected by elements such as gene dosage, which can alter the ratio of Twist family proteins present in the transcription factor pool inside each cell. Phosphoregulation also plays multiple roles in the regulatory process. The phosphorylation of conserved threonine and serine residues within the first α-helix of Twist family bHLH factors has been demonstrated to affect dimerization partner choice and therefore function. Phosphoregulation can also alter intracellular localization of Twist factors thus modifying dimer capacity (Firulli, Howard et al. 2003; Firulli, Krawchuk et al. 2005; Firulli, Redick et al. 2007; Barnes and Firulli 2009). Collectively, these observations establish the need to not only define where and when Twist proteins are expressed, but also understand the potential partners and the post-translational modifications that collectively drive the formation of tissue-specific transcriptional complexes.
Cardiac Neural Crest Cells
Neural crest cells (NCC) arise from ectoderm at the lateral edge of the neural plate, and extend along the dorsal aspect of the neural tube from the mid-diencephalon through the caudal portion of the developing embryo (Creazzo, Godt et al. 1998; Huang and Saint-Jeannet 2004). These ectodermal cells are induced to undergo an epithelial to mesenchymal transformation (EMT) by a combination of Wnt, bone morphogenic protein (BMP), and fibroblast growth factor (Fgf) signaling, resulting in delamination from the neuroepithelium and migration into the caudal pharyngeal arches (Huang and Saint-Jeannet 2004). Many cell types and structures including facial bones, the peripheral nervous system, and endocrine system all receive heavy contributions from NCC and have been extensively reviewed (Gross and Hanken 2008; Creuzet 2009; Heude, Bouhali et al. 2010; Wang, Chan et al. 2011). cNCC are a small vagal subpopulation of NCC and their migration paths feed them directly into the heart (Stoller and Epstein 2005). During heart development, cNCC populations make essential contributions to the heart via migration into the third, fourth, and sixth pharyngeal arches where they will contribute to remodeling of the aortic arch arteries and cardiac outflow tract (Stoller and Epstein 2005). Upon arrival, cNCC differentiate into smooth muscle cells and pericytes, which participate in septation of the embryonic OFT tract into pulmonary and aortic components (Kirby, Gale et al. 1983; Snider, Olaopa et al. 2007; Nelms and Labosky 2010). Subsequently, the majority of neural crest derived cells in the aorticopulmonary septum undergo apoptosis (Poelmann and Gittenberger-de Groot 1999). The remodeling of the aortic arch arteries is a complex programmed process involving the regression of some arterial structures and the persistence of others. Initially, each pharyngeal arch contains a symmetrically branched artery originating from the single vessel of the early OFT. Through the action of cNCC, this symmetrical arterial network is remodeled into the predominantly left sided vascular pattern seen in adults (Snider, Olaopa et al. 2007). This coordinated regression has been associated with Tgfβ2 signaling, as Tgfβ2-/- mice display aberrant apoptosis in both fourth arch arteries, while lacking normal apoptosis in the right dorsal aorta (Molin, DeRuiter et al. 2002).
If a significant portion of the cNCC fail to reach the OFT, as a result of cell death or a defect in migration, a common congenital abnormality known as persistent truncus arteriosus (PTA) results. In PTA, the OFT fails to septate and results in a single large common vessel. The high incidence of PTA in congenital disorders such as Di George Syndrome implicates NCC dysfunction in the pathology of these diseases (Van Mierop and Kutsche 1986). Other NCC-related OFT abnormalities include double outlet right ventricle (DORV) and overriding aorta. These defects result from the incorrect alignment of the aorta with the left ventricle (LV). During proper alignment, the cardiac tube loops to bring the OFT (distal end) into close proximity with the inflow tract (proximal end). The OFT undergoes septation such that the newly formed aorta gets placed between the left and right AVC in a process called wedging, which ultimately results in correct alignment of the aorta over the developing LV and the pulmonary artery over the right ventricle (Allwork and Anderson 1978; Kirby and Waldo 1995). If wedging occurs incorrectly, the aorta is often displaced to the right, resulting in either DORV, where both the pulmonary artery and aorta are connected to the right ventricle, or in overriding aorta, where the aorta partially connects to each ventricle. Experimental evidence suggests that OFT septation must occur properly to avoid looping defects which result in improper wedging (Yelbuz, Waldo et al. 2002). Indeed, the OFT septum in genetic models of cNCC ablation is often either misaligned with the ventricular septum or incompletely developed (Creazzo, Godt et al. 1998).
Although the processes of aortic arch artery and OFT remodeling are still not well understood, genetic loss-of-function models have revealed a number of proteins that are involved in the regulation of cNCC. These include the transcription factor Pax3, which is thought to play a role in NCC progenitor formation (Olaopa, Zhou et al. 2011), the Wnt/Dvl downstream effector Pitx2 (Kioussi, Briata et al. 2002), and bHLH factor Ets1 (Gao, Kim et al. 2010). Loss of these factors results in aortic arch artery and OFT defects such as DORV, PTA, and transposition of the great arteries (TGA). While transcriptional regulation of the genes discussed above clearly affects cNCC movement and function, many other proteins in the surrounding environment have a non-cell autonomous role to play. For instance Tbx1, which is affected by the chromosomal deletion causing Di George Syndrome, is not expressed in NCC, but can be found in tissue of the pharyngeal arches adjacent to NCC (Snider, Olaopa et al. 2007). Despite this fact, haploinsufficient mice display major defects in the development of the aortic arch arteries, indicating that cNCC take important cues relating to migration and morphogenesis from their environment (Garg, Yamagishi et al. 2001; Hutson and Kirby 2003; Snider, Olaopa et al. 2007). Indeed, the forkhead/winged helix transcription factors Foxc1 and Foxc2 are expressed in endothelial tissues surrounding NCCs and have been shown to be essential for proper OFT and arch artery morphogenesis (Hutson and Kirby 2003). These factors most likely act through transcriptional regulation of secreted molecules, such as Endothelin-1, which is produced by endothelial cells neighboring NCCs (Kurihara, Kurihara et al. 1995). Additional factors that mediate cell-cell signaling within NCC populations include the gap junction protein Connexin43 (Cx43) (Reaume, de Sousa et al. 1995), the secreted ligand Semaphorin3c (Sema3C) (Feiner, Webber et al. 2001), and its receptor Plexin A2 (Brown, Feiner et al. 2001; Hutson and Kirby 2003). Research on these factors has demonstrated the critical nature of cell-cell interactions and communication during cNCC development. While this fact is well appreciated, future studies are still needed to fully map out the signaling pathways involved and thereby determine what role the Twist family bHLH factors play.
Twist1 Function in cNCC
Twist was first identified in Drosophila, as a regulator of mesodermal differentiation and myogenesis (Baylies and Bate 1996). Twist and its vertebrate homolog Twist1, which are essential for gastrulation, have different biological functions depending on dimer partner choice (Castanon and Baylies 2002; Firulli, Krawchuk et al. 2005; Firulli, Redick et al. 2007; Firulli and Conway 2008). Twist1 is a dynamic member of the Twist family of bHLH transcription factors, which is broadly expressed during development and marks the extra-embryonic, somitic, limb, and pharyngeal mesenchyme. (Chen and Behringer 1995; Fuchtbauer 1995; Vincentz, Barnes et al. 2008). Given this expression pattern, it is not surprising that genetic models reveal a role for Twist1 in regulating specification and differentiation in developing mesenchyme. Systemic deletion of Twist1 (see Table 1) in mice results in embryonic lethality at E11.5 due to a spectrum of defects that include failure of the neural tube to close, defects in cranial mesenchyme, pharyngeal arches, somites, and limb buds (Chen and Behringer 1995). This phenotype is consistent with a role for Twist1 in regulating mesenchyme morphology and behavior during NCC migration and development. Furthermore, this correlates well with what is known about Twist1 function in neoplastic diseases, where a direct connection between elevated Twist1 expression and EMT has been found (Hoek, Rimm et al. 2004). During EMT, epithelial cells lose polarity and take on an invasive phenotype via loss of cell adhesion due in part to a loss of the calcium dependent transmembrane protein E-cadherin, which is regulated by Twist1 as well as Snail and Zeb1 (Smit and Peeper 2008). In the context of migratory cNCC, current evidence points to N-cadherin as a major mediator of EMT with Snail, Slug, Id2, and Pinch1 expression specifying EMT competence (Duband, Monier et al. 1995; Martinsen and Bronner-Fraser 1998; Snider, Olaopa et al. 2007). It is not currently clear what role, if any, Twist1 plays in cNCC EMT.
