Abstract
Dietary selenium restriction in mammals causes bodily selenium to be preferentially retained in the brain relative to other organs. Almost all of the known selenoproteins are found in brain, where expression is facilitated by selenocysteine-laden selenoprotein P. The brain also expresses selenocysteine lyase, an enzyme that putatively salvages selenocysteine and recycles the selenium for selenoprotein translation. We compared mice with a genetic deletion of selenocysteine lyase to selenoprotein P knockout mice for similarity of neurological impairments, and whether dietary selenium modulates these parameters. We report that selenocysteine lyase knockout mice do not display neurological dysfunction comparable to selenoprotein P knockout mice. Feeding a low-selenium diet to selenocysteine lyase knockout mice revealed a mild spatial learning deficit without disrupting motor coordination. Additionally, we report that the neurological phenotype caused by the absence of selenoprotein P is exacerbated in male versus female mice. These findings indicate that selenocysteine recycling via selenocysteine lyase becomes limiting under selenium deficiency, and suggest the presence of a complementary mechanism for processing selenocysteine. Our studies illuminate the interaction between selenoprotein P and selenocysteine lyase in the distribution and turnover of body and brain selenium, and emphasize the consideration of sex differences when studying selenium and selenoproteins in vertebrate biology.
Keywords: selenoprotein P, selenocysteine lyase, motor control, spatial learning, sex difference, selenium, selenoprotein, selenocysteine
INTRODUCTION
Selenium (Se) is an essential dietary micronutrient with antioxidant properties, and the human health consequences of Se-deficiency are under extensive study (Rayman, 2000). Sex-specific differences are observed in Se status and metabolism, which has complicated this research (Combs et al., 2011, Galan et al., 2005). The functions of Se in biochemical reactions and cellular processes of organisms are principally mediated by selenoproteins that incorporate Se into the amino acid selenocysteine (Sec). Biosynthesis of Sec occurs on its UGA-recognizing tRNA, and is catalyzed by Sec synthetase (SepSecS) in the presence of selenophosphate [reviewed in (Bellinger et al., 2009)]. Recoding the UGA stop codon for Sec incorporation requires a Sec-specific elongation factor (EFSec), an mRNA stem-loop termed a Sec insertion sequence (SECIS), and a SECIS-binding protein (SBP2). Among 25 human selenoproteins, the glutathione peroxidase, thioredoxin reductase, and iodothyronine deiodinase families of enzymatic selenoproteins are relatively well characterized, and crucial for the health of mammals.
Selenoprotein P (Sepp1) uniquely contains up to 10 Sec residues in primates and rodents. During Se-deficiency, brain Se is maintained compared to other organs by tissue-specific uptake of Sepp1 by ApoER2 and Megalin (Burk et al., 2007, Chiu-Ugalde et al., 2010). Mutant mice lacking full-length Sepp1 or the Sec-rich C-terminus show a greater depletion of brain Se than can be achieved through dietary Se deprivation (Hill et al., 2007). Sepp1 gene disruption in mice additionally causes cognitive, motor and sensory symptoms that can be exacerbated by dietary Se restriction and diminished by Se supplementation. These mice present spatial learning deficits, spasticity, and hyperreflexia that coincide with deficient synaptic plasticity and widespread neurodegeneration (Caito et al., 2011, Peters et al., 2006, Valentine et al., 2008, Valentine et al., 2005).
If endocytosis of Sepp1 delivers Se to cells, the Sec residues from Sepp1 must be processed for incorporation into selenoproteins. Sec lyase (Scly) catalyzes the decomposition of Sec into alanine and hydrogen selenide (Esaki et al., 1982), and promotes the production of selenophosphate in the presence of Sec and selenophosphate synthetase (SPS) (Tobe et al., 2009). Scly mRNA and protein are expressed in mouse brain (Mihara et al., 2000), where it is posited to recycle Sec from Sepp1 for selenoprotein synthesis (Schweizer et al., 2005).
We hypothesized that Scly liberates Se from Sepp1 in brain, and that deletion of Scly in mice would cause similar neurological deficits as observed in Sepp1−/− mice. To test this hypothesis, we assessed whether a novel transgenic mouse strain lacking functional Scly develops a phenotype similar to Sepp1-deficient mice. Here we report that, in contrast to Sepp1−/− mice, Scly−/− mice display few neurological abnormalities. However, spatial learning and selenoprotein expression are sensitive to Scly disruption when the mice are challenged with a low-Se diet. In addition, we extensively characterized sex differences in the behavioral phenotype of Sepp1−/− mice, and report that male mice are more dependent on Sepp1 and Se than females for normal brain function.
MATERIALS AND METHODS
Animals
Genetically modified male and female mice on a C57BL/6 background lacking Sepp1 or Scly were bred on commercially available diets containing adequate Se (~0.25 ppm). Animals were given food and water ad libitum on a 12-hour light-cycle and group housed until behavioral experimentation. All behavioral experiments were conducted on single-housed adult mice aged 4–6 months during the light cycle. Male and female mice of all genotypes were used in approximately equal numbers to examine sex differences present in the animals. All animal procedures and experimental protocols were approved by the University of Hawaii Institutional Animal Care and Use Committee.
Generation of Sepp1−/− and Scly−/− mice
Sepp1−/− mice were generated by electroporating a construct into 129S9/SvEvH-derived embryonic stem (ES) cells that were subsequently injected into C57BL/6 blastocysts. The resulting chimeric males were bred with C57BL/6J females (Hill et al., 2003). Mutant mice were backcrossed to C57BL/6J for at least 10 generations before arriving in our lab, and were bred with our C57BL/6J colony to ensure congenic strains (Hoffmann et al., 2007). Sepp1+/− mice were bred to generate littermate pups of Sepp1−/−knockout and Sepp1+/+ control mice. Genotyping of the mice was carried out using methods previously described (Hoffmann et al., 2007).
A targeting vector was generated by the NCRR-NIH supported KnockOut Mouse Project (KOMP) Repository, and included an ~11-kb region of the wild type Scly locus subcloned from a positively identified C57BL/6 BAC clone. The vector was designed with one homology arm extending 5.5 kb 5′ to exon 4, and the other 5.5 kb homology arm terminating 3′ to exon 7. A promoterless trapping cassette (L1L2_gt0) with flanking Flp-recombinase target (FRT) sites was inserted in an intron 5′ of exon 4. Efficient splicing to the reporter cassette results in truncation of the endogenous transcript, causing a constitutive null mutation in the Scly gene. Cre-recombinase target loxP sites were inserted 5′ and 3′ of critical coding exon 4. The total size of the targeting construct, including vector backbone (L3L4_pZero_DTA_kan) and Neo cassette, was 21.642 kb. The targeting vector was transfected into C57BL/6 embryonic stem cells by electroporation. After selection with antibiotic, surviving clones were expanded and analyzed by PCR to identify recombinant ES cell clones. ES cell clones were microinjected into C57BL/6 blastocysts to produce chimeras with one wild-type and one mutant Scly allele, which were then mated to generate Scly−/− mice on a pure C57BL/6 background. Mutant mice were backcrossed to C57BL/6J to ensure genetic comparability with wild-type control C57BL/6J mice. The latter were maintained not more than five generations after arrival from The Jackson Laboratory. Deletion of Scly was confirmed in all offspring using PCR that amplified a 1.2-kb product in the targeted region present in the wild-type allele (forward, 5′-CAC AGG TGC GGC CAT GAG GG-3′; reverse, 5′-CTG GCT GTC CCT GAA CTA GCT TCA TA-3′) and a 233-bp product in the mutant allele (forward, 5′-GAG ATG GCG CAA CGA AAT TAA T-3′; reverse, 5′-CTG GCT GTC CCT GAA CTA GCT TCA TA -3′).
