Abstract
Isothermal titration calorimetry (ITC) is a powerful classical method that enables researchers in many fields to study the thermodynamics of molecular interactions. Primary ITC data comprise the temporal evolution of differential power reporting the heat of reaction during a series of injections of aliquots of a reactant into a sample cell. By integration of each injection peak, an isotherm can be constructed of total changes in enthalpy as a function of changes in solution composition, which is rich in thermodynamic information on the reaction. However, the signals from the injection peaks are superimposed by the stochastically varying time-course of the instrumental baseline power, limiting the precision of ITC isotherms. Here, we describe a method for automated peak assignment based on peak-shape analysis via singular value decomposition in combination with detailed least-squares modeling of local pre- and post-injection baselines. This approach can effectively filter out contributions of short-term noise and adventitious events in the power trace. This method also provides, for the first time, statistical error estimates for the individual isotherm data points. In turn, this results in improved detection limits for high-affinity or low-enthalpy binding reactions and significantly higher precision of the derived thermodynamic parameters.
Keywords: isothermal titration microcalorimetry, singular value decomposition, protein interactions, binding enthalpy
INTRODUCTION
Isothermal titration calorimetry (ITC) is a key technique to measure thermodynamic parameters of molecular interactions with applications in many different fields, including food chemistry1,2, materials science3, supramolecular chemistry and host-guest interactions4–6, thermodynamics of nucleic acids7, peptide and protein interactions8,9, drug discovery10,11, lipid membrane research12,13, and others (for a recent literature survey, see 14). ITC is firmly rooted in physical chemistry, and among the few first-principle techniques that allow the study of molecular interactions between unmodified reactants in free solution. It is unique in directly measuring enthalpy changes, thereby providing direct, unequivocal insight into chemical thermodynamics. During the last decades, calorimeters have become continuously smaller and more sensitive15, resulting in smaller required sample volumes, and data analysis techniques have become more sophisticated. These advances have facilitated many new applications, for example, in protein interactions and for automated higher throughput operation in secondary screening and lead optimization of candidate drugs10,15,16, for which previously sample requirements would have been prohibitive. Smaller sample requirements also facilitate experiments that demand multiple titrations, such as the determination of the heat capacity change (ΔCp) of an interaction8,9, the derivation of cooperativity parameters8,17,18, and the description of binding reactions that are coupled to salt or proton binding19–21.
The primary data of an ITC experiment is a thermogram, which records the time-course of the compensatory power required to maintain a constant vanishing temperature differential between the insulated sample and a reference cell, while known aliquots of a reactant are titrated into the sample cell in a series of injections. Each injection causes a change in the solution composition in the sample cell, and during the relaxation to a new equilibrium the heat of reaction is recorded as a peak in the power trace. Integration of this peak provides the total heat generated or consumed upon changing the solution composition. For a stoichiometric binding reaction, for instance, after modeling the isotherm of integrated heats the reaction enthalpy change (ΔH), the binding affinity, and information on the binding stoichiometry can be inferred.
A critical step in the analysis of an ITC thermogram is the distinction between the net reaction heat and the instrumental baseline power trace. Unfortunately, the baseline power trace cannot be measured separately and is observed only intermittently. This makes the assignment of reaction peak signal intrinsically ambiguous, and limits the precision of the method. Ideally, in an experiment conducted in thermal equilibrium of all calorimeter parts and in the absence of external mechanical or electrical influences, the baseline power would be constant. However, in a real experiment the baseline will always exhibit some degree of drift as well as low- and high-frequency noise components.22 The low-frequency noise is a major source of uncertainty for the integration of peaks of net reaction heats, which propagates as noise into the isotherm to be analyzed. The smaller sample volumes and smaller total heats of modern calorimeters accentuate this source of noise, which has become a significant factor in the precision of the data and is often limiting for practical applications. Low-frequency noise is also a critical factor in the analysis of the time-course of binding or folding reactions by ITC via peak-shape analysis.23,24
Although the importance of baselines has been recognized previously25, the problem of baseline assignment is only very sparsely addressed in the calorimetry literature24,26,27. To the best of our knowledge, the algorithms implemented by instrument manufacturers for default baseline assignment have not been entirely published and thus remain unclear. The necessity to manually improve on automatically designated baselines after visual inspection, an option encouraged and implemented in at least some of the commercial instrument software28,29, is familiar to most in this field, and is indeed the recommended state-of-the-art9,19,30,31. Obviously, such human intervention is unsatisfactory because it can introduce unlimited bias directly into the experimental isotherm data to be modeled. Moreover, it is often a time-consuming procedure and can even become prohibitive in practice in the context of moderate to high-throughput analyses.
Automated peak identification and the assignment of unknown baselines is an important recurring problem in many techniques (including NMR spectroscopy, mass spectrometry, Raman and other optical spectroscopy, chromatography, and analytical ultracentrifugation), and has often been addressed by taking advantage of a specific data structure.32–36 Our goal in the present work was to develop an approach for automatic peak integration of ITC thermograms resting on rational statistical principles and exploiting the specific data structure of ITC thermograms. On the basis of the expected properties of local continuity in the baseline and detailed modeling of the pre- and post-injection baselines, consensus baselines spanning each injection are estimated, and their confidence limits are determined for all peaks. We use an analysis of peak shapes to filter out signals not shared by peaks of most injections. Such features are likely adventitious and are therefore unlikely to represent the reaction of interest and instead can be assigned as refinements to the baseline. In addition, we use the expectation of monotonicity of the binding isotherm for the adaptive determination of integration parameters such as the characteristic reaction time. Compared to standard methods the approach presented here leads to a considerable improvement of the sensitivity and precision of ITC analyses, as demonstrated in the present work by application to different protein interactions. The algorithms described herein are implemented in the software NITPIC.