Table 1.
Cardiovascular defects in select Twist family mouse models
| Cardiovascular Related Expression | Mouse Model | Cardiovascular Related Phenotype | Source | |
|---|---|---|---|---|
| Twist1 | NCC, Mesenchyme, Endocardium | Twist1-/- | Pharyngeal arch defects, aberrant NCC migration | (Chen and Behringer 1995; Vincentz, Barnes et al. 2008) |
| Tie2-Cre inducible Twist1 overexpression (Endothelium) | Abnormal valve remodeling, increased valve thickness | (Chakraborty, Wirrig et al. 2010) | ||
| Hand1 | NCC, Myocardium, Epicardial Precursors | Hand1-/- | Abnormal looping, but early embryonic lethality is due to extra-embryonic defects | (Firulli, McFadden et al. 1998; Riley, Anson-Cartwright et al. 1998; Morikawa and Cserjesi 2004) |
| Wnt1-Cre; Hand1 CKO (NCC) | No defects (genetic interactions with Hand2) | (Barbosa, Funato et al. 2007) | ||
| Hand2 | Myocardium, NCC, Epicardium, Endocardium | Hand2-/- | Single ventricle, failure of aortic arch artery formation, dilation of aortic and pericardial sac | (Srivastava, Thomas et al. 1997) |
| Wnt1-Cre; Hand2 CKO (NCC) | Aortic arch artery defects, DORV, VSD | (Morikawa and Cserjesi 2008; Holler, Hendershot et al. 2010) | ||
| Mef2C-Cre; Hand2 CKO (SHF progenitors, Myocardium, Endocardium) | Tricuspid Atresia, PTA, RV hypoplasia | (Tsuchihashi, Maeda et al. 2011) | ||
| Hand1Cre; Hand2 CKO (NCC, PEO, Epicardium, lateral mesoderm) | OFT septation defects including PTA and DORV, abnormal compaction, and defective epicardialization | (Barnes, Firulli et al. 2011) | ||
| WT1ERT2Cre; Hand2 CKO (Lateral mesoderm, PEO, Epicardium) | Phenocopies Hand1Cre; Hand2 CKO | (Barnes, Firulli et al. 2011) |
NCC, neural crest cells; DORV, double outlet right ventricle; VSD, ventricular septal defect; PTA, persistent truncus arteriosus; OFT, outflow tract; PEO, proepicardium; CKO, conditional knockout
Upon close examination of Twist1-/- mice, it was discovered that the OFT cushions contain amorphic cellular nodules (Vincentz, Barnes et al. 2008). These nodules are of NCC origin, as determined by Wnt1-Cre lineage mapping, and at E11.5 strongly express the Twist family members Hand1 and Hand2 which are expressed within approximately 50% of the NCC mesenchyme within the OFT. In addition, Twist1 null mice exhibited a delay in colonization of the OFT cushions, as well as aberrant placement of NCC derivatives in the pericardium and endocardium. These data are consistent with research on Saethre Chotzen Syndrome (SCS), which is caused by TWIST1 mutations (Howard, Paznekas et al. 1997). The data suggests that Twist1 is essential for guiding cNCC migration and cell adhesion, although aberrant differentiation may also contribute to the phenotype (Vincentz, Barnes et al. 2008). While a role for Twist1 in cNCC migration has been established, the details regarding its function are still being investigated. Additional experimentation will be needed to fully characterize the OFT nodules, determine why Hand-expressing cNCC are prone to forming nodules, as well as which transcriptional programs modulated by Twist1 control cNCC morphological and migratory properties.
Hand Factor Function in cNCC
Hand1 and Hand2 are restricted to both distinct and partially overlapping domains during cardiogenesis, suggesting that these factors have both redundant and unique functions, depending on the spatiotemporal nature of their expression. In mice, prior to E7.5 Hand1 is expressed within extra-embryonic tissues such as the yolk sac, chorion, and mesoderm with this expression being maintained throughout embryonic development (Cross, Flannery et al. 1995; Cserjesi, Brown et al. 1995; Hollenberg, Sternglanz et al. 1995). Robust Hand1 expression can be observed during early embryonic development in the septum transversum. By E8.5 Hand1 expression can also be found within the developing primary heart tube in the region that will form the left ventricle, and pericardium (Barnes and Firulli 2009; Barnes, Firulli et al. 2011). As the heart loops rightward, cardiac expression of Hand1 is maintained in the outer curvature of the left ventricle myocardium, as well as in the pericardium and septum transversum (Cserjesi, Brown et al. 1995; Firulli, McFadden et al. 1998; Barnes, Firulli et al. 2010). During mid-gestation, Hand1 is expressed within the left ventricular myocardium, medial cNCC populating the caudal pharyngeal arches, and approximately 50% of the cNCC mesenchyme within the OFT cushions (Vincentz, Barnes et al. 2011). Hand1 null mice die in utero at E9.5, due to defects in the yolk sac, placenta, vasculature, and heart (Firulli, McFadden et al. 1998; Riley, Anson-Cartwright et al. 1998; Morikawa and Cserjesi 2004). Cardiac defects include failure of complete heart tube fusion and hypoplastic LV, which are likely secondary to placental, yolk sac and vascular defects. Recently, hypomorphic Hand1 alleles have allowed for analysis of the Hand1 deficient phenotype at later stages, generating additional insight into the function of Hand1 during cardiogenesis and further showing the precise genetic balance required for expression of Twist-family proteins. Mice with reduced levels of Hand1 expression (40% of endogenous) display ventricular hypotrabeculation with a thin LV myocardium and abnormally low expression of the left ventricular markers Nppa, Cited1, and Chisel (Firulli, McConville et al. 2010). Taken together with myocardial specific ablation models, these results support a primary role for Hand1 function within cardiomyocytes regulating looping and expansion of the left ventricle (McFadden, Barbosa et al. 2005; Firulli, McConville et al. 2010). Conditional ablation of Hand1 within NCC does not reveal any developmental defects and may reflect a true functional redundancy with Hand2 in these cells (Barbosa, Funato et al. 2007). Indeed, NCC specific loss of Hand1 in Hand2+/- mice results in a dysregulation of Msx2, Pax9, and Prrx2 expression, as well as hypoplasia of certain pharyngeal arch derived structures such as the mandible. This again demonstrates the critical nature of gene dosage for the Twist family of bHLH factors (Barbosa, Funato et al. 2007). Given this data, it seems likely that interactions and redundancy with additional transcription factors may be masking an unknown role for Hand1 in cNCC development.