Diets
For dietary experiments, animals were switched from standard laboratory diets containing ~0.25 mg/kg Se to defined diets at the time of weaning (3–4 wk of age). Mice were fed Open Source Diets purchased from Research Diets containing either 0.08 mg/kg (cat.#D19101) or 1.0 mg/kg (cat.#D05050403) Se. The diets were formulated with purified ingredients and contained 20.3% protein, 66% carbohydrate, and 5% fat. The protein source was casein, which was also the source of Se in the low Se diet. For high Se diet, sodium selenite was added to achieve final Se levels. Multiple lots were independently tested to confirm the Se concentration by inductively coupled plasma-MS (Bodycote) with lot-to-lot variation at or below the detection limit of inductively coupled plasma-MS testing (0.02 mg/kg). Mice were fed the defined diets from weaning until the time of sacrifice. Se-adequate standard lab diets containing ~0.25 mg/kg Se are not completely defined, and may be considered slightly supplemented compared to the rodent RDA of 0.15 mg/kg Se. The low-Se diet containing 0.08 mg/kg Se is marginally deficient, and causes moderate selenium deficiency in mice (Hoffmann et al., 2010). The high-Se diet containing 1.0 mg/kg Se has been shown to prevent many of the neurological symptoms in Sepp1−/− mice (Hill et al., 2003). This range of dietary Se concentration reflects the global spectrum of human Se intake, from deficiency to therapeutic supplementation.
Animal Behavior
Spontaneous activity
Animals were placed in a transparent cylinder (20 cm diameter, 20 cm height) and activity was videotaped for three minutes. The cylinder was situated on clear plexiglass with a mirror placed at an angle underneath for clear view of movement along the ground as well as along the walls of the cylinder. The number of rears, forelimb and hindlimb steps, and time spent grooming were measured. Videotapes were scored in slow motion by an experimenter blind to the mouse genotype. A rear was counted when an animal made a vertical movement with both forelimbs removed from the ground. Forelimb and hindlimb steps were counted when an animal moved both forelimbs or both hindlimbs across the floor of the cylinder. Number of steps, rears, and time spent grooming were compared for wild-type and knockout mice (Fleming et al., 2004).
Open field
Animals were placed in a square box (50 cm sides, 40 cm walls) and monitored by overhead camera linked to computer-assisted tracking software. During the test, the mice were allowed to move freely around the open field and to explore the environment for five minutes. The path of each mouse was automatically recorded, and recordings were then analyzed. Total distance traveled, number of rears, time spent grooming, and center time was compared between groups.
Vertical pole
The vertical pole descent test has been used to assess coordination and basal ganglia related movement disorders in transgenic mice (Fleming et al., 2004). Animals were placed head-up on top of a vertical wooden pole 50 cm long (1.2 cm in diameter). The base of the pole was fixed in plexiglass and put in the home cage. When placed on the pole, animals orient themselves downward and descend the length of the pole back into their home cage. Knockout and wild-type mice received two days of training consisting of five trials per day. On the third day, animals received five trials, and time to orient downward (turn) and total time to descend (total) were measured with a stopwatch. The best performance over the five trials was used for both wild-type and knockout mice.
Inverted grid
The ability to hang upside down is a test of neuromuscular strength (Crawley, 1999). Each mouse was placed on a wire grid (mesh, 12 cm2 with 0.5 cm2 squares) 20 cm above a table top for 120 sec and videotaped. The lid was gently turned upside down, 60 cm above a soft surface to avoid injuries. The latency to fall was timed. Each mouse was given up to two attempts to hold on to the inverted grid for a maximum of 120 seconds and the longest period was recorded.
Morris water maze
A circular pool (2 m in diameter, 1 m deep) surrounded by constant external cues was located in an observation room and filled with 24°C water. White non-toxic paint was added to make the water translucent. Tests were performed under dim light conditions. Lights below the height of the tank, necessary for video capture, also provided light for the swimming mouse. A small circular escape platform (7 cm diameter, located just below the water surface, or protruding just above the water) was placed in a constant location in the center of quadrant 1. Four equally spaced points around the wall of the pool were used as starting points. The mice were given one block of four trials each day with an inter-trial interval of 5 to 10 min. Each trial started from one of four different points, in a semi-random order. The mouse was allowed to swim around until it located the platform or 60 sec, at which point the mouse was placed on the platform by the experimenter and allowed to stay on the platform for 15 sec. The time to locate the platform was recorded as escape latency during training days. After sufficient training a one-minute probe trial was performed, in which the platform was removed and the path of each mouse on each trial was automatically recorded and then analyzed. Time investigating the target, opposite, and adjacent quadrants, platform latency and platform crossings were measured. Average swim speed was calculated from total distance traveled per 60 sec trial. Animals that spent significant time floating, which occurred sporadically in very few animals independent of genotype, were excluded from this analysis.
Quantitative RT-PCR
Animals were sacrificed by CO2 asphyxiation, or deeply anesthetized with tribromoethanol and decapitated, and the brains rapidly excised, washed in PBS and snap-frozen in liquid nitrogen. Tissue was ground into powder, using a mortar and pestle on dry ice, and collected into pre-chilled tubes. Total RNA from tissue was prepared by Trizol extraction (Invitrogen, Carlsbad, CA, USA) followed by purification using the RNeasy kit (QIAgen, Valencia, CA, USA). Concentration and purity of extracted RNA and synthesized cDNA was determined using A260/A280 ratio measured on an ND1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Synthesis of cDNA was carried out using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), with 1 μg RNA per 20 μl reaction. For real-time PCR, 100 ng of the cDNA was used in 5 μl reactions with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). Reactions were carried out in triplicate or quadruplicate in a LightCycler 480 II thermal cycler (Roche, Indianapolis, IN, USA). Cycling conditions followed the manufacturers suggestions in the SYBR Green kit instructions. All qPCR results were normalized to 18S rRNA expression as a housekeeping gene and analyzed using Absolute Quantification Software (Roche).