COMPUTATIONAL METHOD
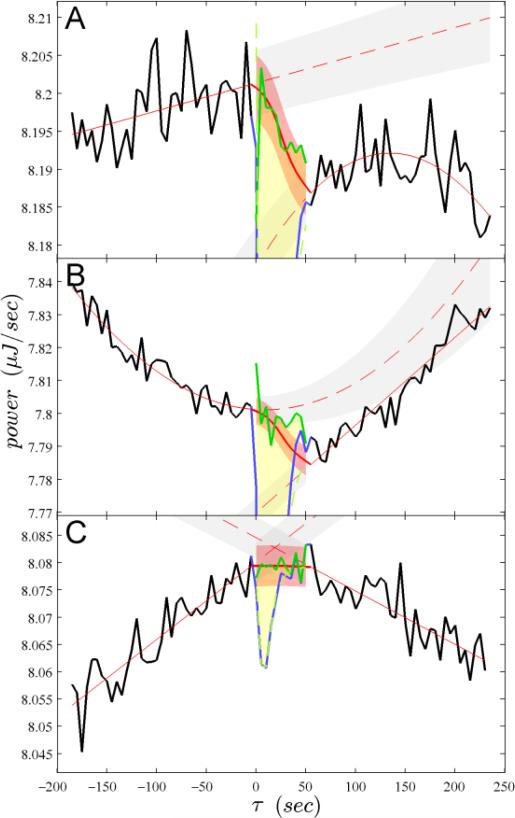
Given a thermogram, let us designate signals from the beginning of each injection j at t0,j to just prior to the start of the next injection as pj(t – t0,j), and let us assume that the reaction peak has sufficiently decayed back to baseline level after time τend, taken to be uniform for all injections (see below). τend allows us to distinguish the pre-injection baseline pj–1(τ>τend) and the post-injection baseline signal pj(τ>τend) surrounding the actual injection signal pj(τ ≤τend). Both baseline segments are fitted by least-squares with a linear function, and, if warranted based on the quality of fit using F-statistics on a certain confidence level (usually taken as 95%), a quadratic term is added37. We can extrapolate these best-fit linear or quadratic functions into the injection region 0 <τ≤τend, termed ej,pre(τ) and ej,post(τ),with their associated confidence bands e(±)j,pre(τ) and e(±)j,post(τ). From these, a smooth connecting function is used to empirically construct a continuous interpolation into the injection region, bj(τ), as well as upper and lower baseline estimates b(±)j(τ), as described in detail in the Supplementary Information (Figure 1).
Figure 1.
Illustration of the principle of baseline interpolation. Data are taken from injections #7 (A), #13 (B), and #44 (C) of the thermogram of Figure 2A, to illustrate different types of injections as well as magnitudes and orientations of baseline drifts. Solid black lines are available experimental baseline data surrounding the injections, and solid blue lines are injection data, respectively. Thin solid red lines through the baseline regions are the least-squares fits to the pre- and post-injection baselines, dashed red lines denote their extrapolations, and gray bands indicate their 68% confidence range. The bold red line in the injection region is the interpolated baseline bj(τ), and the area shaded in light red is the corresponding confidence band, which determines the error of the peak integration. The dashed green line, virtually superimposing on the injection data, is the truncated SVD model using eqs 3 and 5. Attributing residuals from the SVD peak model to the baseline bj(K’) after eq 6, the bold solid green line is the best-estimate baseline during the injection. The heat generated by each injection therefore corresponds to the area between this green line and the blue line of the injection signal, as highlighted with yellow shade, resulting in values of –7.2, –6. 9, and –0.28 μJ, respectively.
In parallel, we can pursue a second strategy for an initial baseline estimate by simply relying on a straight-line connection of anchor points just before and after the injection, bj,straight(τ). For the anchor points, we take the average of the closest navg data points. The virtue of this approach is that it is insensitive to medium-frequency fluctuations in the pre- and post-injection period, but it has the drawback that it will be more sensitive to the high-frequency noise in the data points adjacent to the injection, especially at low navg. Dependent on the particular experiment, this may be an advantage, or it may constitute a neglect of useful information in the baseline shapes. The suitability of the straight-line strategy cannot be known a priori. Therefore, NITPIC compares the performance of both approaches in a model-free assessment of isotherm noise and automatically chooses the better approach.
After initial baseline determination, the integration for the total heat of injection is straightforward
| (1) |
, which is followed by normalization with respect to the molar quantity of injectant added in injection j. The error associated with this integral arising from baseline uncertainty is estimated as
| (2) |
Next, for the analysis of peak shapes, we perform a singular value decomposition (SVD)
| (3) |
to decompose the net injection signal in terms of K characteristic signal components uk(τ), with J being the total number of injections, and the matrix vj,k describing the amplitude of each signal component required to reconstruct a particular injection signal j. The singular values sk are ordered and describe the importance of each component in a sense of a global least-squares fit to all injections. As illustrated in Figure 2, only the first few shape components are relevant, with the higher components essentially only contributing noise. SVD also allows us to calculate the contributions of each singular component to each of the integrated heats:
| (4) |
. At this point, we can attempt to eliminate the noise components and truncate the summation to consider the first K’ components, which leads to a different isotherm, Q̃(1...K '). As a criterion for the choice of K’, NITPIC considers the root-mean-square deviation (rmsd) of the filtered isotherm from the original, and limits K’ to be no smaller than the minimum number necessary for keeping the rmsd within the estimated uncertainty of the integrals (from eq 2):
| (5) |
. Among all K’ satisfying this condition, NITPIC chooses the one that minimizes a model-free measure of the noise in the isotherm (described below), which is usually the smallest K’-value. The signals from the truncated SVD components can be combined with our previously predicted smooth baseline bj(τ) as
| (6) |
for the final best estimate of the baseline bj(K’). All of the computations above depend on the prior assignment of the end-point of the injection τend, taken to be common to all injections (with few exceptions; see Discussion), and determined as described in the Supporting Information.