The expression profile of Hand2 overlaps extensively with Hand1 and Twist1 expression within NCC derivatives. Early embryonic Hand2 expression occurs within the SHF-derived myocardium, OFT NCC, and endocardium. Recent examination of Hand2 expression reveals that early cardiac Hand2 expression is restricted to endocardium and underlying pharyngeal mesoderm (Barnes, Firulli et al. 2011). Hand2 right ventricular myocardial expression correlates with ingress of SHF pharyngeal mesoderm into the expanding heart tube. Hand2 is also expressed within PEO and epicardium. Not surprisingly, Hand2 null mice die in utero from cardiac defects. These embryos display a range of severe cardiovascular malformations including a single ventricular chamber with left-sided properties, a dilated aortic and pericardial sac, and failure of the aortic arch arteries to form, resulting in death at E10.5 (Srivastava, Thomas et al. 1997).
In contrast to Hand1, Hand2 is known to regulate a large number of important processes in cNCC development. While targeted deletion of Hand2 in NCC derived tissues results in embryonic lethality at E12.5, pharmacological rescue with the β-agonist isoproterenol allows for analysis of cardiovascular development to birth, as the primary cause of death is a reduction in catecholamine production within the sympathetic nervous system. In pharmacologically rescued embryos conditional ablation of Hand2 in NCC derived tissues (by crossing with the Wnt1-Cre strain) results in alignment defects of the aortic arch arteries and OFT, resulting in DORV with associated ventricular septal defects (VSD) (Morikawa and Cserjesi 2008; Holler, Hendershot et al. 2010). These phenotypic abnormalities are thought to be the result of defects in cell migration, cell-cell communication/adhesion, and cell cycle regulation. Histological analysis shows that fewer cNCC reach the OFT in mutants, and that more of the migrating cells travel independently, whereas cNCC of the control embryos seem to migrate as a coherent sheet. Interestingly, Hand2 function in zebrafish development is required for proper ECM remodeling and the subsequent migration of lateral plate mesodermal cells. Hand2 is thought to regulate this migratory event by maintaining matrix metalloproteinase (MMP) activity, resulting in the diminishment of laminin deposition and proper gut looping.(Yin, Kikuchi et al. 2010) While it is not currently clear if Hand2 regulates ECM remodeling during NCC migration, a microarray screen of RNA collected from murine E10.5 Wnt1-Cre Hand2 conditional knockout (CKO) whole hearts revealed a set of over 300 differentially regulated genes in the NCC specific Hand2 deletion embryos, and included Mmp14. (Holler, Hendershot et al. 2010). Other candidates related to cell migration included Pdgf, Itga9, Itgα4, Adam19 and Cx40. Cx40 was of particular interest since it is a component of cardiac gap junctions, which are major mediators of cell-cell communication in the developing heart. Interestingly, Cx40 was strongly expressed in wild-type cNCC, but was dramatically down-regulated in Hand2 CKO cNCC. To determine if Hand2 directly binds the Cx40 putative promoter, chromatin immunoprecipitation assays were conducted on a Hand2 expressing rat cardiomyocyte cell line. This pull-down showed that Hand2 binds the proximal Ebox-containing regions of the Cx40 promoter in vitro. Furthermore, a luciferase transactivation assay demonstrated that when co-transfected, Hand2 and its dimer partner E12 can modestly up-regulate Cx40-luciferase expression, suggesting that Hand2 regulates chemical and electrical cell-cell communication through transcriptional control of gap junction components.
In addition to cNCC migration related factors, many cell cycle related genes are differentially expressed in Wnt1-Cre Hand2 CKOs (Holler, Hendershot et al. 2010). The most highly regulated of these genes (up 37 fold) was cyclin B1 interacting protein 1 (Ccnb1ip1), which is an ubiquitin ligase that promotes the degradation of cyclin B. While the details of its function during development remain unclear, recent malignancy related research suggests that Ccnb1ip1 plays a role in coordinating the cell cycle with cell migration and invasion (Singh, Nicolas et al. 2007) – functions required for OFT septation and valve formation. Others included cdk6, a serine/threonine kinase that regulates the G0 to G1 transition by phosphorylating retinoblastoma protein (pRb) (Malumbres and Barbacid 2005), Insm1, a regulator of NCC derived sympathetic neuron development which also interacts with cyclin D1 in the heart (Liu, Wang et al. 2006; Pellegrino, Parrish et al. 2011), and several histones. Interestingly, a number of genes already associated with various aspects of cardiovascular development were found to be differentially regulated in the Wnt1-Cre Hand2 CKO mice. One down-regulated candidate that was particularly interesting was Sox11, since Sox11 null embryos develop DORV and associated VSDs as well as other OFT defects (Sock, Rettig et al. 2004). Other differentially regulated genes include the transcription factor Foxc1, the Notch downstream bHLH affecter and potential Hand dimer partner Hey1 (Firulli, Hadzic et al. 2000; Kokubo, Miyagawa-Tomita et al. 2005; Firulli, Redick et al. 2007), the metalloproteinases MMP14 and Adam19, which are critical for tissue remodeling, and NF-ATc2 which induces NF-ATc1. NF-ATc1 has key functions within the endocardium where Hand2 is also expressed (Zhou, Cron et al. 2002). Collagen type XI a1 (Col11a1), which is expressed in non-cartilaginous heart tissue, has been implicated in valve development (Peacock, Lu et al. 2008), while the latent transforming growth factor beta binding protein 1 (Ltbp1), has also been implicated in various aspects of cardiogenesis, including OFT septation, endocardial cushion EMT, and valve remodeling (Todorovic, Finnegan et al. 2011). Significantly, both Col11a1 and Ltbp1 are down-regulated in the Wnt1-Cre Hand2 CKOs. The Sonic Hedgehog (Shh) signaling repressor Gli3 was also down-regulated. GLI3 mutations in humans are associated with isolated VSDs (Qiu, Gong et al. 2006). Interestingly, Gli3 is downstream of Hand2 within the developing limb (Charite, McFadden et al. 2000). During limb morphogenesis, Hand2 restricts Gli3 expression to anterior mesenchyme, while Gli3 restricts Hand2 to the posterior limb mesenchyme. This genetic interaction establishes a Shh/FGF signaling feedback loop that is essential for proper limb patterning (te Welscher, Fernandez-Teran et al. 2002). Since Shh has been demonstrated to be necessary for normal OFT development (Washington Smoak, Byrd et al. 2005), it is possible that Hand2 plays a similar role within cNCC. Given the number of transcripts regulated and the range of developmental processes they are known to function in, it is clear that Hand2 plays an important role in cNCC development. In order to more precisely define this role, additional experiments are required to determine which genes are regulated directly by Hand2, and which are regulated by as of yet unidentified intermediates.
Other Twist Family Members
While Twist1 and Hand factors are established regulators of cardiac morphogenesis, less is known regarding Twist2, Paraxis, and Scleraxis. Twist2 shares a high level of identity with Twist1, as well as significant spatial overlap in its expression profile. Like Twist1, embryonic Twist2 expression predominantly occurs in mesenchymal cell populations and mesodermally derived cartilage (Li, Cserjesi et al. 1995), and is thought to be involved in EMT (Ansieau, Bastid et al. 2008). However, Twist2 expression temporally follows Twist1, and a significant role in cardiogenesis has not yet been described. Similarly, little is known of Paraxis and its functions during development. Embryonic expression is first observed at E7.5 in cells that will give rise to the paraxial mesoderm. Paraxis is subsequently expressed in the rostral paraxial mesoderm, where it plays an integral role in somitogenesis (Burgess, Cserjesi et al. 1995), but has not been directly linked to cardiogenesis. The final member of the Twist family, Scleraxis, is first expressed in mice at E9.5 in somites and mesenchyme of the body wall and limb buds. As the embryo matures this expression becomes restricted to areas of developing cartilage and connective tissue, including heart valves (Cserjesi, Brown et al. 1995). Indeed, Scleraxis-/- mouse embryos have thickened valve structures, increased expression of cartilage-associated genes such as Sox9, Msx1, and Snail, and a disruption of ECM and collagen fiber organization. These observations have lead researchers to conclude that Scleraxis is involved in valve precursor cell differentiation and ECM organization (Levay, Peacock et al. 2008). However, few direct transcriptional targets of Scleraxis have been identified (Liu, Watanabe et al. 1997; Espira, Lamoureux et al. 2009), and the extent of its role in cardiogenesis is not well defined. Additional investigation will be required to extend our current understanding.