SDS-PAGE and Western blot
Total protein was extracted from powdered mouse tissues by light sonication in CelLytic MT buffer (Sigma, St. Louis, MO, USA), followed by centrifugation according to the manufacturers’ protocol. Protein was added to reduced Laemmli buffer, boiled for 10 minutes, and loaded into 4–20% gradient polyacrylamide gels (Bio-Rad, Hercules, CA, USA). Following electrophoresis, gel contents were transferred to PVDF membranes, which were blocked with undiluted Odyssey Blocking Buffer (Li-Cor Biosciences, Lincoln, NE, USA) for one hour. Membranes were then probed for 90 minutes with the following primary antibodies: Goat-anti-GPX1 (R&D Systems, Minneapolis, MN, USA), Rabbit-anti-GPX4 (AbFrontier, Seoul, Korea), Rabbit-anti-SEPW1 (Rockland, Gilbertsville, PA, USA), and Mouse-anti-alpha Tubulin (Novus, Littleton, CO, USA). After washing with PBS containing 0.05% tween-20 (PBST), membranes were incubated in the dark in secondary antibodies labeled with infrared fluorophores (Li-Cor Biosciences). After further washes in PBST, blots were imaged and quantified with the Odyssey infrared imaging system (Li-Cor Biosciences).
Glutathione peroxidase activity assay
Total glutathione peroxidase activity was measured using the Bioxytech GPx-340 Assay kit (Oxis International, Foster City, CA, USA). Mouse tissues for the assay were collected in the same manner as above. Powdered tissue was homogenized in lysis buffer by sonication and centrifuged at 15,000 × g. The resulting supernatant was serially diluted to determine the linear range of the assay. 5 μl of diluted sample (1:10 for brain, 1:200 for liver) was added to 25 μl of assay buffer and 25 μl of NADPH reagent in a 96-well plate. 25μl of tert-butyl hydroperoxide was added to initiate the reaction, which was monitored for 10 minutes at room temperature by measuring kinetic absorbance at 340 nm on a SpectraMax M3 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Statistical analysis
Data were analyzed using Microsoft Excel (Redmond, WA, USA), and plotted using GraphPad Prism software (San Diego, CA, USA). Two-way ANOVA was used to determine an interaction between genotype and sex for all experiments. If no interaction effect was observed, male and female groups were sometimes combined. Repeated measures ANOVA was used to assess training in the water maze, and genotype and quadrant were the two factors for analyzing quadrant investigation on the probe trial. Post hoc test using Bonferroni correction for multiple comparisons was used to determine significance between individual groups. When male and female groups were combined, unpaired t-tests comparing genotypes were performed for end-point measures in the water maze and for biochemical experiments. The significance criteria were set at p < 0.05 for all statistical measures.
RESULTS
Neurological motor phenotype in Sepp1−/− mice is more pronounced in males
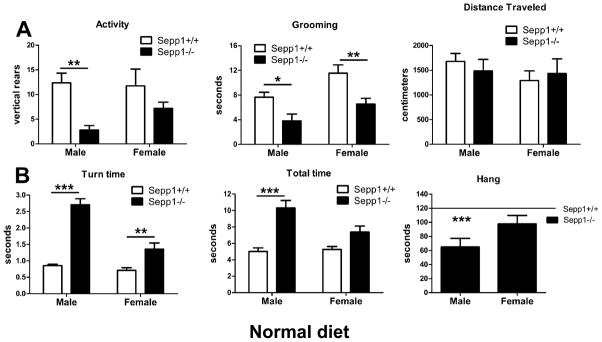
Genetic deletion of Sepp1 in mice results in neuromotor impairments (Hill et al., 2004, Schomburg et al., 2003). We characterized the phenotype of Sepp1-deficient mice raised on a standard lab diet using a battery of behavioral tests to assess motor function in male and female mice. We found that motor coordination was worse in male knockout animals compared to females. Spontaneous locomotor activity, as assessed using the cylindrical chamber, was greatly decreased in the male Sepp1-deficient mice while being slightly decreased in the female knockout animals, when compared to wild-type mice [Fig. 1A]. Two-way ANOVA revealed an effect of genotype on spontaneous rearing (F(1,32)=13.06, p=0.0010) [Fig. 1A, left] and grooming (F(1,32)=16.93, p=0.0003) [Fig. 1A, center]. Post hoc analysis indicated a statistically significant decrease in rearing (t(16)=3.464, p<0.01) and grooming (t(16)=2.532, p<0.05) in male Sepp1−/− mice, whereas only grooming (t(16)=3.287, p<0.01) was decreased in female Sepp1−/− mice, when compared to wild-type. Despite these differences, we did not observe any genotype or sex interaction effects on total distance traveled in the open field [Fig. 1A, right]. To assess motor coordination and strength, animals were subjected to the pole descent and inverted hang tests. Two-way ANOVA revealed an interaction between genotype and sex on time to turn (F(1,42)=16.57, p=0.0002) [Fig. 1B, left] and descend (F(1,42)=5.222, p=0.0274) [Fig. 1B, center] a vertically oriented pole. Genotype strongly affected ability to hang upside down for 120 seconds (F(1,32)=16.10, p=0.0003), however only male Sepp1−/− mice were significantly different from wild-type control mice (t(16)=4.045, p<0.001) [Fig. 1B, right].
Figure 1.
Spontaneous activity and motor coordination is more reduced in male than female Sepp1−/− mice compared to control mice fed a standard diet. (A) Rearing and grooming were measured in the cylinder, and total distance traveled in the open field. Sepp1−/− mice were less active as measured by rearing (left, genotype **p<0.01) and grooming (center, genotype ***p<0.001), but did not show decreased exploration of the open field (right). (B) Motor coordination was measured using the pole test and inverted hang test. Sepp1−/− mice took longer to turn (left, genotype x sex ***p<0.001) and descend the pole (center, genotype x sex *p<0.05), and had reduced ability to suspend themselves for two minutes (right, genotype ***p<0.001). Values are expressed as means ± SEM. n=8–12 per group, *p<0.05, **p<0.01, ***p<0.001, compared with control mice.
