Abstract
A series of N-formyl-α-amino acid esters of β-lactone derivatives structurally related to tetrahydrolipstatin (THL) and O-3841 were synthesized that inhibit human and murine diacylglycerol lipase (DAGL) activities. New ether lipid reporter compounds were developed for an in vitro assay to efficiently screen inhibitors of 1,2-diacyl-sn-glycerol hydrolysis and related lipase activities using fluorescence resonance energy transfer (FRET). A standardized thin layer chromatography (TLC) radioassay of diacylglycerol lipase activity utilizing the labeled endogenous substrate [1″-14C]1-stearoyl-2-arachidonoyl-sn-glycerol with phosphorimaging detection was used to quantify inhibition by following formation of the initial product [1″-14C]2-arachidonoylglycerol and further hydrolysis under the assay conditions to [1-14C]arachidonic acid.
Keywords: diacylglycerol lipase, 2-arachidonoylglycerol, endocannabinoid, labeled, assay, fluorescence resonance energy transfer
The catalytic sites of diacylglycerol lipases (DAGLs) are unique among the lipases and hydrolases in effecting good selectivity for the hydrolysis of the 1-acyl group of 1,2-diacyl-sn-glycerol substrates.1 The DAGLα (120 KDa) and DAGLβ (70 KDa) isoforms1,2 are both present during brain development3 where their presynaptic axonal location suggests an important role in this period of neuritogenesis, rapid cell growth, and plasticity.4,5 However, the ultimate postsynaptic dendritic location of the α-isoform in the adult brain is consistent with the subsequent role of DAGLα in endocannabinoid paracrine retrograde signaling with a lesser role for the DAGLβ isoform.1,5–13 Thus, DAGLs have an important role in the endocannabinoid system as they are primarily responsible for releasing endocannabinoid 2-arachidonoylglycerol (2-AG) from diacylglycerols, including 1-stearoyl-2-arachidonoyl-sn-glycerol that is the principal 1,2-diacyl-sn-glycerol component of brain and nerves,14–16 for signaling at cannabinoid receptors.17–24 Small molecules that inhibit DAGL would have major effects on lipid metabolism. For example, less DAGL-catalyzed biosynthesis of endocannabinoid 2-arachidonoylglycerol (2-AG) may reduce activation of cannabinoid receptors. The resulting reduction of receptor signaling would be distinct from the actions of cannabinoid receptor inverse agonists and may be of pharmacological utility.
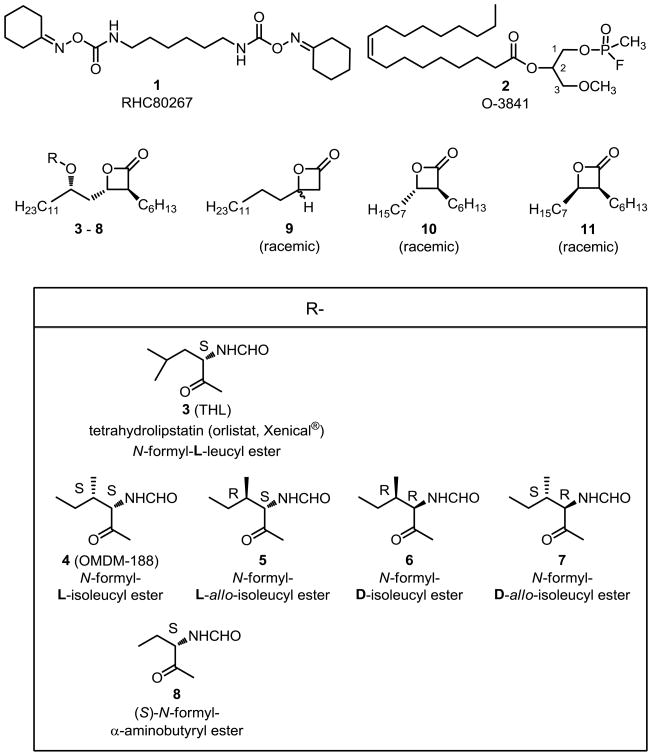
The three lead inhibitors of DAGL activity include bis-oximinocarbamate RHC80267 1,1,25 fluorophosphonate O-3841 2,26 and tetrahydrolipstatin (3, THL)1,27 (Figure 1) that inhibit hDAGLα with apparent IC50 values of 65000 nM, 160 nM, and 60 nM, respectively. The transition-state-mimicking fluorophosphonate group (RP=OOEtF) of FP-fluorescein (see Supplementary data for structure) utilized for the identification of serine hydrolases does not react appreciably with DAGL,27 unlike fluorophosphonate O-3841 2 (ROP=OMeF)26 and its t-butyl analog O-5596.28 Also, DAGL activity is not affected by phenylmethylsulfonylfluoride (PhCH2SO2F).1 There have been several previously reported analytical methodologies for measuring DAGL activities including radio-TLC,1,26,29,30 LC-MS,2,27 and the use of general esterase reporter molecules.2
Figure 1.
Diacylglycerol lipase inhibitors.
We now report our studies of new inhibitors of DAGL activity that generally resemble diglycerides. We will also discuss the DAGL proteins utilized in these studies. Our work on assay conditions using a radiolabeled endogenous diglyceride substrate will be detailed. Finally, the development of novel fluorescent resonance energy transfer (FRET) reporter substrates for the in vitro assay of DAGL activity will be discussed including their utilizations for the assays of related lipases.
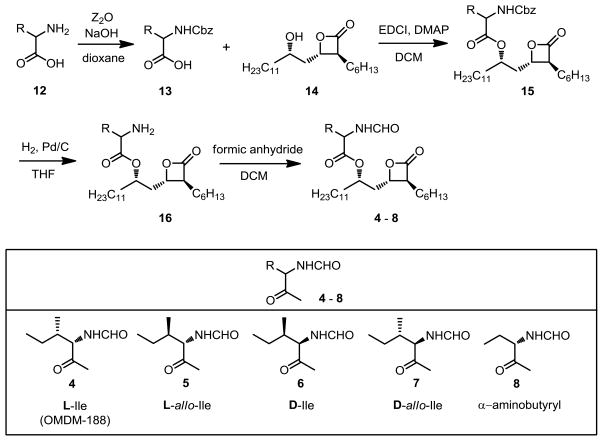
We first prepared new analogs of THL 3 and OMDM-188 4.31 THL 3 has an N-formyl-L-leucyl ester and OMDM-188 4 is the corresponding N-formyl-L-isoleucyl ester. The three other novel isoleucine diastereomers (5, 6, 7) as well as the (S)-α-aminobutyryl ester 8 were prepared via the reported method31 from the corresponding benzyloxycarbonyl protected α-amino acids 13 (see Scheme 1). The β-lactone analog 9 completely lacking both the N-formyl-α-aminoacyloxy and the 2-hexyl groups was prepared by treating racemic 3-hydroxypalmitic acid with N-phenyl-bis(trifluoromethanesulfonimide). The shorter chain separable trans-10 and cis-11 β-lactones were prepared according to the reported method.32 Also, a new series of racemic ether lipids derived from lead compounds RHC80267 1 and O-3841 2 was prepared (see Supplementary data).