Twist Family Members in the Epicardium
The epicardium originates from a cluster of cells at the venous pole of the heart termed the proepicardial organ (PEO). Due to a combination of BMP and FGF signaling, aggregates of cells from within the PEO’s mesothelial projections undergo an EMT event and migrate to the surface of the developing ventricles, ultimately covering the entire surface of the heart in a caudal to cranial fashion (Komiyama, Ito et al. 1987; Lie-Venema, van den Akker et al. 2007). In mice, this process is initiated by E9.0 and is complete by approximately E11.0 (Komiyama, Ito et al. 1987). Once the epicardial cells have enveloped the heart, multiple waves of epicardial cells undergo a secondary EMT and migrate into the adjacent myocardium. Similar to cNCC EMT, this event is regulated by a combination of paracrine growth factors and proteins such as Slug, Snail, and E-cadherin, with good evidence for Ets1, Ets2, α4 integrin, and WT1 involvement (Lie-Venema, van den Akker et al. 2007). Growth factors implicated in epicardial cell EMT include FGF, PDGF, TGFβ, and VEGF (Morabito, Dettman et al. 2001). Upon entering the myocardium, epicardial progenitor derived cells (EPDCs) are guided to their final destinations by signaling mechanisms that are not yet well understood, but are known to involve PDGF-B, PDGFRβ, Tbx5, Thymosin β4, and the Ets transcription factors. EPDCs will ultimately take up position in interstitial spaces of the ventricles and atria, contributing to the cardiac fibroblast lineage, smooth muscle of the coronary vasculature, and cardiac cushions (Lie-Venema, van den Akker et al. 2007). In chick there is some evidence for EPDC contribution to the coronary endothelium (Perez-Pomares, Carmona et al. 2002), although this has not been substantiated in mice (for a review of this topic see Gittenberger-de Groot and Poelmann 2012). While controversial, evidence also exists for limited EPDC contribution to the myocardium (Zhou, Ma et al. 2008).
Currently little is known about Twist family member function in the epicardium and EPDCs. As one might expect, Twist1 is expressed in EPDCs undergoing EMT, but is not expressed within the epicardium (Zhou, von Gise et al. 2010). Hand1 is not expressed within the PEO or epicardium but is expressed within the septum transversum that is directly caudal to the PEO. Interestingly, experiments using the Hand1eGFPCre mutant allele in conjunction with R26R lineage mapping shows that the permanently-marked Hand1 expressing cells within the septum transversum migrate into the forming PEO and subsequently mark the entire epicardium, as well as secondary EMT epicardial-derived cell types such as the coronary smooth muscle, and cardiac fibroblasts (Barnes, Firulli et al. 2010). No endothelial cell contributions were observed, further confirming differences between avian and mammalian epicardiogenesis. Collectively, these experiments identify Hand1 as one of the earliest transcription factors that defines the epicardial lineage. Given the transitory expression of Hand1 in the early precursors of the PEO, it will be interesting to see what role if any it plays in the early specification and or migration of the epicardium, and these experiments are currently underway. As a tool, the Hand1eGFPCre allele allows for conditional gene recombination within the early epicardial lineage (Figure 2). In contrast to Hand1, Hand2 is robustly expressed within the PEO and epicardium during cardiovascular development but is not expressed within epicardial precursors that reside in the septum transversum, placing Hand1 temporally upstream of Hand2 in this tissue. Recently, Hand1eGFPCre was used to conditionally delete Hand2 from Hand1 expressing cells within the septum transversum, as well as their derivatives (Barnes, Firulli et al. 2011). Hand2 CKOs display defects in epicardial EMT, a deficiency of cardiac fibroblasts, and increased epicardial apoptosis which results in an incompetent coronary vasculature that leads to embryonic lethality by E14.5 (Barnes, Firulli et al. 2011). This phenotype is recapitulated by E9.5 deletion of Hand2 using the tamoxifen-inducible WT1ERT2Cre allele, which mediates ablation of Hand2 from the septum transversum, PEO, and lateral mesoderm (Zhou, Ma et al. 2008; Barnes, Firulli et al. 2011). Gene expression analysis of primary epicardial cell cultures from Hand1eGFPCre Hand2 CKOs revealed increased levels of the fibronectin receptor Itgα4 (also shown to be effected in NCC Hand2 ablation studies) and a greatly lowered ratio of PDGFRα to PDGFRβ. Interestingly, platelet derived growth factor receptors have been previously shown to function in cell fate and specification of the epicardium after secondary EMT. Epicardial cells expressing PDGFRα derive into cardiac fibroblasts where PDGFRβ is associated with differentiation into coronary smooth muscle (Smith, Baek et al. 2011) Hand2 CKOs show marked reduction in PDGFRα expression as well as reduced numbers of cardiac fibroblasts. Hand2 regulation of PDGFRα regulation is direct, as luciferase assays demonstrated that Hand2 was able to transactivate the PDGFRα promoter (Barnes, Firulli et al. 2011). These data support a novel role for Hand2 in specification of epicardial cell fates post secondary EMT.
Figure 2. Extracardiac Lineages of the heart.
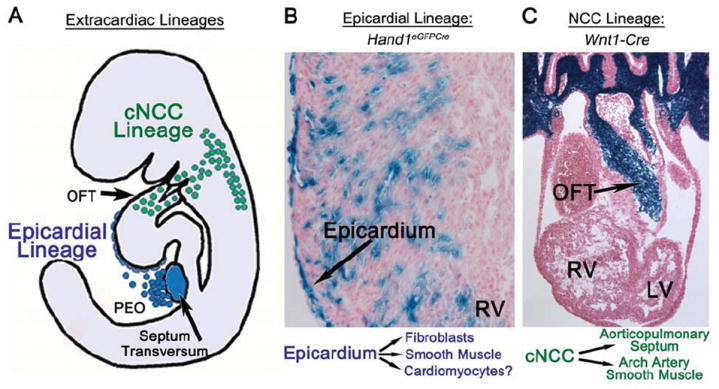
A, diagram of extracardiac lineages. B, Epicardial lineage shown by X-gal staining of Hand1eGFPCre R26R activation in right ventricle at E15.5 showing epicardial, cardiac fibroblast and coronary smooth muscle expression. C, Neural crest cell lineage, shown by X-gal staining of Wnt1-Cre R26R activation in OFT at E11.5. PEO, proepicardial organ; OFT, outflow tract; RV, right ventricle; LV, left ventricle; cNCC, cardiac neural crest cell. Left: diagram of epicardial and cNCC lineage contributions to heart development.