Dietary Se supplementation alleviates motor deficits in Sepp1−/− mice
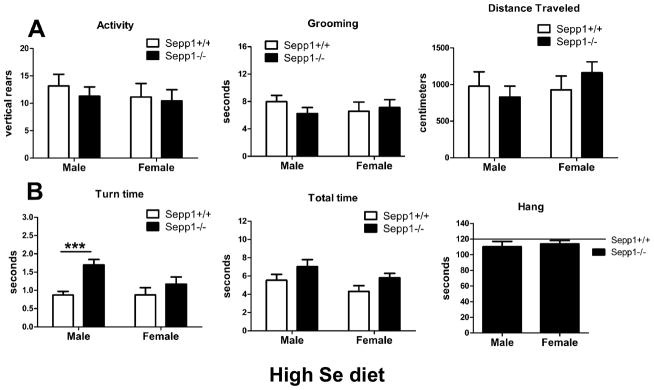
Previous studies have shown that Se supplementation can attenuate neuromotor impairments in Sepp1−/− mice (Schweizer et al., 2004). We determined if gender could influence this attenuation. Animals were supplemented with 1 mg/kg Se in the diet starting at weaning, and assayed in the same manner as those fed a standard diet. We found that Se-supplemented Sepp1−/− mice do not differ from wild-type mice in spontaneous rearing [Fig. 2A, left], grooming [Fig. 2A, center] and distance traveled [Fig. 2A, right]. Performance on the pole descent test also improved in Se-supplemented mice, however a main effect of genotype remained for the time to invert (F(1,33)=12.68, p=0.0011) [Fig. 2B, left] and descend (F(1,33)=4.989, p=0.0324) [Fig. 2B, center] the vertical pole, and post hoc test indicated male Sepp1−/− mice were slower than wild-type littermates in turn time only (t(19)=4.004, p<0.001). Performance on the inverted hang test also improved in the Se-supplemented Sepp1−/− mice, and an effect of genotype did not reach statistical significance (F(1,31)=3.257, p=0.081) [Fig. 2B, right]. In all motor tests, Se-supplemented Sepp1−/− mice showed subtle qualitative behavioral deficits in both sexes, but the male bias was largely eliminated.
Figure 2.
High Se diet improves spontaneous activity and motor coordination more in male than female Sepp1−/− mice compared to control mice. (A) Rearing and grooming were measured in the cylinder, and total distance traveled in the open field. Se-supplemented Sepp1−/− mice were as active as controls when measured by rearing (left), grooming (center), and exploration of the open field (right), and no sex differences were observed. (B) Motor coordination was measured using the pole test and inverted hang test. Se-supplemented Sepp1−/− mice took longer to turn (left, genotype **p<0.01) and to descend the pole (center, genotype *p<0.05) when compared with control mice, but were capable of suspending themselves for two minutes (right). Male and female Sepp1−/− mice performed similarly except for turn time, and an interaction effect between genotype and sex was not statistically evident in any test. Values are expressed as means ± SEM. n=8–10 per group, ***p<0.001, compared with control mice.
Generation and characterization of Scly−/− animals
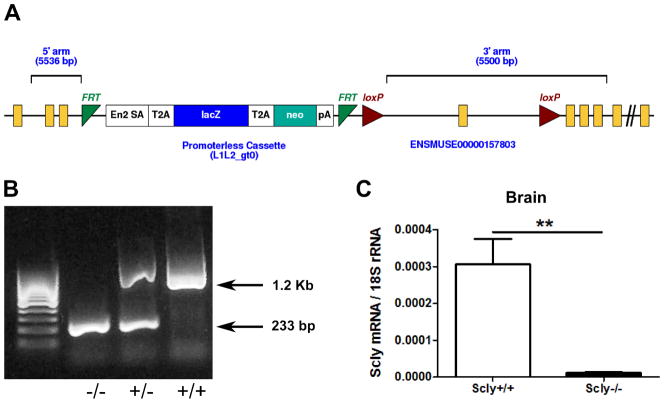
Scly knockout mice were generated by a conditional knockout approach in the event that deletion of Scly caused embryonic lethality. Mice were generated with a trapping cassette inserted upstream of exon 4 of the Scly gene, resulting in a constitutive null mutation in the whole animal [Fig 3A]. Mutation of the targeted region of the Scly gene was con rmed by PCR of mouse tail DNA [Fig. 3B], and Scly mRNA was undetectable by qPCR in all tissues examined from Scly−/− mice, including brain [Fig. 3C]. Scly−/− mice were born at the expected Mendelian ratio, appeared generally healthy, and developed to adulthood. Unlike Sepp1−/− mice, Scly−/−mice were fertile, producing viable offspring that did not differ in size at birth compared to wild-type control mice.
Figure 3.
Generation of Scly-knockout mice. (A) Image from KOMP. A promoterless trapping cassette was inserted upstream of exon 4 of the mouse Scly locus on chromosome 1, causing splicing at the cassette and truncation of the endogenous transcript. The cassette was flanked by FRT sites for conditional excision of the cassette by breeding with FLP-recombinase transgenic mice in case of embryonic lethality. Presence of the loxP sites flanking exon 4 allowed excision of a coding exon critical for enzymatic function by breeding with Cre-recombinase transgenic mice. (B) PCR genotyping of mice tails was performed to detect presence of wild-type allele (1.2 kb) or knockout allele (233 bp) using primers described in Materials and Methods. (C) Quantitative RT-PCR analysis of brains from Scly+/+ and Scly−/−mice indicated no detectable Scly mRNA in homozygous knockout mice.
Neurological motor phenotype is largely absent in Scly−/− mice
Scly is a relatively uncharacterized enzyme thought to be involved in recycling Se from Sec in support of selenoprotein synthesis. Sepp1 contains up to 10 Sec residues and is proposed to be a source of Se for the brain. We hypothesized that mice lacking Scly would be unable to efficiently catalyze Sec degradation, and therefore would manifest a phenotype similar to the Sepp1−/− mice. Contrary to our prediction, we found that Scly−/− animals raised on a Se-adequate diet displayed no neurological phenotype. Spontaneous rearing and movement were unaffected by Scly genotype, and performance on the vertical pole descent was similar between Scly+/+ and Scly−/− mice [data not shown].
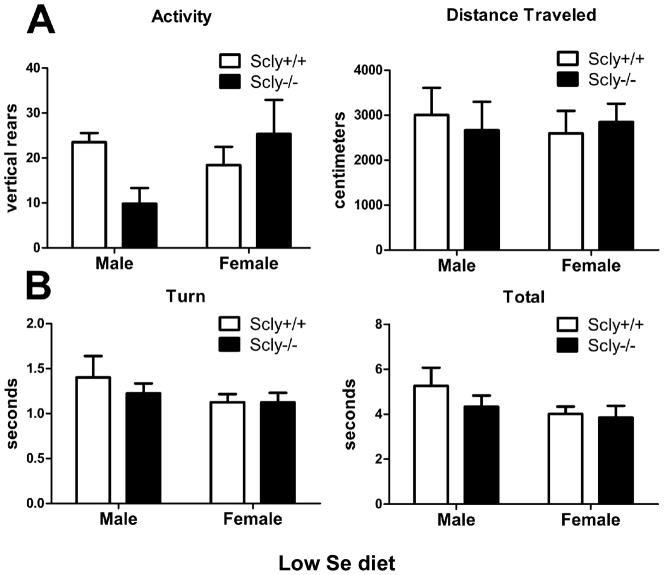
As no apparent behavioral phenotype presented in the Scly−/− mice raised on a standard lab diet, we subjected the animals to a marginally low Se diet, containing 0.08 mg/kg Se, to assess whether the animals display enhanced sensitivity to Se deficiency as measured by locomotor behavior. Total distance traveled in the open field was unaffected by Scly genotype under low-Se conditions, however two-way ANOVA revealed an interaction between Scly genotype and sex on rearing activity (F(1,22)=4.937, p=0.0369) [Fig. 4A]. Motor coordination assayed by turn and total time in the vertical pole descent test indicated no difference due to genotype in Se-deficient Scly−/− animals [Fig. 4B].