Figure 2.
Example of a thermogram with low-frequency noise and the SVD of the raw injection data. (A) Experimental thermogram titrating 389 μM CAII into 40 μM TFMSA in 0.5 μL injections; peaks highlighted in red correspond to those magnified in Figure 1. (B) Singular values sk for all shape components. (C) Shape components uk(τ) scaled by their corresponding singular value. Colors are equivalent to those in Panel B.
An empirical measure of noise in the isotherm is based on the expectation that at over most of the range of the titration the Qj values should vary smoothly with solution composition. For each injection j we fit a second-order polynomial to the set of surrounding isotherm points {Qj-m, Qj-m+1,...Qj--1, Qj-+1,...,Qj-+m}, typically with m = 4. The quality of fit determines a weight wj for the confidence of the prediction. For example, the polynomial fit is poor in very steep transitions, in which case the polynomial prediction is replaced by a spline. When applied to all injections, a weighted root-mean-square of all deviations (wrmsd) of ΔQj between the predicted and measured values, calculated as , provides a model-free measure of the overall noise of the isotherm. The wrmsd can be used to optimize some overall parameters affecting the baseline determination of all points, for example, the navg for the anchored straight-line baseline, and judge whether overall to use the anchored straight-line approach with bj,straight(τ) or the extrapolation approach with bj(τ). Furthermore, we can identify individual outliers, which may arise from adventitious signals invalidating the assumption in the baseline-prediction approach. Dependent on a pre-set option, NITPIC may be allowed the replacement of the particular ΔQj with ΔQj(straight), or vice versa, whichever method shows higher robustness by displaying a smaller deviation from the polynomial or spline prediction, respectively, for that injection.
Adjustments to the above procedure are applied when NITPIC recognizes high-quality bimodal thermograms with less than three injections in the transition between the regime of stoichiometric binding and the saturation plateau, such as generated at high c-values (c-values being customarily defined as the ratio between the concentration of reactant in the cell and Kd38–40). Recognizing that these injections often exhibit slower return to baseline and may have somewhat different shape, 20% longer injection times are allowed for these injections and they are excluded from SVD truncation. Furthermore, high-quality bimodal isotherms with ~10 or fewer injections in the transition, such as achieved often at moderately high c-values, occasionally exhibit slightly different injection shapes from those in the stoichiometric and saturation plateau. A possible common residual shape component in the transition region not represented well in the overall SVD is added to the injection model in eq 3 as the calculated first SVD terms of the residuals .
The approach above was implemented in the program NITPIC as a compiled MATLAB script and will be made available as PYTHON-based stand-alone executable for reading .itc data from MicroCal titration calorimeters; extensions to other data formats can be added on request. NITPIC is a standalone program that will save a table of the calculated heats and their error estimates for all injections into ASCII files for any further processing by the user. Optionally, for convenience it can also write configuration files for the global multi-method modeling software SEDPHAT18,42, and on request spawn a copy of SEDPHAT for data analysis in this platform. NITPIC can be obtained from http://biophysics.swmed.edu/MBR/software.html. It can be run in a fully automated manner or optionally provides a graphical user interface for visual inspection of the baseline and integration results. However, other than the pre-defined criteria described above, which govern the baseline prediction across the board for all injections and can be customized for specific processes, NITPIC does not offer the possibility to manually change the integration for individual injections.
EXPERIMENTAL
Lyophilized carbonic anhydrase II from bovine erythrocytes (CAII; Sigma–Aldrich, Munich, Germany) was dissolved in 50 mM phosphate buffer, 50 mM NaCl, pH 7.0. The final protein concentration (typically ~400 μM) was determined spectrophotometrically using ε280 = 55,100 M–1cm–1.41 Prior to ITC experiments, the protein solution was supplemented with dimethyl sulfoxide (DMSO) to a final concentration of 0.5% (v/v). A 20 mM stock solution of trifluoromethanesulfonamide (TFMSA) was prepared in DMSO and diluted in the same buffer to a final DMSO concentration of 0.5% (v/v) DMSO. Experiments were performed at 25°C with a MicroCal iTC200 (GE Healthcare, Freiburg, Germany) having a sample cell volume of 197.1 μL. In different experiments, injections of 0.5 – 1.0 μL of CAII at concentrations between 50 – 400 μM were made into the cell containing 5 – 40 μM TFMSA, with time intervals of 240 s, a stirrer speed of 500–1000 rpm, a filter period of 5 s, and a reference power of 4.2–8.4 μJ/s. All integration of thermograms in the present work was carried out with the same default NITPIC parameters. For comparison purposes, baseline adjustment and peak integration were also accomplished using ORIGIN version 7.0 (OriginLab, Northampton, USA) using default settings (24 points per injection, medium smoothness)28.