In addition to modified PDGFR signaling and cell fate, Hand2 plays a role in the ECM by modulating fibronectin assembly. Fibronectin is normally assembled into a complex structural network, which anchors cell and ECM adhesion molecules such as integrins (Pae, Dokic et al. 2008). In wild type epicardial explants fibronectin is organized into neat bundles that facilitate these interactions, influencing actin dynamics, EMT, and the cell cycle. Additionally, fibronectin regulates the stability and availability of extracellular matrix related factors, such as Tgfβ proteins and growth factors (Sottile, Hocking et al. 1998; Leiss, Beckmann et al. 2008). Although transcriptional and protein levels of fibronectin are unaltered in Hand2 epicardial CKO mice, fibronectin organization in mutant explants is abnormally uniform and sheet-like (Barnes, Firulli et al. 2011). Furthermore, the fibronectin cell surface receptor Itgα4 is significantly upregulated in Hand2 CKO explant cultures. Itgα4 is thought to play a role in adhesion of the epicardium to the underlying myocardium via interaction with vascular cell adhesion molecule 1 (VCAM-1), which is reciprocally expressed with Itgα4 during cardiogenesis (Kwee, Baldwin et al. 1995; Yang, Rayburn et al. 1995; Pinco, Liu et al. 2001). This suggests that the combination of fibronectin disarray and transcriptional dysregulation of downstream signaling and adhesion components could be a key mediator of the Hand2 epicardial CKO phenotype. Studies in zebrafish have complemented these observations. In wild type zebrafish embryos, fibronectin deposition is restricted to the basal surface of myocardial precursors, while in Hand2 mutants, fibronectin assembly is disorganized and no longer localized to a single surface (Trinh, Yelon et al. 2005). Unfortunately, how Hand2 regulates fibronectin assembly is not yet known. Recent research in zebrafish demonstrated that fibronectin deposition at the midline is initially required for temporally correct migration of myocardial precursors (Trinh and Stainier 2004), and that Hand2 transcriptionally downregulates fibronectin expression, to create an environment conducive to fusion of these precursors at the midline (Garavito-Aguilar, Riley et al. 2010). These studies indicate that Hand2 plays an important non-cell autonomous role in zebrafish cardiac development by transcriptionally modulating fibronectin expression and deposition.
Twist Family Members in the Endocardium
The endocardium and myocardium both derive from the anterior lateral splanchnic mesoderm which forms the cardiac crescent (Smith and Bader 2007). The heart tube, which consists of an inner endocardial layer and an outer myocardial layer overlying the intervening cardiac jelly, will grow and expand due to SHF and cNCC contributions. Paracrine signaling originating from the myocardium induces endocardial cells to undergo EMT and migrate into the AVC and OFT, forming the endocardial cushions. Simultaneously, signals originating from the endocardium cue expansion and proper patterning of the myocardium. These signaling pathways include VEGF, Notch, BMP, Wnt, and Neuregulin (Armstrong 2004). Post-EMT, the ECM and mesenchymal cells of the cushions are remodeled from their unorganized primitive state into the highly ordered structures that constitute mature heart valves (Combs and Yutzey 2009). Given the complex combination of EMT, ECM secretion, and remodeling that must occur in a precise spatiotemporal manner for correct valve development, it is not surprising that valve related deficiencies are present in a majority of all heart defects (Barnett and Desgrosellier 2003).
While little is known of Twist family function in the epicardium, perhaps less is known regarding the endocardium and Twist factors in the above-mentioned steps of valvulogenesis. The limited information that is currently available primarily pertains to Twist1, and the role it plays in the endocardial cushions during valve formation. While not detectable in ventricular endocardium, Twist1 is robustly expressed in endocardial cushions of the AVC during murine mid-gestation (Fig 2), although this expression is down-regulated as the cushions are remodeled into valves. Indeed, by E17.5 Twist1 expression is either completely absent, or nearly so (Chakraborty, Wirrig et al. 2010). Loss-of-function and gain-of-function experiments in transduced chick endocardial cushion explants have revealed multiple roles for Twist1 during cushion formation and subsequent valve development. Twist1 can induce cell proliferation in these cushion explants, promote cushion migration, and affect differentiation marker genes (Shelton and Yutzey 2008). These proposed functions correlate well with information gathered from persistent and overexpression of Twist1 in mice. In these studies a CAG-CAT-Twist1 transgene was induced by the endothelial specific Tie2-Cre, resulting in stable and persistent overexpression of Twist1 in the endocardium, and endocardium derived cushions (Chakraborty, Wirrig et al. 2010). Despite increases in area, length, and thickness of AV and OFT valve leaflets, double transgenic animals were viable. Detailed phenotypic analysis revealed abnormally high levels of proliferation in these valves, possibly due to a significant increase in Tbx20 expression. Indeed, Tbx20 has previously been associated with proliferation via induction of N-myc (Cai, Zhou et al. 2005). Twist1 overexpression was shown to affect several ECM related genes, possibly accounting for phenotypic abnormalities in cushion remodeling and migration. Dysregulated genes include Col2a1, Mmp2, and Mmp13, and Postn – all of which are significantly upregulated (Chakraborty, Wirrig et al. 2010). Furthermore, a Col2a1 regulatory element is bound by Twist1 in vivo, and can be transactivated in vitro. This binding occurs at a conserved E-box in the Col2a1 first intron. Similarly, Twist1 has been demonstrated to be capable of binding an E-box in the Postn promoter and trans-activating a reporter construct (Oshima, Tanabe et al. 2002). This information correlates well with the increased levels of Postn expression observed at E17.5 in Twist1 overexpressing mice. Together, these results support a role for endocardial Twist1 in promoting proliferation of cardiac valve progenitors, and subsequently guiding their organization as the endocardial cushions remodel, through regulation of critical ECM proteins.
Hand1 is not expressed in the endocardium or endocardial cushions, and Hand1-lineage analysis shows no endocardial contributions of any cells that have expressed Hand1 (Barnes, Firulli et al. 2010; Vincentz, Barnes et al. 2011). However, myocardial specific ablation of Hand1 surprisingly results in hyperplastic endocardial cushions that mature into abnormally thick AV valves (McFadden, Barbosa et al. 2005). This would suggest that Hand1 is involved in a myocardium derived signaling pathway that regulates endocardial cushion cells. However, Bmp2, Bmp4, Smad6, Smad7, and Tgfβ expression levels are all normal in Hand1 myocardial specific CKOs, leaving the details of such a pathway undiscovered (McFadden, Barbosa et al. 2005). Unlike Hand1 and similar to Twist1, Hand2 is strongly expressed within the endocardium and mesenchymal cells of the endocardial cushions during midgestation. However, no endocardial Hand2 functions have currently been defined. In Nkx2.5-Cre Hand2 CKO embryos, the cardiac specific transgene mediates deletion of Hand2 in a ventricular subset of cells primarily consisting of cardiomyocytes. Interestingly, death occurs two days later than in Hand2-/- embryos, suggesting that the earlier lethality seen in Hand2-/- embryos is due to loss of Hand2 function in non-cardiomyocyte cell populations. This idea is reinforced by the fact that approximately one third of embryos featuring cardiomyocyte specific loss of Hand2 via the cTnT-Cre survive until E12.5, as compared to 100% lethality by E10.5 in Hand2-/- embryos (Morikawa and Cserjesi 2008). When the expression profile of E9.0 Hand2-/- whole hearts was compared to that of Nkx2.5-Cre Hand2 CKO hearts, it was reported that Gata4, Has2, and Bmp5 were downregulated in the systemic knockout, but not in the Nkx2.5-Cre Hand2 CKO. These differentially regulated genes could be Hand2 targets in non-cardiomyocyte cell populations, which contribute to the two-day difference in embryonic viability. Interestingly, all of these genes have been linked to endocardial cushion formation and remodeling. Conditional ablation of Gata4 within endothelial cells results in an EMT defect that leads to hypoplastic cushions. EMT can be rescued by expression of a mutant Gata4 that is deficient for interaction with Fog cofactors, but septation of the common AV canal fails (Rivera-Feliciano, Lee et al. 2006). Significantly, Gata factors directly regulate Hand2 expression during RV development via two conserved consensus sites in a Hand2 cardiac enhancer (McFadden, Charite et al. 2000), possibly suggesting a positive feedback mechanism maintains Gata4 expression in the endocardium. Similarly, Has2-/- mice have abnormal endocardial cushion formation, with AV septal defects (Camenisch, Spicer et al. 2000), while endocardial cushions fail to form entirely in Bmp5-/-;Bmp7-/- double mutant mice (Solloway and Robertson 1999). Furthermore, many of the genes identified as Hand2 targets in cNCC and the epicardium are also expressed in the endocardium and mesenchymal cells of the endocardial cushions. Given this information, it is tempting to speculate that Hand2 may regulate the expression of these targets during the initiation of EMT, AV canal septation and valvulogenesis. Indeed, the extracardiac Hand2 targets Cx40, Ltbp1, col11a1, Adam19, Pdgfrα, and Hey1 have all been implicated in these processes (Gu, Smith et al. 2003; Zhou, Weskamp et al. 2004; Kokubo, Miyagawa-Tomita et al. 2005; Kokubo, Tomita-Miyagawa et al. 2007; Peacock, Lu et al. 2008; Bleyl, Saijoh et al. 2010; Todorovic, Finnegan et al. 2011). Additional tissue specific loss and gain-of-function experiments will need to be completed to determine if Hand2 similarly interacts with or indirectly regulates these targets in the endocardium.