Scheme 1.
Synthesis of analogs of THL 3 and OMDM-188 4.
We used diacylglycerol lipase (DAGL) protein from either cell lysates or membrane fractions that were prepared according to the previously reported method.1,33 The Cravatt group at Scripps provided hDAGLα, mDAGLα, and mDAGLβ from overexpression by transient infection of HEK293T cultures in addition to cell lysate of the empty vector HEK293T control. Some experiments used a second commercially prepared plasmid to provide additional human α-isoform overexpressed in HEK293T using the same methodology. All attempts to overexpress human DAGLα were disappointing. The lipase activity of hDAGLα expressed in the human cell line was still sufficient for assay of newly synthesized inhibitors. At least 100 μg of total protein from crude cell lysates or at least 10 μg of total protein from membrane preparations was required per well. The proteins were utilized such that the substrate hydrolysis would proceed to the extent of about 5% in 20 min. The more readily expressed mDAGLβ isoform (that has a 79% homology1 with the human isoform) or mDAGLα isoform (that has a 97% homology1 with the human isoform) were also used to confirm inhibition of DAGL activities.
It was of particular importance for biological relevance and inhibitor evaluation that assays of DAGL activities utilize pure endogenous substrate such as radiolabeled [1″-14C]1-stearoyl-2-arachidonoyl-sn-glycerol ([14C]SAG).29,34 Any radiolabeled 1-stearoyl-3-arachidonoyl-sn-glycerol present in the substrate, due to rearrangement of [14C]SAG to labeled 1,3-diglyceride, would be readily hydrolyzed by other enzymes in the relatively crude enzyme preparations and the result misinterpreted to be due to DAGL activity. The 1,2-diglyceride substrate [14C]SAG that we used contained less than 0.5% of the 1,3-diglyceride isomer (see Supplementary data).
Although the [14C]SAG substrate has previously been used for TLC-based assays of DAGL activity,1,26,30 details of these TLC assays were not fully described. Also, the apparent IC50 of THL 3 for hDAGLα has been reported by the Di Marzo group to range from 60 nM1,25,26 to 1000 nM,31 postulated to be due to the effect of DAGL protein concentration. Therefore, we developed a standardized assay that contained 10% DMSO.35,37 Higher DMSO concentrations did not increase rates of substrate conversion to fluorescent products in any of the in vitro assays. Non-denaturing detergents n-heptyl-βD-thioglucopyranoside and Triton X-100 at concentrations up to 0.2% were also used in some experiments (see Tables 1 and 3). Detergent use dramatically increases apparent DAGL specific activities and also decreases the apparent IC50 for DAGL inhibitory compounds. The DAGL enzyme suspensions had a 15 minute preincubation period of covalent quasi-irreversible inactivation by THL 3 or other potential inhibitors. The substrate [14C]SAG was then added (20 μM final concentration) and residual DAGL activity was quantified after 20 min by quenching the reaction with 2:1 chloroform/methanol and vortexing to denature the protein and move all lipids out of the aqueous phase. Very little rearrangement of [14C]SAG (1,2-diglyceride) to 1,3-diglyceride occurred under the reaction and workup conditions as assessed by TLC using boric acid treated silica gel plates.
Table 1.
Assays of the inhibition of hDAGLα (radio-TLC assay with [14C]SAG substrate),37 rFAAH,57 and hMAGL63 enzyme activities
| Compound Number | Detail of N-formyl- α-amino ester | hDAGLα inhibition at 10 μM detergent free or (with Triton X-100) (%) | rFAAH inhibition at 10 μM (%) | hMAGL inhibition at 10 μM (%) |
|---|---|---|---|---|
| 3 (THL) | L-leucyl | 100, (100A) | 6 | 47 |
| 4 (OMDM-188) | L-isoleucyl | (98A) | 7 | 16 |
| 5 | L-allo-isoleucyl | (95A) | 3 | 1 |
| 6 | D-isoleucylC | (78A) | 28 | 39 |
| 7 | D-allo-isoleucyl | (86A) | 0 | 42 |
| 8 | (S)-α-aminobutyryl | (99A) | 12 | 45 |
| 9 | none | (25A, 45B) | 10 | 4 |
| trans-10 | none | (37B) | 79 | 55 |
| cis-11 | none | (25B) | 94D | 100D |
| JZL184 | NA | (37B) | 97E | 100F |
| URB597 | NA | ND | 100E | 18 |
| n-C16H33SO2F | NA | ND | 100E | 82 |
| PMSF | NA | 19 | 100E | 5 |
| RHC80267 (1) | NA | 30 | 95E | 22 |
| SD41 | NA | <0* | 10 | 13 |
NA, not applicable; ND, not done
0.05% Triton X-100 present
0.015% Triton X-100 present
5% impurity in D-isoleucyl analog due to D-allo-isoleucyl analog
cis-11 inhibits rFAAH only 13% at 1 μM, but inhibits hMAGL 66% at 100 nM
| JZL184 | 974 nM (784–1210) |
| URB597 | 4.9 nM (4.1–6.0) |
| n-C16H33SO2F | 6.3 nM (4.5–8.7) |
| PMSF | 833 nM (746–931) |
| RHC80267 (1) | 2,240 nM (2010–2500) |
8 pt hMAGL IC50 (95% confidence)
| JZL184 | 57 nM (53–62) |
protein was pretreated with JZL184 to completely inhibit MAGL activity
Table 3.