Fitting of the resulting isotherms excluding the first data point was performed with the software SEDPHAT18,42 version 10.36, which allows global weighted least-squares fitting of ITC data, taking advantage of the error bars provided by NITPIC for the individual data points. The model for 1:1 binding was implemented based on standard expressions for mass action law and total reaction heat, accounting explicitly for baseline offsets and binding incompetent fractions18, using an unmixed neck model28. Statistical analysis was performed according to 43. Projection maps of the error surface were calculated by constraining the parameter of interest sequentially to different values, while allowing all other unknowns to compensate for the constraint.
RESULTS
NITPIC was applied to a large number of thermograms from different types of biomolecular binding reactions. After initial adjustment of the algorithm and default parameters, NITPIC could subsequently be applied without user intervention. Usually the baseline extrapolation from linear or quadratic fits to the pre- and post-injection baselines yielded better results than the approach of interpolating a straight line between box-average anchor points. Upon visual inspection, the predicted baseline bj(τ) was found to be smooth, slowly changing, and robust with regard to adventitious pre- and post-injection baseline shapes, as illustrated in Figure 1.
Figure 2 shows an example for the analysis of a non-optimal thermogram, generating initial heats of ~–7.2 μJ per injection, with considerable low-frequency noise (in this case likely caused by fluctuations in the room temperature). Typical in our experience for any type of thermogram is the presence of only a few relevant shape components derived from the SVD analysis (see Methods). The third singular value is often an order of magnitude smaller than the first, with a characteristic ‘L’ shape on a log-scale that bends between the 5th and 10th singular value. The uk(τ) components essentially are noise beyond this point. These results justify the technique of using SVD to filter out components that contribute only noise to the heats, as well as those that might represent adventitious shape components unrelated to the reaction. Usually, NITPIC selects the smallest K’ satisfying the constraints of eq 5, which is consistent with the notion that those components with smaller singular values contribute essentially only noise to the integrals. Despite the fact that, with the condition of eq 5, SVD usually filters only subtly, the number of components is typically only 3 to 5, although it can be much higher for data with very high heat signals. When the residuals between the truncated SVD fit and the measured injection data are attributed to be baseline noise, the resulting baselines bj(K’) usually appear plausible (Figure 1).
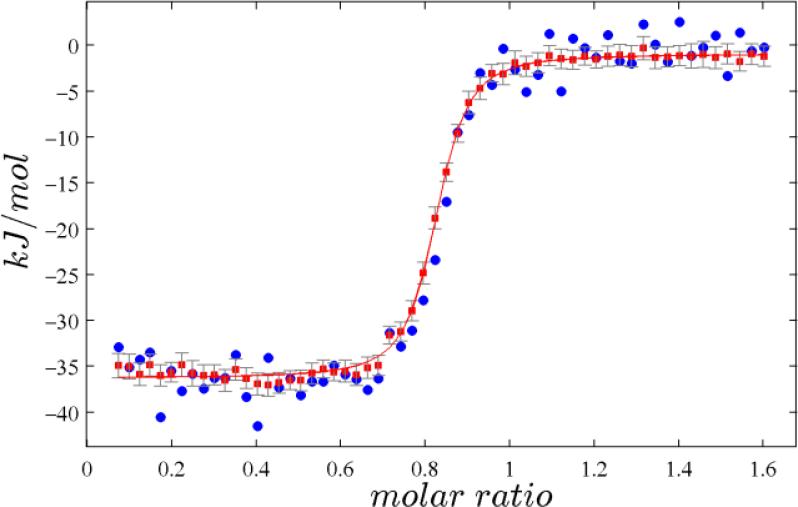
The effect of SVD filtering is immediately apparent for low-heat injections with low signal/noise ratio, as can be discerned, for example, in the second half (25 sec < τ < 50 sec) of the injection in Figure 1C. No significant shape component corresponds to this oscillating signal pattern, such that it is attributed to baseline noise and does not contribute to the integrated heat. Similarly, adventitious events with no similarity to any injection and no plausible relation to reaction heats will be filtered out, as indicated in some examples in the Supporting Information. We also found SVD filtering effective in compensating for imperfections in the baseline bj(τ), such as shown in Figure 1A,B. Figure 3 shows the isotherm as derived from the data in Figure 2. For comparison, the data points generated from ORIGIN automated integration are shown in blue. It can be discerned that for a large number of injections the ORIGIN derived points are outside the confidence intervals calculated by NITPIC. A fit to a simple 1:1 binding model has residuals with 3.4-fold smaller root-mean-square deviation (rmsd)with NITPIC than with ORIGIN. Typically, this factor was observed to fall into the range of 2–5. More examples with different types of thermograms can be found in the Figures SI-2 and SI-3 (Supporting Information). Exceptions where NITPIC was not superior were thermograms exhibiting few baseline disturbances when simultaneously the total heat per injection was very large (>50 μJ), so that uncertainties in the baseline determination are essentially irrelevant, and thermograms where less than ~ 30% of the time between injections represents baseline (see Discussion).
Figure 3.
Titration isotherm resulting from the data in Figure 2 after automated thermogram processing using ORIGIN (blue circles) or NITPIC (red squares) with associated error bars (grey) and 1:1 binding model fitted to the NITPIC isotherm (red line).