Summary
Twist family factors are widely expressed during cardiogenesis, and systemic deletion has convincingly demonstrated their essential nature. However, the heart is composed of several distinct cell populations which must integrate during development. The tissue specific nature of Twist protein interactions, and how these interactions affect signaling cascades to facilitate integration, is only just beginning to be explored. Our lab, in conjunction with others, is currently undertaking some of the tissue specific loss and gain-of-function experiments that are necessary to fully understand this dynamic family of transcription factors. A firm understanding of the lineage specific roles that Twist proteins play during embryonic development will be indispensable when analyzing how transcription factor dysfunction leads to congenital disease.
Figure 1. Expression Profile of Twist family members.
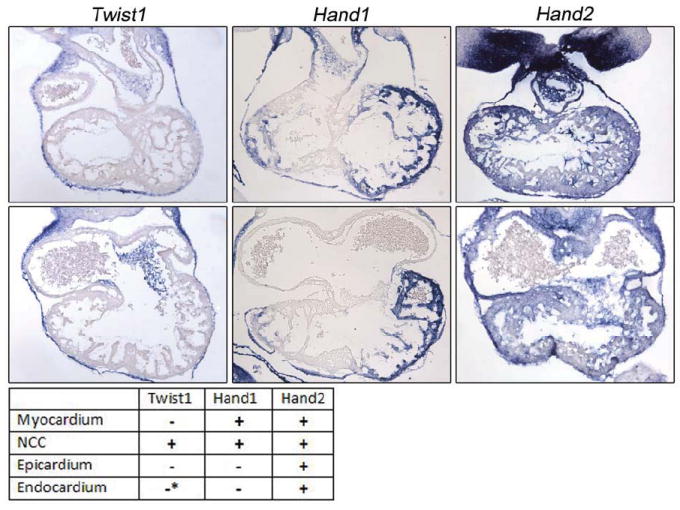
in situ hybridization of Twist1, Hand1 and Hand2 at E10.5 reveal partially overlapping expression. Twist1 is expressed in the cardiac neural crest, endocardial-derived cushions of the AVC, and the overlying endothelium as well as pericardium. Hand1 expression is coexpressed with Twist1 in the cardiac neural crest, the myocardium of the left ventricle, myocardial cuff, and is coexpressed in the pericardium with Twist1 (and Hand2). Hand2 expression robustly marks the endocardium, cushions of the AVC (coexpressed with Twist1), cardiac neural crest (coexpressed with Twist1 and Hand1) and within the epicardium.
Acknowledgments
We thank all of the Researchers that have contributed valuable insights into Hand biology. We also thank members of the Firulli lab and Riley Heart Research Center for critical input on this manuscript. Infrastructural support at the Herman B Wells Center is partially supported by the Riley Children’s Foundation and Division of Pediatric Cardiology. Grant support for this work was provided by: NIH R01HL061677-12, and 1P01HL085098-05
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allwork S, Anderson R. Remodeling of the membranous septum throughout life and its relevance to defective ventricular septation. British Heart Journal. 1978;40:463. [Google Scholar]
- Ansieau S, Bastid J, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14(1):79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Armstrong EJ. Heart Valve Development: Endothelial Cell Signaling and Differentiation. Circulation Research. 2004;95(5):459–470. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94(10):5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Funato N, et al. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Biol. 2007;310(1):154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. The International Journal of Developmental Biology. 2009;53(7):909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, et al. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Developmental Dynamics. 2010;239(11):3086–3097. doi: 10.1002/dvdy.22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, et al. Hand2 Loss-of-Function in Hand1-Expressing Cells Reveals Distinct Roles in Epicardial and Coronary Vessel Development. Circulation Research. 2011;108(8):940–949. doi: 10.1161/CIRCRESAHA.110.233171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett JV, Desgrosellier JS. Early events in valvulogenesis: a signaling perspective. Birth Defects Res C Embryo Today. 2003;69(1):58–72. doi: 10.1002/bdrc.10006. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M. twist: a myogenic switch in Drosophila. Science. 1996;272(5267):1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- Bleyl SB, Saijoh Y, et al. Dysregulation of the PDGFRA gene causes inflow tract anomalies including TAPVR: integrating evidence from human genetics and model organisms. Hum Mol Genet. 2010;19(7):1286–1301. doi: 10.1093/hmg/ddq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258(1):1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- Brown CB, Feiner L, et al. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128(16):3071–3080. doi: 10.1242/dev.128.16.3071. [DOI] [PubMed] [Google Scholar]
- Burgess R, Cserjesi P, et al. Paraxis: a basic helix-loop-helix protein expressed in paraxial mesoderm and developing somites. Dev Biol. 1995;168(2):296–306. doi: 10.1006/dbio.1995.1081. [DOI] [PubMed] [Google Scholar]
- Cai CL, Zhou W, et al. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132(10):2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch TD, Spicer AP, et al. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106(3):349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon I, Baylies MK. A Twist in fate: evolutionary comparison of Twist structure and function. Gene. 2002;287(1-2):11–22. doi: 10.1016/s0378-1119(01)00893-9. [DOI] [PubMed] [Google Scholar]
- Castanon I, Von Stetina S, et al. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development. 2001;128(16):3145–3159. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Wirrig EE, et al. Twist1 promotes heart valve cell proliferation and extracellular matrix gene expression during development in vivo and is expressed in human diseased aortic valves. Dev Biol. 2010;347(1):167–179. doi: 10.1016/j.ydbio.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charite J, McFadden DG, et al. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127(11):2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9(6):686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105(5):408–421. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creazzo TL, Godt RE, et al. Role of cardiac neural crest cells in cardiovascular development. Annu Rev Physiol. 1998;60:267–286. doi: 10.1146/annurev.physiol.60.1.267. [DOI] [PubMed] [Google Scholar]
- Creuzet SE. Neural crest contribution to forebrain development. Semin Cell Dev Biol. 2009;20(6):751–759. doi: 10.1016/j.semcdb.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Cross JC, Flannery ML, et al. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development. 1995;121(8):2513–2523. doi: 10.1242/dev.121.8.2513. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, et al. Scleraxis: a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development. 1995;121(4):1099–1110. doi: 10.1242/dev.121.4.1099. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, et al. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev Biol. 1995;170(2):664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- DeLaughter DM, Saint-Jean L, et al. What chick and mouse models have taught us about the role of the endocardium in congenital heart disease. Birth Defects Res A Clin Mol Teratol. 2011;91(6):511–525. doi: 10.1002/bdra.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband JL, Monier F, et al. Epithelium-mesenchyme transition during neural crest development. Acta Anat (Basel) 1995;154(1):63–78. doi: 10.1159/000147752. [DOI] [PubMed] [Google Scholar]
- Espira L, Lamoureux L, et al. The basic helix-loop-helix transcription factor scleraxis regulates fibroblast collagen synthesis. J Mol Cell Cardiol. 2009;47(2):188–195. doi: 10.1016/j.yjmcc.2009.03.024. [DOI] [PubMed] [Google Scholar]
- Feiner L, Webber AL, et al. Targeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruption. Development. 2001;128(16):3061–3070. doi: 10.1242/dev.128.16.3061. [DOI] [PubMed] [Google Scholar]