Results of in vitro FRET-based screening using reporter compound 17 of the inhibition of lipase activities
| Compound Number | Detail of N-formyl- α-amino ester | hDAGLα inhibition at 10 μM detergent free or (0.05% Triton X-100) (%) | Lipoprotein Lipase (Bacterial) inhibition at 10 μM (%) | Pancreatic Lipase (Porcine) inhibition at 10 μM (%) | ||
|---|---|---|---|---|---|---|
| 3 (THL) | L-leucyl | 92, | 86* | (99) | 92 | 98 |
| 4 (OMDM-188) | L-isoleucyl | 92 | (99) | 84 | 99 | |
| 5 | L-allo-isoleucyl | 92 | (99) | 61 | 98 | |
| 6 | D-isoleucylA | 90 | (98) | 86 | 99 | |
| 7 | D-allo-isoleucyl | 90 | (98) | 68 | 95 | |
| 8 | (S)-α-aminobutyryl | 93 | (100) | 80 | 97 | |
| 9 | none | 14 | (72) | <0 | 38 | |
| trans-10 | none | 69* | 67 | 96 | ||
| cis-11 | none | 65* | 33 | 94 | ||
| JZL184 | NA | 59, | 44* | 36 | 36 | |
| URB597 | NA | 22, | 7* | 21 | 68 | |
| n-C16H33SO2F | NA | 40, | 94* | 8 | 29 | |
| PMSF | NA | 8* | <0 | 17 | ||
| RHC80267 (1) | NA | 70, | 40* | <0 | 78 | |
| SD41 | NA | 17, | 7* | <0 | <0 | |
| JZL195 | NA | 84, | 55* | <0 | 43 | |
NA, not applicable
5% impurity in D-isoleucyl analog due to D-allo-isoleucyl analog
protein was pretreated with JZL184 to completely inhibit MAGL activity
TLC DAGL assays with [14C]SAG substrate (or LC-MS assays with pure unlabeled SAG substrate) could result in significant errors if subsequent hydrolysis of 2-AG was not considered. It was critical that evaluations of enzymatic hydrolysis of [14C]SAG substrate include the sums of radiolabeled 2-AG and free radiolabeled arachidonic acid released. The crude cell preparations that were used had considerable monoacylglycerol lipase (MAGL), fatty acid amide hydrolase (FAAH), and other lipase activities which further degraded the radiolabeled 2-AG as it was formed under the assay conditions. It was very clear from the controls and from experiments with poor inhibitors that the release of labeled 2-AG was followed by further hydrolysis to labeled arachidonic acid. Thus DAGL activity was calculated via the sum of [14C]2-AG plus [14C]AA released divided by the sum of [14C]2-AG, [14C]AA, and final [14C]SAG concentrations for each lane. Also, DAGL activity in this human cell line (HEK293T) was not adjusted for hDAGLα and hDAGLβ activities demonstrated to be present in cell lysates following the mock infections.
We observed that the conversion of [14C]2-AG to [14C ]AA was reduced in modified radio-TLC assays by pre-treatment of cell lysate with the highly selective MAGL inhibitor JZL184 at 10 μM for 15 min. Then, tenfold dilution to 1 μM for screening assay use in some experiments as noted in the data tables gave little JZL184 interference with hDAGLα activities in radioassays and fluorescent assays.
The use of 10 μM THL 3 resulted in complete inhibition of (human) hDAGLα activity for all protein preparations (Table 1). Using TLC under basic conditions1,26 and phosphorimaging analysis,34,41 the radioassays consistently showed the apparent IC50 of THL 3 to be in the range of 10 to 100 nM. Analogs 4 – 8 were also all extremely potent inhibitors of hDAGLα in radio-TLC assays. The β-lactones 9 – 11 and other compounds including JZL184, PMSF, and RHC80267 1 were poor inhibitors of hDAGLα. The β-lactone SD41 and ether lipid analogs of O-3841 we synthesized (see Supplementary data) did not inhibit hDAGLα or mDAGLβ at 10 μM screening concentrations.
The β-lactones THL 3, OMDM-188 4, and new analogs 5 – 9, were assayed for the inhibition of (murine) mDAGLα at 10 nM to 10 μM concentrations (Table 2). The D-isoleucyl- and D-allo-isoleucyl-analogs (6 and 7) were clearly less potent than analogs of (S)-α-amino acids. Several analogs (THL 3, OMDM-188 4, and the new L-allo analog 5) showed good selectivity for diacylglycerol lipase over monoacylglycerol lipase and fatty acid amide hydrolase enzymes. Future studies can include shorter alkyl chain analogs similar to the fatty acid synthase inhibitors from the Romo group42 that might have better solubility and membrane penetration properties.
Table 2.
Radio-TLC assay with [14C]SAG substrate of the inhibition of mDAGLα activity
| Compound Number | Detail of N-formyl-α-amino ester | mDAGLα inhibition at 10 nM (%) | mDAGLagr; inhibition at 100 nM (%) | mDAGLagr; inhibition at 1000 nM (%) | mDAGLagr; inhibition at 10000 nM (%) |
|---|---|---|---|---|---|
| 3 (THL) | L-leucyl | 55 | 72 | 92 | 98 |
| 4 (OMDM-188) | L-isoleucyl | 69 | 80 | 96 | 96 |
| 5 | L-allo-isoleucyl | 60 | 61 | 88 | 100 |
| 6 | D-isoleucylA | 33 | 29 | 69 | 85 |
| 7 | D-allo-isoleucyl | 19 | 51 | 75 | 90 |
| 8 | (S)-α-aminobutyryl | 50 | 68 | 96 | 100 |
| 9 | NA | ND | ND | ND | <0 |
| JZL184 | NA | ND | ND | ND | 3 |
| URB597 | NA | ND | ND | ND | <0 |
| n-C16H33SO2F | NA | ND | ND | 18 | 64 |
| JZL195 | NA | ND | ND | ND | 10 |
NA, not applicable; ND, not done
5% impurity in D-isoleucyl analog due to D-allo-isoleucyl analog
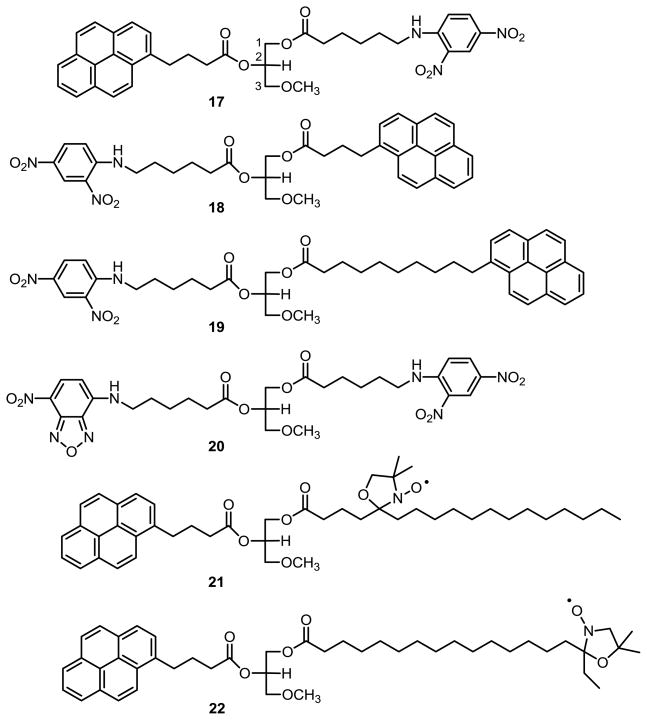
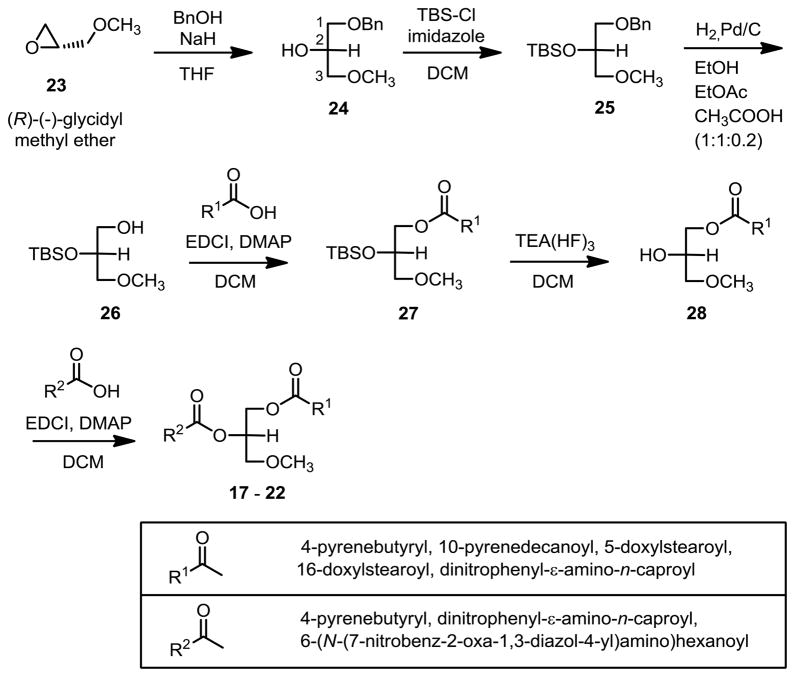