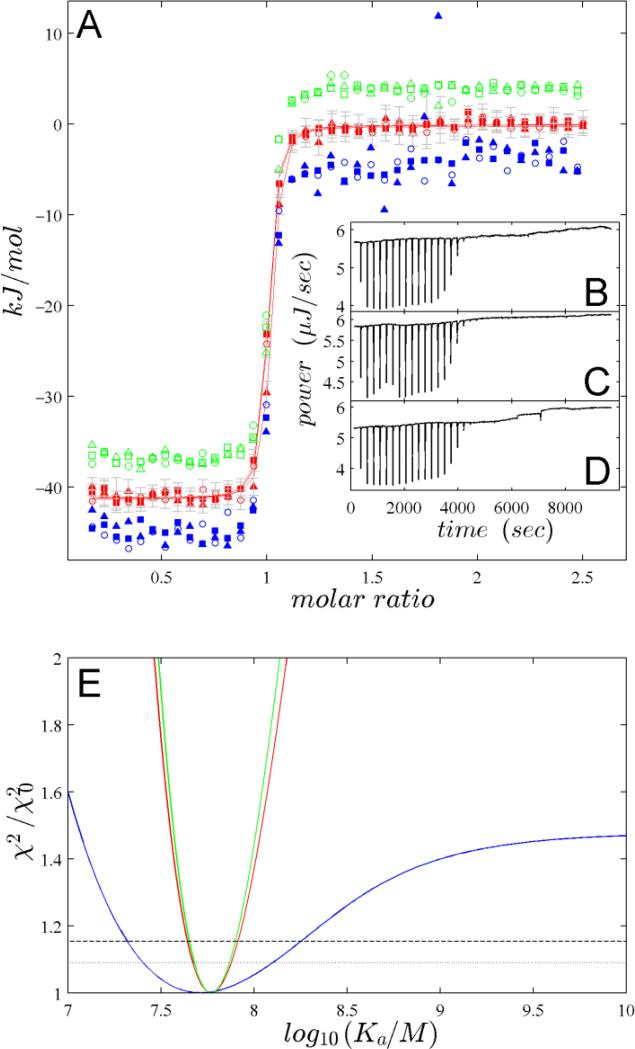
Next, we studied how the lower noise translates into more accurate estimates of thermodynamic binding parameters. Figure 4 shows a titration experiment with initial heats of ~ 18 μJ, carried out in triplicate. With few exceptions, the heats determined from NITPIC processing of the three thermograms at equivalent molar ratios exhibit a variation consistent with their assigned error bars (Figure 4A, red symbols). When each of the isotherms is fitted separately to a 1:1 binding model, the resulting rmsd values are similar with 451, 459, and 574 J/mol. The best-fit Kd-values and 95% confidence intervals are consistent with estimates of 20 (13 – 28) nM, 22 (13 – 34) nM, and 16 (10 – 24) nM, respectively. For an overall best-fit Kd-value we can include all isotherms into a global analysis with thermodynamic binding parameters representing the global parameters, and small baseline offsets and incompetent fractions representing parameters local to each experiment, leading to a global Kd estimate of 17 (12 – 23) nM, with a well-determined minimum of the error surface (Figure 4E). For comparison, ORIGIN processed thermograms (Figure 4, blue symbols) resulted in a Kd estimate of 19 (6–49) nM, with an rmsd of 1736J/mol. Thus, NITPIC allowed narrowing the uncertainty for the estimate of ΔG by a factor ~3.2, and similarly, the error interval of ΔH was reduced by a factor 3.7.
Figure 4.
Isotherm analyses of triplicate experiments titrating 443 μM CAII into 40 μM TFMSA. (A) Integrated heats calculated by ORIGIN (blue, for clarity offset by –4 kJ/mol), NITPIC (red), and manually determined baselines (green, offset by +4 kJ/mol). Best-fit isotherms to the NITPIC data are shown as red lines. (B-D) Raw thermograms corresponding to the isotherms shown as circles (B), squares (C), and triangles (D), respectively. (E) Error projection maps of the global analysis of the triplicate experiments showing the relative increase of the global χ2 over the best-fit as a function of parameter values for Kd. The error projection maps are from isotherms after thermogram processing by ORIGIN (blue), NITPIC (red), or manually determined baselines (green). Also indicated is the relative increase of χ2 associated with the 68% and 95% confidence levels (gray dotted and black dashed lines, respectively).
In the present context very interesting features are two adventitious baseline jumps at ~ 6100 and ~7200 sec of the thermogram of one the replicates shown in Figure 4D. These data are highlighted in Figure SI-1 (Supporting Information). The second of these artifacts is associated with a “false peak” (an unexplained adventitious baseline spike not resulting from an injection) only 5 s prior to the scheduled injection time. The discontinuities cause broadening of the confidence band b(±)j(τ) and bias the predicted smooth baselines bj(τ). However, since the shape components in the injection data pj(τ) –bj(τ) implied by these artifacts are unique, SVD compensates for these features in the net baseline bj(K’), resulting in very little heat being assigned by NITPIC to these injections. In contrast, strong artifacts arise in the integrated heats for the injections flanking such spikes (data not shown) when the thermogram is integrated automatically by the instrument software ORIGIN. Similar artifacts of varying magnitude are not uncharacteristic for experimental thermograms, and SVD filtering in NITPIC was found to routinely recover their detrimental effects on the baseline prediction, such as shown in Figure SI-1.