Figure 4.
Spontaneous activity and motor coordination is normal in male and female Scly−/− mice fed a low Se diet. (A) Rearing and total distance traveled was measured in the open field. Scly−/−mice were as active as controls when measured by exploration of the open field (right), however an interaction between genotype and sex affected rearing activity (left, genotype x sex *p<0.05). (B) Motor coordination was measured using the pole test. Scly−/− mice and control mice performed similarly to turn (left) and descend the pole (right). Male and female mice performed similarly, and no genotype effects were present. Values are expressed as means ± SEM. n=6–7 per group.
Spatial learning is sensitive to disruptions in Se availability
The Morris water maze is a paradigm for assessing spatial learning and memory in rodents (Morris, 1984). Sepp1 knockout mice raised on a Se-supplemented diet have mild impairments in learning measured with this paradigm, despite having a large deficit in long-term potentiation, a cellular model for learning and memory (Peters et al., 2006). As Se supplementation attenuates neurological impairments in Sepp1 knockout mice, we questioned if learning deficits would be greater for Sepp1−/− mice raised on a normal Se diet. Mice were initially trained over several days to locate a hidden platform in a large tank of water. Subsequently the platform was removed for one final trial, in which the amount of time the animal investigated the area where the platform used to be was measured. We compared male and female Sepp1−/− and wild-type animals raised on standard laboratory chow with adequate Se for learning deficits.
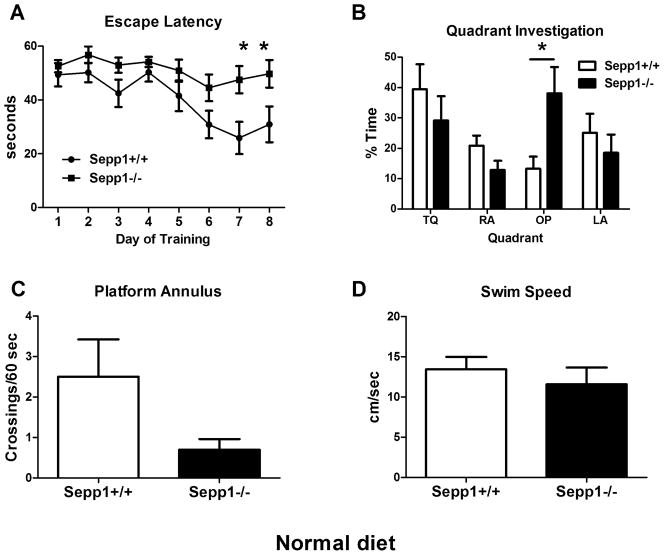
In contrast to our findings of gender differences in neuromotor function, we did not observe significant learning differences between Sepp1−/− male and female mice. Therefore male and female groups were combined after eliminating sex as an interacting variable. We found that learning was impaired in Sepp1−/− mice fed a standard diet [Fig. 5A]. Two-way ANOVA revealed an interaction between genotype and training (F(7,112)=2.361, p=0.0275). Escape latency over time was not substantially reduced in Sepp1−/− as compared to control mice, suggesting the mice did not learn the spatial location of the platform. The probe trial results are ambiguous since the training was ineffective in Sepp1−/− mice; however a genotype x quadrant interaction effect was observed (F(3,64)=3.277, p=0.0266) during quadrant investigation [Fig. 5B]. Sepp1−/−mice spent significantly more time than controls investigating the opposite quadrant, which was not due to uncoordinated swimming, and likely indicates failure to learn the platform location. There was a strong trend towards reduced number of platform crossings (t(16)=2.064, p=0.0557) [Fig. 5C], but swim speed (t(14)=0.7262, p=0.48) [Fig. 5D] was not significantly different between genotypes by t-test. Similar to published work on Se-supplemented Sepp1−/−mice in the water maze, we found a genotype difference during training, but not on the probe trial in Sepp1−/− mice fed a high Se diet [data not shown].
Figure 5.
Spatial learning and memory is disrupted in Sepp1−/− mice fed a standard diet. (A) Average escape latency per training day over time was measured in the Morris Water Maze. An interaction between Sepp1 genotype and training was present (*p<0.05) and Sepp1−/− mice had significantly longer latency on days 7 and 8 (*p<0.05). (B) In the probe trial, Sepp1−/− spent more time exploring the opposite (OP) quadrant (*p<0.05) and we found a significant interaction effect (genotype x quadrant *p<0.05). They also had non-significant trends toward fewer platform crossings (p=0.0557) (C) and a reduced swim speed (p>0.05) (D). Values are expressed as means ± SEM. n=8–10 per group.
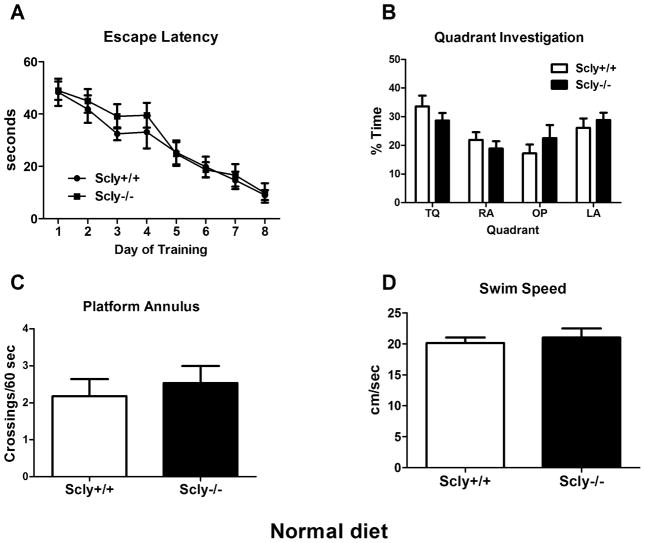
Although we found little difference between Scly−/− and control mice in locomotor activity, we assayed the Scly−/− animals using the Morris water maze for comparison with Sepp1−/− mice. Scly−/− mice fed standard chow performed like wild-type mice during training [Fig. 6A], as the only effect observed by two-way ANOVA was for training day (F(7,154)=24.07, p<0.0001). Similarly, in the probe trial we found an effect of quadrant (F(3,88)=5.78, p=0.0012), but no interaction with genotype [Fig. 6B]. Platform crossings (t(22)=0.5415, p=0.59) [Fig. 6C], latency to platform location [data not shown], and swim speed (t(22)=0.4955, p=0.63) [Fig. 6D] were not significantly different between genotypes when assessed by t-test. Scly−/− mice showed a trend towards delayed learning on days three and four. The lack of spatial learning impairment in Se-adequate Scly−/− mice starkly contrasted with Sepp1−/− mice fed the same diet.