Ether lipid substrates 17 – 22 (Figure 2) for in vitro fluorescence resonance energy transfer (FRET) assay of DAGL and related lipase activities were developed from related lipase activity reporter molecules.43–47 This series of novel ether lipid molecules was synthesized (Scheme 2) with the biomimetic stereochemistry at sn-2 and incorporated terminally functionalized sn-1 and sn-2 fatty acyl groups. The epoxide of (R)-(-)-glycidyl methyl ether (23) was opened with benzyl alcohol and sodium hydride followed by silyl protection of the secondary alcohol 24, and hydrogenolysis of the benzyl ether 25 in ethanol/ethyl acetate/acetic acid (1:1:0.2). Subsequent acylation of the primary hydroxyl group of 26, deprotection of the secondary alcohol t-butyldimethylsilyl group, followed immediately by acylation of the secondary hydroxyl of 28 gave ether lipid products 17 – 22 with only 5% impurity from acyl rearrangement.
Figure 2.
Newly synthesized reporter substrates for in vitro FRET-based DAGL and related lipase assays.
Scheme 2.
Synthesis of new ether lipid reporter compounds 17 – 22 for in vitro assays of DAGL and related lipase activities.
The FRET pairs initially studied were the pyrene and nitrobenzoxadiazole (NBD) fluorophors with either the dinitrophenyl or nitroxyl group quenchers. Excitation of the pyrene or NBD results in radiationless energy transfer to the quenchers when close enough and sufficiently well oriented. The fully extended distances between the fluorophors and quenchers were estimated (Schrödinger Suite 2010, in an aqueous environment with a dielectric constant of 80) to be 18 Å for 17, 18, and the NBD analog 20; and, to be 24 Å for the pyrenedecanoyl analog 19. The assays with NBD analog 20 had too much baseline instability as this fluorescent group is quite sensitive to the polarity of its environment. The pyrene was estimated to be 15 Å and 24 Å from the nitroxyl stable free radical quenching groups48 of the 5-doxylstearoyl analog 21 and the 16-doxylstearoyl analog 22, respectively, and both compounds were poor substrates for enzymatic hydrolysis. The pyrene-dinitrophenyl FRET pairings 17 and 18 were stable to uncatalyzed hydrolysis at neutral pH, were the best enzyme substrates, and were readily utilized in a 96-well format. A convenient in vitro fluorometric esterase assay utilizing the BioTek Instruments Synergy HT Multi-Mode Microplate 96-well reader was developed that measured nanomolar concentrations of fluorescent reaction product.49 Cell lysate or membrane preparations containing overexpressed DAGL catalyzed the hydrolysis of the reporter substrate, and a fluorescence response increased at a nearly linear rate for over two hours. Wells that had a 15 minute pre-incubation with DAGL inhibitors and that showed a concentration dependent attenuation of fluorescence response were identified as “hits.” At a screening concentration of 10 μM, compounds 3–8 were identified as potent inhibitors of hDAGLα (Table 3). However, any potential DAGL inhibitor identified from the in vitro FRET assay should then be submitted to the TLC assay with the radiolabeled endogenous diglyceride substrate to identify any false positives for DAGL inhibition. Fluorescence results could reflect inhibition of other hydrolytic enzymes in the crude cell preparations that hydrolyze the ether lipid substrates 17 – 22 to a much greater extent than the endogenous 1,2-diacyl-sn-glycerol substrate. Using highly selective enzyme inhibitors JZL184, URB597, and others, the enzymes responsible for false positives include monoacylglycerol lipase (MGL) and fatty acid amide hydrolase (FAAH). The ether lipid FRET-substrate 17 will be most useful for assays with DAGLs purified to homogeneity.
Additional assays were performed to establish the selectivity of DAGL inhibitors. Compounds 3 – 11 were assayed for binding to the CB1 (rat brain preparation) and CB2 (mouse or human receptor expressed in HEK293),50 and none had a Ki below 1 μM in these competition binding experiments. Compounds 3 – 11 were also assayed for inhibition of endocannabinoid hydrolytic enzymes in fluorescence-based assays (Table 1). Assays of the inhibition of fatty acid amide hydrolase (rFAAH) used the reported coumarin amide reporter compound,56,57 and assays of the inhibition of monoacylglycerol lipase (hMGL) used the 7-hydroxy-6-methoxy analog62,63 of the reported coumarin ester.66 The assays were validated with standard compounds including the selective FAAH inhibitor URB59759 and the selective MAGL inhibitor JZL184.36 Preliminary studies with [1″-14C] SAG using the purified rFAAH and hMAGL endocannabinoid hydrolytic enzymes in these fluorescent assays showed low activities of these enzymes for the 1,2-diglyceride substrate. Also, preliminary studies with ether lipid substrate 17 in the in vitro FRET-based assay confirmed the potent inhibition by THL 367,68 of commercially available homogeneous bacterial lipoprotein lipase and porcine triacylglycerol lipase enzymes (Table 3).69 This new FRET-based methodology should be suitable for the assay of new inhibitors of human recombinant proteins including lipoprotein lipase, triacylglycerol lipase, and other related hydrolases to determine DAGL selectivity in vitro.