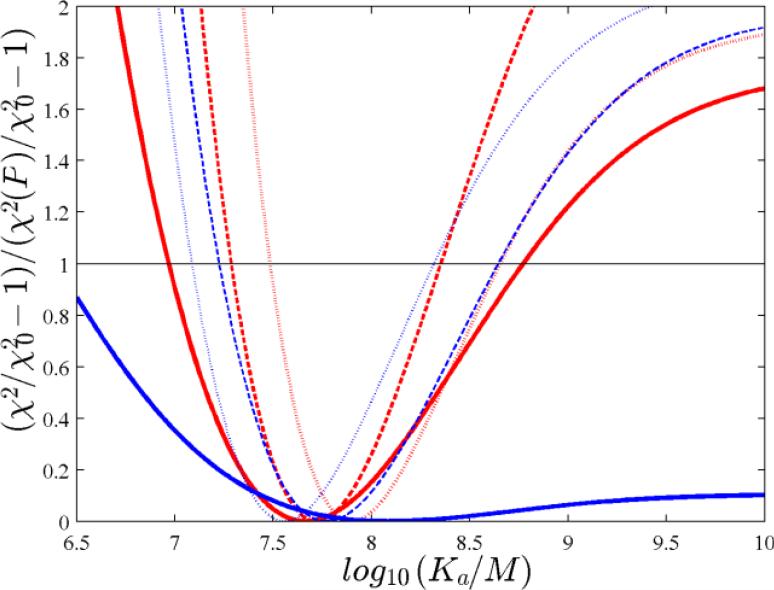
Finally, we hypothesized that the higher precision afforded by NITPIC thermogram integration might allow one to conduct experiments at lower reactant concentration, producing smaller reaction peaks. To this end, we extended the experiments with the high-affinity system of Figure 4 and performed a series of titrations with two-fold dilution of reactant concentrations, under otherwise identical experimental design, generating initial peaks with absolute heats of ~8.2, ~4.2, and ~2.1 μJ. At each dilution step, titrations were carried out in triplicate or quadruplicate and subjected to a global analysis. After thermogram integration with NITPIC, the best-fit values for Kd are consistent and well-determined even for the lowest concentration examined (Figure 5), conditions at which the ORIGIN analysis failed to produce well-determined parameters. An example for even lower reaction heats of < 0.6 μJ is shown in Supporting Figure SI-3. In contrast to the default ORIGIN thermogram processing, from which no binding reaction was discernible, the application of NITPIC resulted in a well-defined binding isotherm confirming high-affinity binding.
Figure 5.
Analyses of ITC experiments conducted in a dilution series. Error projection map from the analysis of (I) triplicate experiments titrating 200 μM CAII into 20 μM TFMSA with 40 injections of 1 μl each generating initial heats of ~-8.2 μJ (dotted lines); (II) quadruplicate experiments titrating 100 μM CAII into 10 μM TFMSA with 40 injections of 1 μl each generating initial heats of ~-4.2 μJ (short dashed lines); (III) triplicate experiments titrating 50 μM CAII into 5 μM TFMSA with 40 injections of 1 μl each generating initial heats of ~ -2.1 μJ (solid lines). The error projection maps shows the value of the global χ2 as a function of parameter values for Kd, scaled such as to have a minimum at ordinate values of 0 corresponding to the best-fit χ2 and values of 1 at a χ2 corresponding to a 95% confidence level (black horizontal line).Global analyses are shown from NITPIC processing of thermograms (red) and ORIGIN processing (blue). In set I, one outlier data point was excluded in the ORIGIN analysis, and in set III one outlier data point was excluded in both NITPIC and ORIGIN analyses.
DISCUSSION
As sample volumes of isothermal titration calorimeters become smaller and instruments more sensitive11,15, adventitious events in the power trace as well as medium- and low-frequency noise components, all of which cannot be addressed simply by longer data acquisition windows, appear to become limiting. Experimentally contributing factors often remain unclear, although bubbles, bent needles, and external mechanical or electronic events have been identified as sources of noise and poorly reproducible signals9.Unfortunately, techniques for the analysis of thermograms with imperfectly constant baselines have rarely been addressed in the scientific literature. Strategies such as a spline model or other smooth interpolation functions for long-term drifts 24,26,44, as well as linear interpolation between average pre- and post-injection baselines27 have been reported. The examples of Figure 1 and Figure S1 illustrate where these approaches would clearly not be detailed enough and introduce errors. Thus, the visual inspection of the power trace and manual assignment of the baseline and peak integration limits have become the standard procedure in most laboratories and is recommended in standard protocols 9,19,30,31. Although subjective choices undoubtedly are at play in the data acquisition of other techniques, this approach obviously detracts from the rigor of the method, and it would be desirable to avoid. While this subjectivity is no serious problem for reactions producing large heats and correspondingly high signal/noise ratio, the relevance of this factor is exacerbated for experiments with small measured heat signatures.
The basic dilemma of thermogram analysis is that the baselines cannot be measured independently. However, we know a great deal about the data structure of a thermogram, which we aimed to exploit: (1) the power peaks resulting from chemical reactions start approximately at pre-determined times, are continuous, largely similar in shape, and asymptotically relax back to the baseline; (2) the baseline should have no discontinuities and change slowly relative to the signal change at the peak of the injection, with isolated exceptions; (3) there is a finite time after which we can consider the reaction heats to be negligible and the power signal to reflect the baseline. We added to the above assumptions the expectation on the isotherm that the relationship between subsequent injections is smooth in most regions. Some operational aspects of the NITPIC algorithm remain empirical, such as the choice of weights for pre- and post-injection baselines and the choice of the transition function and its parameter, and reflect our attempt to distill our experimental intuition that currently goes into the manual baseline assignment. Yet, in a fundamental difference to the manual assignment, NITPIC automatically applies all procedures with the same criteria and same parameters to all injections and all thermograms. It thus eliminates individual operator bias and injection-to-injection variation in the application of these principles, while automation enables consistency and high-throughput processing.
A new aspect of NITPIC that cannot be exploited by manual processing is the shape similarity of the injections, which via truncated SVD allows the filtering of unusual shape components from the signals during the injections. This allows us to filter out effects of unique adventitious events during the injection, such as false peaks or spurious baseline fluctuations. It also can provide a fail-safe mechanism for the baseline interpolation, for example, if the baseline interpolation is biased by imperfections in between the injections, to the extent that the erroneous baselines imply an unusual shape of the net injection data. Furthermore, it allows us to effectively estimate the short-term noise components in the interpolated baseline, significantly enhancing the signal/noise ratio of the resulting isotherm.