- Firulli AB, Conway SJ. Phosphoregulation of Twist1 provides a mechanism of cell fate control. Curr Med Chem. 2008;15(25):2641–2647. doi: 10.2174/092986708785908987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, et al. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18(3):266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Hadzic DB, et al. The basic helix-loop-helix transcription factors dHAND and eHAND exhibit dimerization characteristics that suggest complex regulation of function. J Biol Chem. 2000;275(43):33567–33573. doi: 10.1074/jbc.M005888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Howard MJ, et al. PKA, PKC, and the protein phosphatase 2A influence HAND factor function: a mechanism for tissue-specific transcriptional regulation. Mol Cell. 2003;12(5):1225–1237. doi: 10.1016/s1097-2765(03)00425-8. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Krawchuk D, et al. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37(4):373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, McConville DP, et al. Analysis of a Hand1 hypomorphic allele reveals a critical threshold for embryonic viability. Dev Dyn. 2010;239(10):2748–2760. doi: 10.1002/dvdy.22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Redick BA, et al. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem. 2007;282(37):27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchtbauer EM. Expression of M-twist during postimplantation development of the mouse. Dev Dyn. 1995;204(3):316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- Gao Z, Kim GH, et al. Ets1 is required for proper migration and differentiation of the cardiac neural crest. Development. 2010;137(9):1543–1551. doi: 10.1242/dev.047696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavito-Aguilar ZV, Riley HE, et al. Hand2 ensures an appropriate environment for cardiac fusion by limiting Fibronectin function. Development. 2010;137(19):3215–3220. doi: 10.1242/dev.052225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Yamagishi C, et al. Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev Biol. 2001;235(1):62–73. doi: 10.1006/dbio.2001.0283. [DOI] [PubMed] [Google Scholar]
- Gitelman I. Evolution of the vertebrate twist family and synfunctionalization: a mechanism for differential gene loss through merging of expression domains. Mol Biol Evol. 2007;24(9):1912–1925. doi: 10.1093/molbev/msm120. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Poelmann RE. The epicardium in development, disease and repair. Differentiation. 2012 doi: 10.1016/j.diff.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Winter EM, et al. Epicardium-derived cells (EPDCs) in development, cardiac disease and repair of ischemia. J Cell Mol Med. 2010;14(5):1056–1060. doi: 10.1111/j.1582-4934.2010.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Hanken J. Review of fate-mapping studies of osteogenic cranial neural crest in vertebrates. Dev Biol. 2008;317(2):389–400. doi: 10.1016/j.ydbio.2008.02.046. [DOI] [PubMed] [Google Scholar]
- Gu H, Smith FC, et al. High incidence of cardiac malformations in connexin40-deficient mice. Circ Res. 2003;93(3):201–206. doi: 10.1161/01.RES.0000084852.65396.70. [DOI] [PubMed] [Google Scholar]
- Heude E, Bouhali K, et al. Jaw muscularization requires Dlx expression by cranial neural crest cells. Proc Natl Acad Sci U S A. 2010;107(25):11441–11446. doi: 10.1073/pnas.1001582107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek K, Rimm DL, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64(15):5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, et al. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15(7):3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler KL, Hendershot TJ, et al. Targeted deletion of Hand2 in cardiac neural crest-derived cells influences cardiac gene expression and outflow tract development. Dev Biol. 2010;341(1):291–304. doi: 10.1016/j.ydbio.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15(1):36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev Biol. 2004;275(1):1–11. doi: 10.1016/j.ydbio.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Hutson MR, Kirby ML. Neural crest and cardiovascular development: a 20-year perspective. Birth Defects Res C Embryo Today. 2003;69(1):2–13. doi: 10.1002/bdrc.10002. [DOI] [PubMed] [Google Scholar]
- Kelly RG. Molecular inroads into the anterior heart field. Trends Cardiovasc Med. 2005;15(2):51–56. doi: 10.1016/j.tcm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Briata P, et al. Identification of a Wnt/Dvl/bCatenin - Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111(5):673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Gale TF, et al. Neural crest cells contribute to normal aorticopulmonary septation. Science. 1983;220(4601):1059–1061. doi: 10.1126/science.6844926. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77(2):211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- Kokubo H, Miyagawa-Tomita S, et al. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Developmental Biology. 2005;278(2):301–309. doi: 10.1016/j.ydbio.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Kokubo H, Tomita-Miyagawa S, et al. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134(4):747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- Komiyama M, Ito K, et al. Origin and development of the epicardium in the mouse embryo. Anat Embryol (Berl) 1987;176(2):183–189. doi: 10.1007/BF00310051. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, et al. Aortic arch malformations and ventricular septal defect in mice deficient in endothelin-1. J Clin Invest. 1995;96(1):293–300. doi: 10.1172/JCI118033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee L, Baldwin HS, et al. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development. 1995;121(2):489–503. doi: 10.1242/dev.121.2.489. [DOI] [PubMed] [Google Scholar]
- Leiss M, Beckmann K, et al. The role of integrin binding sites in fibronectin matrix assembly in vivo. Curr Opin Cell Biol. 2008;20(5):502–507. doi: 10.1016/j.ceb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Levay AK, Peacock JD, et al. Scleraxis is required for cell lineage differentiation and extracellular matrix remodeling during murine heart valve formation in vivo. Circ Res. 2008;103(9):948–956. doi: 10.1161/CIRCRESAHA.108.177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cserjesi P, et al. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev Biol. 1995;172(1):280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- Lie-Venema H, van den Akker NM, et al. Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. ScientificWorldJournal. 2007;7:1777–1798. doi: 10.1100/tsw.2007.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WD, Wang HW, et al. INSM1 functions as a transcriptional repressor of the neuroD/beta2 gene through the recruitment of cyclin D1 and histone deacetylases. Biochem J. 2006;397(1):169–177. doi: 10.1042/BJ20051669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Watanabe H, et al. Overexpression of a single helix-loop-helix-type transcription factor, scleraxis, enhances aggrecan gene expression in osteoblastic osteosarcoma ROS17/2.8 cells. J Biol Chem. 1997;272(47):29880–29885. doi: 10.1074/jbc.272.47.29880. [DOI] [PubMed] [Google Scholar]
- Lyons GE. Vertebrate heart development. Curr Opin Genet Dev. 1996;6(4):454–460. doi: 10.1016/s0959-437x(96)80067-0. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30(11):630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Martinsen BJ, Bronner-Fraser M. Neural crest specification regulated by the helix-loop-helix repressor Id2. Science. 1998;281(5379):988–991. doi: 10.1126/science.281.5379.988. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20(2):429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, et al. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132(1):189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Charite J, et al. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127(24):5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- Molin DG, DeRuiter MC, et al. Altered apoptosis pattern during pharyngeal arch artery remodelling is associated with aortic arch malformations in Tgfbeta2 knock-out mice. Cardiovasc Res. 2002;56(2):312–322. doi: 10.1016/s0008-6363(02)00542-4. [DOI] [PubMed] [Google Scholar]
- Morabito CJ, Dettman RW, et al. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev Biol. 2001;234(1):204–215. doi: 10.1006/dbio.2001.0254. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development. 2004;131(9):2195–2204. doi: 10.1242/dev.01091. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ Res. 2008;103(12):1422–1429. doi: 10.1161/CIRCRESAHA.108.180083. [DOI] [PubMed] [Google Scholar]
- Nakajima Y. Second lineage of heart forming region provides new understanding of conotruncal heart defects. Congenit Anom (Kyoto) 2010;50(1):8–14. doi: 10.1111/j.1741-4520.2009.00267.x. [DOI] [PubMed] [Google Scholar]
- Nelms BL, Labosky PA. Transcriptional Control of Neural Crest Development. Morgan & Claypool Life Sciences Publishers; 2010. [PubMed] [Google Scholar]