In summary, our structure-activity relationship (SAR) studies have demonstrated molecular features of inhibitors that result in inactivation of human and murine DAGLs at nanomolar inhibitor concentrations. The importance of a small (S)-N-formyl-α-amino group as a structural feature in targeting DAGLs was clearly demonstrated. The β-lactone was the most active of the covalently reactive quasi-irreversible inactivating functional groups that we tested for DAGL inhibition. In pilot experiments, we have looked at many factors that may affect assay results including protein concentration, substrate structure and concentration, length of the incubation period for enzyme inactivation, and the presence of co-solvent and detergent. An in vitro FRET-based screen was established for rapidly identifying inhibitors of DAGL activity. Although false positives can occur due to inhibition of other hydrolytic enzymes present in the cell lysate or membrane preparations used, the assay will be suitable for the preliminary screening of compound libraries. An improved and detailed radio-TLC assay of DAGL activity with the labeled endogenous substrate [1″-14C]1-stearoyl-2-arachidonoyl-sn-glycerol was utilized to unambiguously distinguish DAGL activity from the activities of MAGL and FAAH. Thus, methodologies were established to determine α/β-subtype selectivity as well as selective inhibition of DAGL over esterases, amidases, and other lipases.
Supplementary Material
Acknowledgments
This work was supported with NIH research grant R03-DA-24842 (RID, Jr.) and by the Office of Science (BER) of the U. S. Department of Energy with research grant DE-SC0005251 (SJG). The first author (MJ) was supported with NIH training grant T32-DA-07312 (AM). We are extremely grateful to Jacqueline Blankman, Ku-Lung (Ken) Hsu, and Benjamin Cravatt at The Scripps Research Institute for generously providing rFAAH, hDAGLα, mDAGLβ, and mDAGLα proteins. We wish to thank the office staffs of NU Environmental Health and Safety, the Department of Pharmaceutical Sciences, and the Center for Drug Discovery. We are also grateful to JodiAnne Wood, Aneetha Halikhedkar, Kirin Vemuri, Jianxin Guo, David Budil, Vidyanand Shukla, Sonyuan Lin, Frank Desarnaud, Christopher Kearns, Yan Peng, and Haotian Wang for their consultations and assistance here at NU.
Abbreviations
- AA
arachidonic acid
- DAGL
diacylglycerol lipase
- FAAH
fatty acid amide hydrolase
- FRET
fluorescence resonance energy transfer
- MAGL
monoacylglycerol lipase
- NBD
nitrobenzoxadiazole
- [14C]SAG
[1″-14C]1-stearoyl-2-arachidonoyl-sn-glycerol
- THL
tetrahydrolipstatin
Footnotes
Supplementary data (the structures and names of the standard chemicals used, structures and synthetic schemes for newly synthesized ether lipid analogs of O-3841, western blot of hDAGLα and mDAGLβ proteins, TLC of [14C]SAG substrate for radioassays, radio-TLC assay data for inhibitors 3 – 8 (at 100 nM and 10 nM) of mDAGLα, the comparison of the effect of DMSO concentration on hDAGLα activity, the comparison of hDAGLα activities on reporter compound substrates 17 – 22 at 10% DMSO, the inhibition of hDAGLα activity by THL 3 using reporter compound 17 with 10% DMSO conditions, and the characterizations including 1H NMR spectra of 3 – 11, 17, and 18) associated with this article can be found in the online version, at http://.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Meghan Johnston, Email: meghanryan.johnston@gmail.com.
Shachi R. Bhatt, Email: shachi.r.bhatt@gmail.com.
Surina Sikka, Email: surinasikka4u@gmail.com.
Richard W. Mercier, Email: r.mercier@neu.edu.
Jay M. West, Email: j.west@neu.edu.
Alexandros Makriyannis, Email: a.makriyannis@neu.edu.
S. John Gatley, Email: s.gatley@neu.edu.
References and notes
- 1.Bisogno T, Howell F, Williams G, Minassi A, Cascio MG, Ligresti A, Matias I, Schiano-Moriello A, Paul P, Williams EJ, Gangadharan U, Hobbs C, Di Marzo V, Doherty P. J Cell Biol. 2003;163:463. doi: 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedicord DL, Flynn MJ, Fanslau C, Miranda M, Hunihan L, Robertson BJ, Pearce BC, Yu XC, Westphal RS, Blat Y. Biochem Biophys Res Commun. 2011;411:809. doi: 10.1016/j.bbrc.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 3.Harkany T, Guzman M, Galve-Roperh I, Berghuis P, Devi LA, Mackie K. Trends Pharmacol Sci. 2007;28:83. doi: 10.1016/j.tips.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Fride E. J Neuroendocrinol. 2008;20(Suppl 1):75. doi: 10.1111/j.1365-2826.2008.01670.x. [DOI] [PubMed] [Google Scholar]
- 5.Walker DJ, Suetterlin P, Reisenberg M, Williams G, Doherty P. J Neurosci Res. 2010;88:735. doi: 10.1002/jnr.22251. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida T, Fukaya M, Uchigashima M, Miura E, Kamiya H, Kano M, Watanabe M. J Neurosci. 2006;26:4740. doi: 10.1523/JNEUROSCI.0054-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katona I, Urban GM, Wallace M, Ledent C, Jung KM, Piomelli D, Mackie K, Freund TF. J Neurosci. 2006;26:5628. doi: 10.1523/JNEUROSCI.0309-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung KM, Astarita G, Zhu C, Wallace M, Mackie K, Piomelli D. Mol Pharmacol. 2007;72:612. doi: 10.1124/mol.107.037796. [DOI] [PubMed] [Google Scholar]