Figure 6.
Spatial learning and memory is not disrupted in Scly−/− mice fed a standard diet. (A) Average escape latency per training day over time was measured in the Morris Water Maze. Scly−/− and control mice showed strongly reduced latency over time, and no interaction between genotype and training was apparent. (B) In the probe trial at the end of training, Scly−/− and control mice spent equal time exploring the target (TQ), left (LA) and right adjacent (RA), and the opposite (OP) quadrants (p>0.05). Both genotypes had similar platform crossings (C) and swim speed (D) (p>0.05). Values are expressed as means ± SEM. n=11–13 per group.
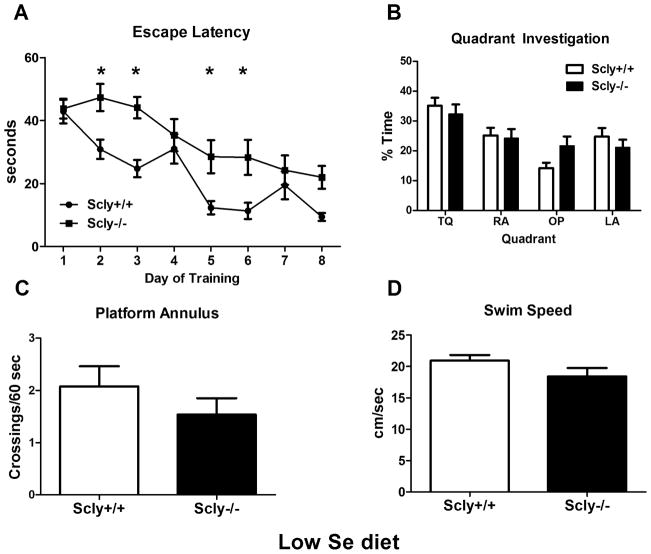
The trend toward mild learning deficits in Scly−/− mice led us to question if a restricted Se diet would result in greater learning impairments in these animals. When Scly−/− animals fed a Se-deficient diet were assessed, two-way ANOVA revealed an interaction between genotype and training (F(5,120)=2.496, p=0.0345), while post hoc tests indicated longer latency times on days 2 (t(24)=2.942, p<0.05), 3 (t(24)=3.467, p<0.01), 5 (t(24)=2.895, p<0.05), and 6 (t(24)=3.037, p<0.05) in the knockout mice [Fig. 7A], which suggests reduced learning. However the mice learned the platform location and the deficit was not as severe as in non-supplemented Sepp1−/−animals. Scly−/− animals performed as well as wild-type animals in the probe trial. We found a main effect on quadrant investigation (F(3,96)=11.19, p<0.0001) but no Scly genotype interaction [Fig. 7B]. We found no difference in platform crossings (t(24)=1.089, p=0.29) [Fig. 7C], latency [data not shown], or average swim speed (t(22)=1.60, p=0.124) [Fig. 7D] in Scly−/−mice on a low Se diet compared to control mice.
Figure 7.
Spatial learning is mildly impaired in Scly−/− mice fed a low Se diet. (A) Average escape latency per training day over time was measured in the Morris Water Maze. An interaction between Scly genotype and training was present (*p<0.05), and Scly−/− mice had significantly longer latency on days 2, 3, 5 and 6 (*p<0.05). (B) In the probe trial at the end of training, Scly−/− and control mice spent equal time exploring the target (TQ), adjacent (LA, RA) and opposite (OP) quadrants (p>0.05). Both genotypes had similar platform crossings (C) and swim speed (D) (p>0.05) in the probe trial. Values are expressed as means ± SEM. n=13 per group.
Selenoprotein expression and Glutathione Peroxidase activity
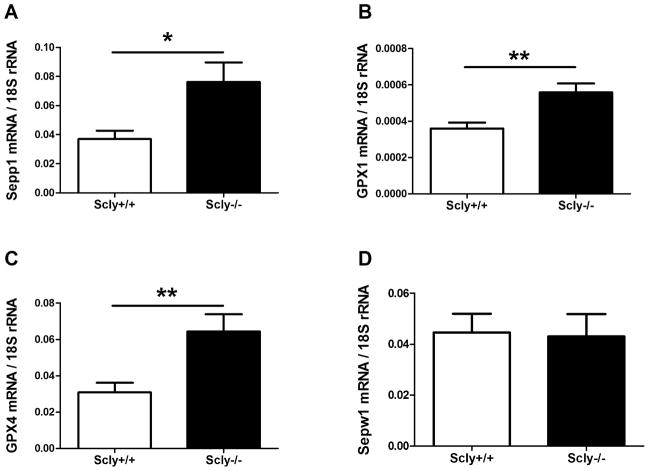
When dietary Se is restricted in mammals, Sec-enriched Sepp1 helps maintain selenoprotein expression in the brain (Hill et al., 2007). We therefore investigated the expression of selenoproteins in brains of Scly−/− animals by quantitative RT-PCR, western blot, and glutathione peroxidase (GPX) activity. We did not find a significant change in the mRNA for Sepp1 (t(9)=1.536, p=0.1589, n=5–6), GPX1 (t(10)=1.056, p=0.3157, n=6), GPX4 (t(10)=0.6899, p=0.5060, n=6), or selenoprotein W (Sepw1) (t(8)=2.149, p=0.0639, n=5) in brains of Scly−/−mice fed normal chow [data not shown]. Thus Scly−/− mice on a Se-adequate diet show neither a behavioral phenotype, nor any major changes in selenoprotein mRNA expression in brain. Therefore western blotting and GPX activity assays to assess the severity of Se-deficiency were performed in mice fed a low-Se diet only.
Scly−/− animals on a low-Se diet displayed significantly elevated Sepp1 mRNA expression in brain compared to control mice (t(14)=2.686, p=0.0177) [Fig. 8A]. GPX1 (t(12)=3.40, p=0.0053) [Fig. 8B] and GPX4 (t(14)=3.093, p=0.0079) [Fig. 8C] mRNA were also significantly increased in brain, while Sepw1 was not changed (t(14)=0.1380, p=0.89) [Fig. 8D]. In the same mice, we did not detect an increase in mRNA expression of Nfs1 [(t(14)=0.7154, p=0.4861, not pictured], a cysteine desulfurase enzyme with Sec lyase activity.
Figure 8.