When compared against manually adjusted baselines, we found the performance of NITPIC to be generally close and sometimes even slightly better. For example, when the thermograms of the triplicate experiments in Figure 4 were manually analyzed (largely blind to the subsequent isotherm analysis), a global Kd value of 17 (13–22) nM was determined, which is comparable to the estimate of 17 (12–23) nM from NITPIC. When inspecting the NITPIC-generated baselines(e.g., those in Figure 1), we did not always find them in agreement with the choice an individual operator might have made subjectively; importantly, however, we cannot identify any features of remaining systematic disagreement across all injections. Furthermore, we noticed discrepancies of similar magnitude among the subjective preferences of different human interpreters of these traces.
A fundamental limitation of the SVD truncation approach is that it will not perform well with thermograms featuring single peaks that have unique shapes, such as the broader peaks often encountered in the transition region of very steep isotherms with very high ‘c-value’, such as in Figure 4. Currently, we have addressed this problem by adding a routine to NITPIC for the automated recognition of such isotherms and the modification or elimination of SVD truncation specifically for the transition points. A possible future improvement could consist in a global peak shape analysis jointly of thermograms from replicate experiments. Another current limitation of NITPIC rests in the local least-squares baseline interpolation, which works best if at least 30%–40% of the time interval between injections reflects baseline signal. For shorter baseline segments, algorithms that utilize estimates of the global baseline drift throughout the entire experiment will likely perform better. On the other hand, providing for sufficient spacing of injections is experimentally trivial and sound practice. Comparing the sub-optimal thermogram in Figure 2 with those of Figure 4, it appears that when local baseline interpolation of NITPIC can be applied, the baseline drifts become essentially irrelevant. This is not to advocate poor experimental practice; however, these results demonstrate that NITPIC is able to generate satisfactory binding isotherms even from data acquired under less than optimal experimental conditions.
Given the ambiguity of baseline determination in some cases, it is conceptually satisfying that the NITPIC algorithm assigns error bars to the integrated isotherm data points quantifying this uncertainty for each injection. Including the integration errors enables a more realistic statistical analysis of the ITC isotherm that takes into account the non-uniformity of these errors and allows the de-emphasis of the statistical weight of data points where the baselines are more ambiguous. This strategy prevents over-interpretation of the data. Even in cases where the errors are uniform, the error estimates translate at least one quality of the raw thermogram into an attribute of the isotherm that is subject to be modeled, which may prevent over- or under-fitting of the isotherm. A suitable extension of the public-domain multi-method analysis software SEDPHAT18 is available to take advantage of this richer information.
Finally, the automated generation of isotherms of similar quality as manual thermogram interpretation can eliminate a time-consuming step in the analysis of ITC data and provide significant improvement in data quality for higher throughput studies that currently rely on existing automated data processing, equivalent to a several-fold increase in instrumental sensitivity.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Stiftung Rheinland-Pfalz für Innovation (Grant 961-386261/969 to S.K.) and by the Intramural Research Program of NIBIB and NHLBI, National Institutes of Health. The authors thank Drs. Ranjit Deka, Michael V. Norgard, and Zhaochun Chen for providing materials for the production of the Supplemental Figures.
Footnotes
Supporting Information Available. Supplementary Computational Methods; Figure SI-1 showing a detailed view of individual peak analyses of Figure 4; and Figures SI-2 and SI-3 showing two additional examples of the application of the NITPIC algorithm to different types of thermograms. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Galleano M, Verstraeten SV, Oteiza PI, Fraga CG. Arch. Biochem. Biophys. 2010;501:23–30. doi: 10.1016/j.abb.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Poncet-Legrand C, Gautier C, Cheynier V, Imberty A. J. Agric. Food Chem. 2007;55:9235–40. doi: 10.1021/jf071297o. [DOI] [PubMed] [Google Scholar]
- 3.Wadsö L. Cem. Concr. Res. 2010;40:1129–1137. [Google Scholar]
- 4.Yang SK, Ambade AV, Weck M. J. Am. Chem. Soc. 2010;132:1637–45. doi: 10.1021/ja908429e. [DOI] [PubMed] [Google Scholar]
- 5.Willerich I, Gröhn F. J. Am. Chem. Soc. 2011;133:20341–56. doi: 10.1021/ja207565m. [DOI] [PubMed] [Google Scholar]