- 9.Uchigashima M, Narushima M, Fukaya M, Katona I, Kano M, Watanabe M. J Neurosci. 2007;27:3663. doi: 10.1523/JNEUROSCI.0448-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludanyi A, Eross L, Czirjak S, Vajda J, Halasz P, Watanabe M, Palkovits M, Magloczky Z, Freund TF, Katona I. J Neurosci. 2008;28:2976. doi: 10.1523/JNEUROSCI.4465-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matyas F, Urban GM, Watanabe M, Mackie K, Zimmer A, Freund TF, Katona I. Neuropharmacology. 2008;54:95. doi: 10.1016/j.neuropharm.2007.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Vasilyev DV, Goncalves MB, Howell FV, Hobbs C, Reisenberg M, Shen R, Zhang MY, Strassle BW, Lu P, Mark L, Piesla MJ, Deng K, Kouranova EV, Ring RH, Whiteside GT, Bates B, Walsh FS, Williams G, Pangalos MN, Samad TA, Doherty P. J Neurosci. 2010;30:2017. doi: 10.1523/JNEUROSCI.5693-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanimura A, Yamazaki M, Hashimotodani Y, Uchigashima M, Kawata S, Abe M, Kita Y, Hashimoto K, Shimizu T, Watanabe M, Sakimura K, Kano M. Neuron. 2010;65:320. doi: 10.1016/j.neuron.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Keough KMW, MacDonald G, Thompson W. Biochim Biophys Acta. 1972;270:337. doi: 10.1016/0005-2760(72)90198-1. [DOI] [PubMed] [Google Scholar]
- 15.Eichberg J, Zhu X. Adv Exp Med Biol. 1992;318:413. doi: 10.1007/978-1-4615-3426-6_37. [DOI] [PubMed] [Google Scholar]
- 16.Nicolaou A, Kokotos G. In: Bioactive Lipids. Nicolaou A, Kokotos G, editors. The Oily Press; Bridgwater, UK: 2004. p. 294. [Google Scholar]
- 17.Sugiura T, Dondo S, Sukagawa A, Nakane S, Shinoda A, Kiyoko I, Yamashita A, Waku K. Biochem Biophys Res Comm. 1995;215:89. doi: 10.1006/bbrc.1995.2437. [DOI] [PubMed] [Google Scholar]
- 18.Sugiura T, Kobayashi Y, Oka S, Waku K. Prostaglandins, Leukotrienes Essent Fatty Acids. 2002;66:173. doi: 10.1054/plef.2001.0356. [DOI] [PubMed] [Google Scholar]
- 19.Piomelli D. Nat Rev Neurosci. 2003;4:873. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
- 20.Bisogno T, Ligresti A, Di Marzo V. Pharmacol Biochem Behav. 2005;81:224. doi: 10.1016/j.pbb.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 21.Sugiura T, Kishimoto S, Oka S, Gokoh M. Prog Lipid Res. 2006;45:405. doi: 10.1016/j.plipres.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Hashimotodani Y, Ohno-Shosaku T, Watanabe M, Kano M. J Physiol. 2007;584:373. doi: 10.1113/jphysiol.2007.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrasco S, Merida I. Trends Biochem Sci. 2007;32:27. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Hashimotodani Y, Ohno-Shosaku T, Maejima T, Fukami K, Kano M. Neuropharmacology. 2008;54:58. doi: 10.1016/j.neuropharm.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Szabo B, Urbanski MJ, Bisogno T, Di Marzo V, Mendiguren A, Baer WU, Freiman I. J Physiol. 2006;577:263. doi: 10.1113/jphysiol.2006.119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisogno T, Cascio MG, Saha B, Mahadevan A, Urbani P, Minassi A, Appendino G, Saturnino C, Martin B, Razdan R, Di Marzo V. Biochim Biophys Acta. 2006;1761:205. doi: 10.1016/j.bbalip.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Hoover HS, Blankman JL, Niessen S, Cravatt BF. Bioorg Med Chem Lett. 2008;18:5838. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bisogno T, Burston JJ, Rai R, Allara M, Saha B, Mahadevan A, Razdan RK, Wiley JL, Di Marzo V. ChemMedChem. 2009;4:946. doi: 10.1002/cmdc.200800442. [DOI] [PubMed] [Google Scholar]
- 29.Majerus PW, Prescott SM. Methods Enzymol. 1982;86:11. doi: 10.1016/0076-6879(82)86162-4. [DOI] [PubMed] [Google Scholar]
- 30.Quistad GB, Sparks SE, Casida JE. Toxicol Appl Pharmacol. 2001;173:48. doi: 10.1006/taap.2001.9175. [DOI] [PubMed] [Google Scholar]
- 31.Ortar G, Bisogno T, Ligresti A, Morera E, Nalli M, Di Marzo V. J Med Chem. 2008;51:6970. doi: 10.1021/jm800978m. [DOI] [PubMed] [Google Scholar]
- 32.Wedler C, Kleiner K, Kunath A, Schick H. Liebigs Ann Chem. 1996:881. [Google Scholar]
- 33.Ortar G, Cascio MG, Moriello AS, Camalli M, Morera E, Nalli M, Di Marzo V. Eur J Med Chem. 2008;43:62. doi: 10.1016/j.ejmech.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 34.Duclos RI, Jr, Gatley SJ, Bhatt SR, Johnston M. J Label Compd Radiopharm. 2009;52:324. doi: 10.1002/jlcr.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.General. THL 3, JZL184, URB597, and other inhibitors used (see Supplementary data for structures) were commercially available and used as freshly prepared DMSO solutions. All arachidonates were maintained under argon or in argon-degassed solvents. Glass-backed silica gel 60 TLC plates were used, and long delays between spotting and elution were avoided. All solvent ratios are by volume. All data are reported as the mean of triplicate experiments except n = 1 or 2 for radio-TLC assays and FRET screenings with DAGLs, lipoprotein lipase, and pancreatic lipase. Increased enzymatic activity (rather than inhibition) is indicated by <0% inhibition, and is likely due to detergent or other effects at higher compound (>10 μM) assay concentrations as has previously been observed in DAGL assays.36 All IC50 data discussed and reported from the literature are actually apparent IC50 as all inhibitors in this investigation undergo covalent reactions with the hydrolytic enzymes.