Expression of selenoprotein transcripts is increased in Se-deficient Scly−/− mice brains. (A) Sepp1 mRNA level was measured in brains of Scly−/− mice (n=8). We additionally measured GPX1 (n=7) (B), GPX4 (n=8) (C), and Sepw1 (n=8) (D) mRNA expression in brain of Scly−/−mice fed a low Se diet. Sepp1, GPX1, and GPX4 were increased (*p<0.05, **p<0.01), while Sepw1 was unchanged. Samples were assayed in triplicate or quadruplicate and values were normalized to 18s rRNA and expressed as means ± SEM.
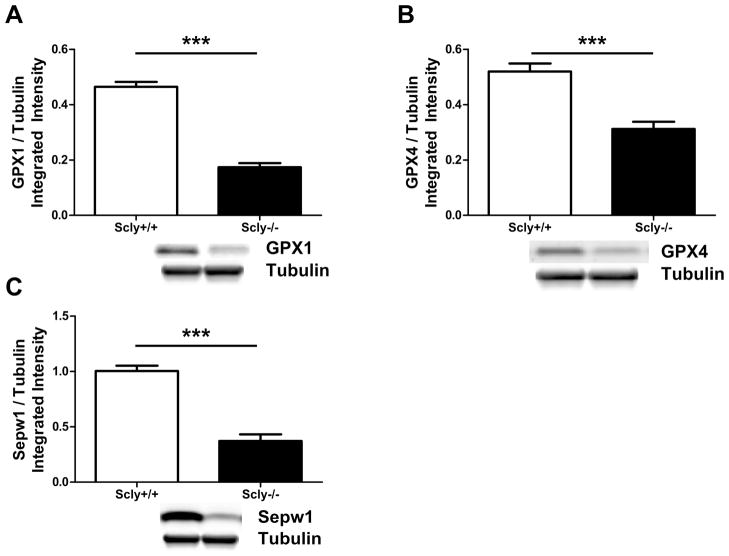
In contrast to the selenoprotein mRNA expression data, the corresponding protein levels assessed by western blot are consistently decreased in the brains of Se-deficient Scly−/− mice. Both GPX1 (t(14)=12.39, p<0.0001) [Fig. 9A] and Sepw1 (t(14)=8.294, p<0.0001)) [Fig. 9C] are expressed at ~37% of the wild-type level, while GPX4 expression (t(14)=5.365, p<0.0001) [Fig. 9B] is reduced to ~60% compared to control brains. These results confirm that Scly contributes to selenoprotein expression in brain during Se-deficiency.
Figure 9.
Expression of selenoproteins is decreased in Se-deficient Scly−/− mice brains. (A) GPX1, (B) GPX4, and (C) Sepw1 protein was measured by western blot and quantified by integrated intensity (n=8). Below each graph is a representative sample of the blot including loading equivalence determined by tubulin. GPX1, GPX4, and Sepw1 were drastically reduced in Scly−/− mice (***p<0.0001). All selenoprotein values are normalized to relative amounts of tubulin, and expressed as means ± SEM.
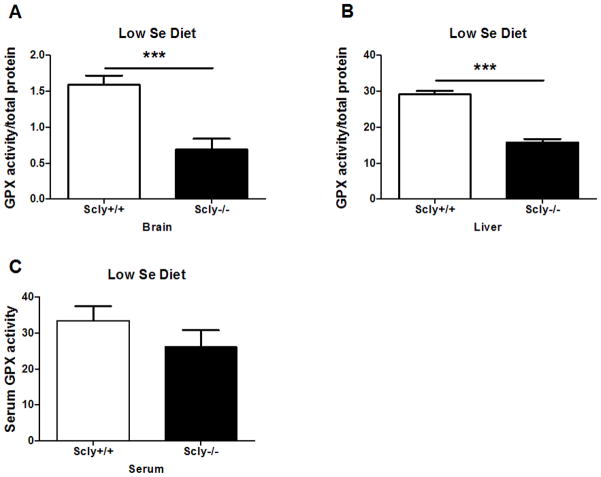
GPX activity in brain was reduced to 43% of the wild-type control level (t(14)=4.495, p=0.0005) [Fig. 10A], and was reduced to 54% in liver (t(14)=9.9297, p<0.0001) [Fig. 10B] of Scly−/− mice fed a low-Se diet. However, GPX activity in serum was not significantly reduced [Fig. 10C]. These data indicate that Scly helps maintain selenoprotein expression and activity when dietary Se availability is limiting. They further suggest that Se status in tissues may be more affected than plasma in the absence of Scly. The contrast of increased selenoprotein mRNA expression in the Scly−/− mouse brain despite reduced selenoproteins and enzyme activity suggests that Scly is a significant contributor to the Se pool for selenoprotein translation.
Figure 10.
Glutathione peroxidase activity is decreased in Se-deficient Scly−/− mice brains. Total GPX activity in brain (n=8) (A), liver (n=8) (B), and serum (n=12–14) (C) was assayed with a coupled reaction measuring NADPH oxidation. Mice were fed the low Se diet from weaning until time of sacrifice, when tissues were harvested for analysis. Although both brain and liver GPX activity were reduced by approximately half (***p<0.001), serum GPX activity was not significantly reduced. Values are standardized to total protein concentration (brain, liver) or volume of serum and expressed as means ± SEM.
DISCUSSION
The results reported herein describe the first characterization of a novel mouse strain deficient in Scly. These mice exhibit minimal neurological deficits, an unexpected finding given the phenotypic effects of Sepp1 knockout and the proposed role of Scly in recycling the essential trace element Se from Sec. Unlike Sepp1−/− animals, deletion of Scly does not result in neuromotor impairments or spatial learning deficits, except under low-Se conditions. We also report sex differences in the motor phenotype of mice with genetic deletion of Sepp1, highlighting the importance of considering gender on studies addressing the biological functions of Se or selenoproteins in mammals.
Sepp1 is a plasma protein that is considered to be the physiological transporter of Se from liver to brain. Sepp1 is additionally found in grey and white matter and cerebrospinal fluid, and may store Se within brain (Scharpf et al., 2007). Sepp1 and ApoER2 maintain a high Se concentration in the testes, and prioritize Se to brain albeit at a lower concentration (Burk & Hill, 2009). Most tissues produce Sepp1 and Scly, and the whole body turnover rate of Sepp1 is high, fluxing a significant proportion of bodily Se even when dietary availability is limiting (Burk & Hill, 2005). Brain Se level is not dependent on hepatic Sepp1 in Se-adequate adult animals (Schweizer et al., 2005). However Se-deficiency directs Se from liver-derived Sepp1 to the brain (Nakayama et al., 2007, Renko et al., 2008). The turnover rate of Sepp1 within the nervous system and the interaction with the circulation are uncertain.