- Olaopa M, Zhou HM, et al. Pax3 is essential for normal cardiac neural crest morphogenesis but is not required during migration nor outflow tract septation. Dev Biol. 2011;356(2):308–322. doi: 10.1016/j.ydbio.2011.05.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Srivastava D. Molecular pathways controlling heart development. Science. 1996;272(5262):671–676. doi: 10.1126/science.272.5262.671. [DOI] [PubMed] [Google Scholar]
- Oshima A, Tanabe H, et al. A novel mechanism for the regulation of osteoblast differentiation: transcription of periostin, a member of the fasciclin I family, is regulated by the bHLH transcription factor, twist. J Cell Biochem. 2002;86(4):792–804. doi: 10.1002/jcb.10272. [DOI] [PubMed] [Google Scholar]
- Pae SH, Dokic D, et al. Communication between integrin receptors facilitates epicardial cell adhesion and matrix organization. Dev Dyn. 2008;237(4):962–978. doi: 10.1002/dvdy.21488. [DOI] [PubMed] [Google Scholar]
- Peacock JD, Lu Y, et al. Temporal and spatial expression of collagens during murine atrioventricular heart valve development and maintenance. Dev Dyn. 2008;237(10):3051–3058. doi: 10.1002/dvdy.21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MJ, Parrish DC, et al. Cytokines inhibit norepinephrine transporter expression by decreasing Hand2. Mol Cell Neurosci. 2011;46(3):671–680. doi: 10.1016/j.mcn.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pomares JM, Carmona R, et al. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46(8):1005–1013. [PubMed] [Google Scholar]
- Pinco KA, Liu S, et al. alpha4 integrin is expressed in a subset of cranial neural crest cells and in epicardial progenitor cells during early mouse development. Mech Dev. 2001;100(1):99–103. doi: 10.1016/s0925-4773(00)00503-7. [DOI] [PubMed] [Google Scholar]
- Poelmann RE, Gittenberger-de Groot AC. A subpopulation of apoptosis-prone cardiac neural crest cells targets to the venous pole: multiple functions in heart development? Dev Biol. 1999;207(2):271–286. doi: 10.1006/dbio.1998.9166. [DOI] [PubMed] [Google Scholar]
- Prall OW, Menon MK, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128(5):947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu GR, Gong LG, et al. Association of the GLI gene with ventricular septal defect after the susceptibility gene being narrowed to 3.56 cM in 12q13. Chin Med J (Engl) 2006;119(4):267–274. [PubMed] [Google Scholar]
- Reaume AG, de Sousa PA, et al. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267(5205):1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, et al. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18(3):271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Rivera-Feliciano J, Lee KH, et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133(18):3607–3618. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton EL, Yutzey KE. Twist1 function in endocardial cushion cell proliferation, migration, and differentiation during heart valve development. Dev Biol. 2008;317(1):282–295. doi: 10.1016/j.ydbio.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Nicolas E, et al. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene. 2007;26(33):4825–4832. doi: 10.1038/sj.onc.1210282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit MA, Peeper DS. Deregulating EMT and senescence: double impact by a single twist. Cancer Cell. 2008;14(1):5–7. doi: 10.1016/j.ccr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Smith CL, Baek ST, et al. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res. 2011;108(12):e15–26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TK, Bader DM. Signals from both sides: Control of cardiac development by the endocardium and epicardium. Seminars in Cell & Developmental Biology. 2007;18(1):84–89. doi: 10.1016/j.semcdb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider P, Olaopa M, et al. Cardiovascular development and the colonizing cardiac neural crest lineage. ScientificWorldJournal. 2007;7:1090–1113. doi: 10.1100/tsw.2007.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Rettig SD, et al. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24(15):6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solloway MJ, Robertson EJ. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126(8):1753–1768. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- Sottile J, Hocking DC, et al. Fibronectin matrix assembly enhances adhesion-dependent cell growth. J Cell Sci. 1998;111(Pt 19):2933–2943. doi: 10.1242/jcs.111.19.2933. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16(2):154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Stoller JZ, Epstein JA. Cardiac neural crest. Semin Cell Dev Biol. 2005;16(6):704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Sucov HM. Molecular insights into cardiac development. Annu Rev Physiol. 1998;60:287–308. doi: 10.1146/annurev.physiol.60.1.287. [DOI] [PubMed] [Google Scholar]
- Tapanes-Castillo A, Baylies MK. Notch signaling patterns Drosophila mesodermal segments by regulating the bHLH transcription factor twist. Development. 2004;131(10):2359–2372. doi: 10.1242/dev.01113. [DOI] [PubMed] [Google Scholar]
- te Welscher P, Fernandez-Teran M, et al. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 2002;16(4):421–426. doi: 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovic V, Finnegan E, et al. Long form of latent TGF-beta binding protein 1 (Ltbp1L) regulates cardiac valve development. Dev Dyn. 2011;240(1):176–187. doi: 10.1002/dvdy.22521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6(3):371–382. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Trinh LA, Yelon D, et al. Hand2 regulates epithelial formation during myocardial diferentiation. Curr Biol. 2005;15(5):441–446. doi: 10.1016/j.cub.2004.12.083. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi T, Maeda J, et al. Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev Biol. 2011;351(1):62–69. doi: 10.1016/j.ydbio.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mierop LH, Kutsche LM. Cardiovascular anomalies in DiGeorge syndrome and importance of neural crest as a possible pathogenetic factor. Am J Cardiol. 1986;58(1):133–137. doi: 10.1016/0002-9149(86)90256-0. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, et al. Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011;91(6):485–494. doi: 10.1002/bdra.20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, et al. An absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev Biol. 2008;320(1):131–139. doi: 10.1016/j.ydbio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chan AK, et al. Analysis of the sacral neural crest cell contribution to the hindgut enteric nervous system in the mouse embryo. Gastroenterology. 2011;141(3):992–1002 e1001-1006. doi: 10.1053/j.gastro.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Washington Smoak I, Byrd NA, et al. Sonic hedgehog is required for cardiac outflow tract and neural crest cell development. Dev Biol. 2005;283(2):357–372. doi: 10.1016/j.ydbio.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Yang JT, Rayburn H, et al. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development. 1995;121(2):549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- Yelbuz TM, Waldo KL, et al. Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation. 2002;106(4):504–510. doi: 10.1161/01.cir.0000023044.44974.8a. [DOI] [PubMed] [Google Scholar]
- Yin C, Kikuchi K, et al. Hand2 regulates extracellular matrix remodeling essential for gut-looping morphogenesis in zebrafish. Dev Cell. 2010;18(6):973–984. doi: 10.1016/j.devcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S, Kelly New development in the second heart field. Differentiation. 2012 doi: 10.1016/j.diff.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Zhou B, Cron RQ, et al. Regulation of the murine Nfatc1 gene by NFATc2. J Biol Chem. 2002;277(12):10704–10711. doi: 10.1074/jbc.M107068200. [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454(7200):109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, von Gise A, et al. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev Biol. 2010;338(2):251–261. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HM, Weskamp G, et al. Essential role for ADAM19 in cardiovascular morphogenesis. Mol Cell Biol. 2004;24(1):96–104. doi: 10.1128/MCB.24.1.96-104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