- 36.Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, Cravatt BF. Nat Chem Biol. 2009;5:37. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radio-TLC assay of DAGL inhibition. All cells, lysates (fresh or stored), and membrane preparations were probe sonicated for 5 periods of 3 s with ice bath cooling immediately prior to use. To the protein (100 μg) suspensions containing DAGL in 90 μL of buffer (50 mM Tris, pH 7.4, 10 mM CaCl2) in screwtop eppendorfs (with O-rings) was added 5 μL of pure DMSO (for the control, 0% inhibition) or the inhibitors in 5 μL of DMSO to be assayed at their appropriate concentrations. As a positive control, 5 μL of a stable solution of lipoprotein lipase (0.23 μg, Sigma, from Pseudomonas sp.,30 0.2% n-heptyl-β-D-thioglucopyranoside, 10 mM CaCl2, 100 mM NaCl, 50 mM Tris, pH 7.4) was always used. Each vial was hand mixed briefly then incubated for 15 min in a sand bath at 37 °C. Then, [14C]SAG substrate (304,000 dpm, specific activity 55 Ci/mol) in 5 μL of DMSO was added by microcap to all vials. An extra 5 μL sample of [14C]SAG in DMSO was always checked by scintillation counting to evaluate substrate concentration. After a brief hand mixing and 20 min incubation at 37 °C, the reaction was terminated by adding 200 μL of 2:1 CHCl3/MeOH and vortexing for 1 min. Centrifugation for 2 min at 10,000 x g gave a 150 μL bottom phase that was predominantly chloroform, a small protein interphase, and an upper phase that was predominantly water. The upper phase and protein interface contained negligible (twice background) radioactive material by scintillation counting. Using a 200 μL pipette tip, approximately 100 μL of each bottom phase was transferred to new eppendorf vials. Then, 5 μL samples of the bottom phases (approximately 10,000 dpm) were spotted for TLC, and 5 μL samples were also checked by scintillation counting. The silica gel 60 TLC plates were eluted with chloroform/methanol/aqueous ammonium hydroxide. Though literature reports range from 85:15:0.11,26,28 to 85:15:1,31 an optimized ratio of 86:14:0.75 elutes substrate [14C]SAG (Rf 0.88), [14C]2-AG (Rf 0.59), and [14C]arachidonic acid (Rf 0.11). In addition to the characteristic decompositions under the basic conditions of radiolabeled arachidonic acid and diglyceride substrate [14C]2-AG, there generally appeared to be more degradation of radiolabeled substrate if it did not contain carrier lipids from cell extraction. The air dried TLC plates were apposed to Perkin Elmer multisensitive screens for 12 h. Raw data as gross digital light units (DLU) were obtained from the Perkin Elmer Cyclone phosphorimaging system for quantitative analysis (OptiQuant software version 5.0).38,39 Percent inhibition was calculated following background subtraction. The protein specific activities were obtained from the controls. The specific activities of the hDAGLα were in the range of 0.003 to 0.01 nmol/mg-min for the cell lysates (specific activity was 0.06 in the presence of 0.05% Triton X-100) and up to 0.1 nmol/min-mg for membrane (10,000 × g fraction) preparations. DAGL activities of protein from empty plasmid transfections were 0.003 to 0.01 nmol/min-mg. Thus, the specific activities of hDAGLα were only 1.5 to 3 times the activities of mock infections with empty plasmid. The radio-TLC analyses of mDAGLβ inhibition (data not shown) used 20 μg of protein from cell lysate with a specific activity above 0.1 nmol/min-mg. The radio-TLC analyses of mDAGLα inhibition (Table 2) used 8.8 μg of protein from a membrane preparation with a specific activity above 0.3 nmol/min-mg. The specific activity of the lipoprotein lipase positive control under the assay conditions was >400 nmol/mg-min. Other standard compounds from prior literature that were used, JZL184,36 PMSF,1 and RHC80267,1,25,40 have been reported to be poor inhibitors of DAGL activity.
- 38.Johnston RF, Pickett SC, Barker DL. Electrophoresis. 1990;11:355. doi: 10.1002/elps.1150110503. [DOI] [PubMed] [Google Scholar]
- 39.Liberatore GT, Wong JYF, Krenus D, Jeffreys BJ, Porritt MJ, Howells DW. Biotechniques. 1999;26:432. doi: 10.2144/99263bm13. [DOI] [PubMed] [Google Scholar]
- 40.Moriyama T, Urade R, Kito M. J Biochem. 1999;125:1077. doi: 10.1093/oxfordjournals.jbchem.a022389. [DOI] [PubMed] [Google Scholar]
- 41.Duclos RI, Jr, Johnston M, Vadivel SK, Makriyannis A, Glaser ST, Gatley SJ. J Org Chem. 2011;76:2049. doi: 10.1021/jo102277q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson RD, Ma G, Oyola Y, Zancanella M, Knowles LM, Cieplak P, Romo D, Smith JW. J Med Chem. 2008;51:5285. doi: 10.1021/jm800321h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zandonella G, Haalck L, Spener F, Faber K, Paltauf F, Hermetter A. Eur J Biochem. 1995;231:50. [PubMed] [Google Scholar]
- 44.Duque M, Graupner M, Stutz H, Wicher I, Zechner R, Paltauf F, Hermetter A. J Lipid Res. 1996;37:868. [PubMed] [Google Scholar]
- 45.Stradner A, Mayer B, Sottmann T, Hermetter A, Glatter O. J Phys Chem B. 1999;103:6680. [Google Scholar]
- 46.Yang Y, Babiak P, Reymond JL. Org Biomol Chem. 2006;4:1746. doi: 10.1039/b601151a. [DOI] [PubMed] [Google Scholar]
- 47.Rosseto R, Tcacenco CM, Ranganathan R, Hajdu J. Tetrahedron Lett. 2008;49:3500. doi: 10.1016/j.tetlet.2008.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu P, Clamme JP, Deniz AA. Biophys J: Biophys Letters. 2005;89:L37. doi: 10.1529/biophysj.105.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.In vitro FRET-based DAGL assays. Both configurations of the FRET pairing of pyrene donor and dinitrophenyl acceptor 17 and 18 were satisfactory, though the 2-pyrenyl analog 17 was used for all in vitro FRET assays. The fluorescent assays were run in the same Tris buffer with calcium used for the radiochemical assays except that 200 μL final volumes were needed for efficient reading. The lipoprotein lipase standard (0.23 μg) was again used for a positive control. The freshly sonicated HEK cell lysate (100 μg total) protein containing DAGL was used as a suspension for each assay. A 15 min period of gentle shaking at ambient temperature was used following the addition of 10 μL of pure DMSO (for the control) or the 10 μL DMSO solutions of inhibitors. The ether lipid substrate (25 μM final concentration) was then added in DMSO (10 μL) to all wells, and after 2 min of shaking at 37 °C, an initial reading was taken with excitation at 320 nm and emission observed at 400 nm. Every 14 min, another 1 min of shaking would precede the fluorescence readings. The readings were followed over 2 h, but the timepoint of 1.5 h was used to calculate percent inhibitions. The inhibition of hydrolysis of the ether lipid reporter compound 17 by the DAGL-containing protein preparations with the compounds under investigation screened at 10 μM was then compared with 10 μM THL control, which gives complete inhibition of all human and murine DAGLs tested with the radiolabeled endogenous substrate [14C]SAG. Using the pyrene-dinitrophenyl reporter compound 17, the apparent IC50 of THL was always approximately 10 nM with hDAGLα using this in vitro FRET-based assay.