- 6.Hughes AD, Anslyn EV. Proc. Nat. Acad. Sci. USA. 2007;104:6538–43. doi: 10.1073/pnas.0609144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khakshoor O, Wheeler SE, Houk KN, Kool ET. J. Am. Chem. Soc. 2012;134:3154–63. doi: 10.1021/ja210475a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leavitt S, Freire E. Curr. Opin. Struct. Biol. 2001;11:560–6. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 9.Pierce MM, Raman CS, Nall BT. Methods (San Diego, Calif.) 1999;19:213–21. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 10.Ladbury JE, Klebe G, Freire E. Nat. Rev. Drug Discovery. 2010;9:23–7. doi: 10.1038/nrd3054. [DOI] [PubMed] [Google Scholar]
- 11.Peters WB, Frasca V, Brown RK. Com. Chem. High T. Scr. 2009;12:772–790. doi: 10.2174/138620709789104889. [DOI] [PubMed] [Google Scholar]
- 12.Heerklotz H, Tsamaloukas AD, Keller S. Nat. Protoc. 2009;4:686–97. doi: 10.1038/nprot.2009.35. [DOI] [PubMed] [Google Scholar]
- 13.Krylova OO, Jahnke N, Keller S. Biophys. Chem. 2010;150:105–11. doi: 10.1016/j.bpc.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Ghai R, Falconer RJ, Collins BM. J. Mol. Recognit. 2012;25:32–52. doi: 10.1002/jmr.1167. [DOI] [PubMed] [Google Scholar]
- 15.Torres FE, Recht MI, Coyle JE, Bruce RH, Williams G. Curr. Opin. Struct. Biol. 2010;20:598–605. doi: 10.1016/j.sbi.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladbury JE. Biochem. Soc. Trans. 2010;38:888–93. doi: 10.1042/BST0380888. [DOI] [PubMed] [Google Scholar]
- 17.Brown A. Int. J. Mol. Sci. 2009;10:3457–77. doi: 10.3390/ijms10083457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houtman JC, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE. Protein Sci. 2007:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velazquez-Campoy A, Freire E. Nat. Protoc. 2006;1:186–91. doi: 10.1038/nprot.2006.28. [DOI] [PubMed] [Google Scholar]
- 20.Baker BM, Murphy KP. Biophys. J. 1996;71:2049–55. doi: 10.1016/S0006-3495(96)79403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coussens NP, Schuck P, Zhao H. J. Chem. Thermodyn. 2012 doi: 10.1016/j.jct.2012.02.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Rivera MR, Socorro F. J. Therm. Anal. Calorim. 2005;80:769–773. [Google Scholar]
- 23.Fanghänel J, Wawra S, Lücke C, Wildemann D, Fischer G. Anal. Chem. 2006;78:4517–23. doi: 10.1021/ac052040x. [DOI] [PubMed] [Google Scholar]
- 24.Burnouf DY, Ennifar E, Guedich S, Puffer B, Hoffmann G, Bec G, Disdier F, Baltzinger M, Dumas P. J. Am. Chem. Soc. 2011;134:559–65. doi: 10.1021/ja209057d. [DOI] [PubMed] [Google Scholar]
- 25.Wadsö I, Wadsö L. J. Therm. Anal. Calorim. 2005;82:553–558. [Google Scholar]
- 26.Krishnamoorthy J, Mohanty S. J. Mol. Recognit. 2011;24:1056–66. doi: 10.1002/jmr.1154. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz FP, Reinisch T, Hinz H-J, Surolia A. Pure Appl. Chem. 2008;80:2025–2040. [Google Scholar]
- 28.MicroCal . ITC Data Analysis in Origin ® Tutorial Guide. MicroCal, LLC; Northampton, MA: 2004. [Google Scholar]
- 29.TA Instruments - Waters, LLC . NanoAnalyze Software(TM) Getting Started Guide. New Castle, DE: 2008. [Google Scholar]
- 30.Roselin LS, Lin M-S, Lin P-H, Chang Y, Chen W-Y. Biotechnol. J. 2010;5:85–98. doi: 10.1002/biot.200900092. [DOI] [PubMed] [Google Scholar]
- 31.Velázquez-Campoy A, Ohtaka H, Nezami A, Muzammil S, Freire E. Curr. Protoc. Cell Biol. 2004:17.8.1–17.8.24. doi: 10.1002/0471143030.cb1708s23. [DOI] [PubMed] [Google Scholar]
- 32.Barkauskas D. a, Rocke DM. Anal. Chim. Acta. 2010;657:191–7. doi: 10.1016/j.aca.2009.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izquierdo-García JL, Villa P, Kyriazis A, del Puerto-Nevado L, Pérez-Rial S, Rodriguez I, Hernandez N, Ruiz-Cabello J. Prog. Nucl. Magn. Reson. Spectrosc. 2011;59:263–70. doi: 10.1016/j.pnmrs.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Devos O, Mouton N, Sliwa M, Ruckebusch C. Anal. Chim. Acta. 2011;705:64–71. doi: 10.1016/j.aca.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Schuck P. Anal. Biochem. 2010;401:280–287. doi: 10.1016/j.ab.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komsta L. Chromatographia. 2011;73:721–731. doi: 10.1007/s10337-011-1962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. Mc-Graw-Hill; New York: 1992. [Google Scholar]
- 38.Wiseman T, Williston S, Brandts J, Lin L. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 39.Turnbull WB, Daranas AH. J. Am. Chem. Soc. 2003;125:14859–66. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 40.Broecker J, Vargas C, Keller S. Anal. Biochem. 2011;418:307–9. doi: 10.1016/j.ab.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 41.Nyman PO, Lindskog S. Biochem Biophys Acta. 1964;85:462–474. doi: 10.1016/0926-6569(64)90310-4. [DOI] [PubMed] [Google Scholar]
- 42.Schuck P. https://sedfitsedphat.nibib.nih.gov/software/default.aspx.
- 43.Johnson ML. Anal. Biochem. 1992;225:215–225. doi: 10.1016/0003-2697(92)90356-c. [DOI] [PubMed] [Google Scholar]
- 44.Chrencik JE, Patny A, Leung IK, Korniski B, Emmons TL, Hall T, Weinberg RA, Gormley JA, Williams JM, Day JE, Hirsch JL, Kiefer JR, Leone JW, Fischer HD, Sommers CD, Huang H-C, Jacobsen EJ, Tenbrink RE, Tomasselli AG, Benson TE. J. Mol. Biol. 2010;400:413–33. doi: 10.1016/j.jmb.2010.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.