- 50.Cannabinoid receptor binding. Assays were performed by the previously reported methods.51,52 Palmitylsulfonyl fluoride (n-C16H33SO2F) had an apparent IC50 of 440 nM, which correlates well with the literature report53 for rCB1 (520 nM). All other standards (including RHC80267 and SD41) did not bind to the cannabinoid receptors, analogous to the previous reports for THL 3,25,31 JZL184,36 URB597,54,55 and PMSF.53
- 51.Khanolkar AD, Lu D, Ibrahim M, Duclos RI, Jr, Thakur GA, Malan TP, Jr, Porreca F, Veerappan V, Tian X, George C, Parrish DA, Papahatjis DP, Makriyannis A. J Med Chem. 2007;50:6493. doi: 10.1021/jm070441u. [DOI] [PubMed] [Google Scholar]
- 52.Lu D, Guo J, Duclos RI, Jr, Bowman AL, Makriyannis A. J Med Chem. 2008;51:6393. doi: 10.1021/jm8005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deutsch DG, Lin S, Hill WA, Morse KL, Salehani D, Arreaza G, Omeir RL, Makriyannis A. Biochem Biophys Res Commun. 1997;231:217. doi: 10.1006/bbrc.1997.6072. [DOI] [PubMed] [Google Scholar]
- 54.Kathuria S, Gaetani S, Fegley D, Valino F, Duranti A, Tontini A, Mor M, Tarzia G, La Rana G, Calignano A, Giustino A, Tattoli M, Palmery M, Cuomo V, Piomelli D. Nat Med. 2003;9:76. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 55.Piomelli D, Tarzia G, Duranti A, Tontini A, Mor M, Compton TR, Dasse O, Monaghan EP, Parrott JA, Putman D. CNS Drug Rev. 2006;12:21. doi: 10.1111/j.1527-3458.2006.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramarao MK, Murphy EA, Shen MWH, Wang Y, Bushell KN, Huang N, Pan N, Williams C, Clark JD. Anal Biochem. 2005;343:143. doi: 10.1016/j.ab.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 57.Inhibition of rat FAAH. The N-terminal his-tagged rFAAH deletion sequence used was expressed in an E. coli cell line provided by the Cravatt group.58 The rFAAH coumarin ester substrate fluorescence assay demonstrated URB597 to have an apparent IC50 of 4.9 nM. This is comparable to inhibition of rat membrane preparations used for the hydrolysis of tritiated anandamide.54,59,60 Palmitylsulfonyl fluoride (n-C16H33SO2F) had an apparent IC50 of 6.3 nM in the fluorescent rFAAH assay (approximately 2 μM in the hMAGL assay detailed below) which correlates well with the IC50 of 7 nM using the radiolabeled N-arachidonoylethanolamine (anandamide) substrate.53 All other standards THL 3,25,27,31 JZL184,36 PMSF,53,61 and RHC8026727 have been reported to be poor inhibitors of FAAH activity.
- 58.Patricelli MP, Lashuel HA, Giang DK, Kelly JW, Cravatt BF. Biochemistry. 1998;37:15177. doi: 10.1021/bi981733n. [DOI] [PubMed] [Google Scholar]
- 59.Mor M, Rivara S, Lodola A, Plazzi PV, Tarzia G, Duranti A, Tontini A, Piersanti G, Kathuria S, Piomelli D. J Med Chem. 2004;47:4998. doi: 10.1021/jm031140x. [DOI] [PubMed] [Google Scholar]
- 60.Paylor B, Holt S, Fowler CJ. Pharmacolog Res. 2006;54:481. doi: 10.1016/j.phrs.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 61.Deutsch DG, Omeir R, Arreaza G, Salehani D, Prestwich GD, Huang Z, Howlett A. Biochem Pharmacol. 1997;53:255. doi: 10.1016/s0006-2952(96)00830-1. [DOI] [PubMed] [Google Scholar]
- 62.Zvonok N, Williams J, Johnston M, Pandarinathan L, Janero DR, Li J, Krishnan SC, Makriyannis A. J Proteome Res. 2008;7:2158. doi: 10.1021/pr700839z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inhibition of human MAGL. The N-terminal his-tagged full length human monoacylglycerol lipase used was expressed in E. coli.62 The coumarin substrate fluorescence assay demonstrated JZL184 to have an apparent IC50 of 57 nM that is comparable to human recombinant MAGL expressed in COS7 cells where the IC50 of JZL184 was reported to be 2 to 6 nM with the endogenous substrate 2-AG.36,64 All other standards, THL 3,25–27,31 JZL184,36 URB597,54,65 palmitylsulfonyl fluoride (n-C16H33SO2F),65 PMSF,65 and RHC80267,27 were reported to be poor inhibitors of MAGL activity.
- 64.Long JZ, Nomura DK, Cravatt BF. Chem Biol. 2009;16:744. doi: 10.1016/j.chembiol.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saario SM, Savinainen JR, Laitinen JT, Järvinen T, Niemi R. Biochem Pharmacol. 2004;67:1381. doi: 10.1016/j.bcp.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Chanda P, Jones PG, Kennedy JD. Assay Drug Dev Technol. 2008;6:387. doi: 10.1089/adt.2007.122. [DOI] [PubMed] [Google Scholar]
- 67.Lookene A, Skottova N, Olivecrona G. Eur J Biochem. 1994;222:395. doi: 10.1111/j.1432-1033.1994.tb18878.x. [DOI] [PubMed] [Google Scholar]
- 68.Hadvary P, Sidler W, Meister W, Vetter W, Wolfer H. J Biol Chem. 1991;266:2021. [PubMed] [Google Scholar]
- 69.Other novel in vitro FRET-based assays. The bacterial lipoprotein lipase assays (0.46 μg of high specific activity lipase as described above) and the porcine pancreatic lipase type II assays (1.0 μg suspended in 1:9 DMSO/water, Sigma L3126, labeled 100–400 units/mg with olive oil substrate) also used pyrene-dinitrophenol FRET reporter compound 17 (25 μM in 200 μL volumes), which was a better substrate than the isomeric 18. The apparent IC50 of THL was approximately 100 nM with lipoprotein lipase, but less than 1 nM with the porcine pancreatic lipase using this FRET assay.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.