Unlike most trace element transporters, Sepp1 cannot rapidly load and unload cargo because Se is covalently incorporated as the amino acid Sec, which must be degraded to supply Se. Biosynthesis and incorporation of Sec is a protracted, energy intensive process that requires organized interaction of specific proteins (SPS, SepSecS, EFSec, SBP2) and nucleic acids (tRNA(Sec), SECIS-containing mRNA), in addition to ATP, pyridoxal phosphate, and the translation machinery. Receptor-mediated uptake by ApoER2 facilitates entry of Sepp1 into cells, but recycling of the Sec residues would depend on the lysosome or proteasome to hydrolyze peptide bonds followed by liberation of Se from Sec, presumably by Scly.
To test the hypothesis that Scly recycles Sec from Sepp1 in the brain, we investigated whether Scly−/− mice manifest a phenotype similar to Sepp1−/− mice. Surprisingly, very little neurological dysfunction was present in the Scly−/− mice, even when fed a diet low in Se. The lack of behavioral changes in Scly−/− mice, compared to Sepp1−/− mice, could be due to Nfs1 catalyzing Sec to selenide conversion for selenoprotein synthesis (Lacourciere et al., 2000). Although we did not detect increased Nfs1 mRNA in Scly−/− mice brains, the normal activity of the enzyme might be compensating for the absence of Scly. Alternatively, Sepp1 may have an acute function in brain not strictly related to Se delivery. Disrupted synaptic plasticity in Se-supplemented Sepp1−/− mice (Peters et al., 2006) supports the notion that Sepp1 has a role in cell signaling via its receptor, ApoER2.
ApoER2 is found at synaptic sites (Beffert et al., 2005), in cultured astrocytes, oligodendrocytes, and microglia (Fan et al., 2001), and in the brain vasculature (Korschineck et al., 2001), suggesting that all brain cells can take up Sepp1. In adult brain, mRNA for Sepp1 is apparent in glia (Lein et al., 2007) but protein expression is dominant in neurons (Bellinger et al., 2008, Scharpf et al., 2007). Sepp1 mRNA and protein are abundant in choroid plexus epithelium (Bellinger et al., 2008, Steinert et al., 1998, Zhang et al., 2008). Scly mRNA appears enriched in grey matter and neurons of mouse brain (Lein et al., 2007). Further, a mouse proteomics study identified Scly in synaptoneurosomes (Filiou et al., 2010).
We found that mRNA for Scly was not significantly changed in brain of Sepp1−/− mice fed a standard diet. Since Sepp1−/− mice have depressed Se in brain, this finding suggests that Scly is not Se-regulated, and the enzyme is minimally affected by dietary Se or tissue Se levels (Deagen et al., 1987). The mRNA expression of Sepp1 trended up in Scly−/− mice fed normal chow, and was significantly increased in brain of Scly−/− mice fed low-Se chow. Se-deficient Scly−/− mice also had increased GPX1 and GPX4 mRNA, while that of Sepw1 was unchanged. However, GPX protein and activity in brain of the low-Se Scly−/− mice were dramatically reduced compared to wild-type animals. Liver GPX activity was similarly reduced, while serum activity was less affected. Therefore Scly supports selenoprotein expression and function under conditions of dietary Se deficiency. Increased GPX mRNA despite reduced protein and activity in the Se-deficient Scly−/− brain could be a compensatory mechanism to boost inefficient selenoprotein translation. These findings extend a recent study, which demonstrated reduced GPX1 expression and reduced incorporation of Se derived from radiolabeled Sepp1, in cells with Scly knocked down by siRNA (Kurokawa et al., 2011). In addition, we observe that tissues are more reliant on Scly than blood.
Our finding that Se-deficient Scly−/− mice manifest a subtle spatial learning deficit in the water maze corresponds with results on Se-supplemented Sepp1−/− mice (Peters et al., 2006). Spatial learning requires the hippocampus, which is more dependent on Sepp1 for optimal Se concentration than other brain regions (Nakayama et al., 2007). Therefore Scly, Sepp1, and probably other selenoproteins in the hippocampus support spatial learning. It is likely that the kinetics of selenoprotein degradation and synthesis are even more disrupted in Scly−/− mice than the steady-state mRNA and protein expression levels. Brain regions with high metabolism or cellular turnover could be more dependent on a putative Sepp1-Scly recycling mechanism, while other cell populations might efficiently utilize an alternate Se source.
These results are the most extensive characterization of behavioral sex differences in Sepp1−/− mice to date. It has been suggested that the phenotype of Sepp1−/− mice is sex-dependent (Riese et al., 2006), however studies on Sepp1−/− mice have focused on males and data regarding behavioral sex differences are limited. Despite variations in behavioral testing paradigms, our results showing impaired motor performance in male Sepp1−/− mice are in agreement with previous reports (Hill et al., 2004, Renko et al., 2008, Schweizer et al., 2004). We additionally assessed motor impairment in female Sepp1−/− mice, and found it to be minimal compared to males. We also report that the spatial learning deficit in Sepp1−/− mice on a Se-adequate diet is worse than in Se-supplemented mice, building on a previous study that used only male mice on a high-Se diet (Peters et al., 2006).
Selenoprotein expression is modulated by sex in mammals (Meplan et al., 2007, Riese et al., 2006, Stoedter et al., 2010), and Sepp1 is an androgen responsive gene (Takahashi et al., 2006). Additionally, testosterone secretion declines during Se-deficiency in male rats (Behne et al., 1996). Male mice displayed increased sensitivity to Sepp1 deletion, and male Sepp1−/−mice greatly improved when given supranutritional dietary Se, indicating that males have a higher demand than females for Sepp1 and Se in the nervous system. However the phenotype is not exclusive to males, suggesting that some aspect of metabolism or development that is more prominent in males is dependent on Se.
Male gender is a risk factor for poor neurodevelopmental outcome after premature birth. Cerebral palsy and related developmental disorders are more common in males than females (Johnston & Hagberg, 2007). The phenotype of Sepp1−/− mice resembles cerebral palsy in that the developmental onset of spasticity and ataxia often presents with intellectual impairment and seizures. Moreover, this phenotype is not rapidly progressive and remains stable in adulthood when given adequate Se. Perinatal infection and hypoxia-ischemia are synergistic risk factors for cerebral palsy (Johnston & Hagberg, 2007, Mayoral et al., 2009), while Sepp1 is known to modulate immunity (Bosschaerts et al., 2008) and metabolism (Misu et al., 2010). Metabolically demanding brain regions and cells, with a presumably higher rate of selenoprotein synthesis, are susceptible to neurodegeneration in Sepp1−/− mice (Valentine et al., 2008). Moreover, an autosomal-recessive human disease termed progressive cerebellocerebral atrophy has been linked to mutations in SepSecS that globally disrupt selenoprotein synthesis (Agamy et al., 2010). The sequelae of these patients, including mental retardation, spasticity and seizures, are similar to those found in Sepp1−/− mice and emphasize the importance of selenoproteins in the function and health of the nervous system.
In conclusion, these results indicate that a novel mouse strain lacking Scly does not develop a neurological phenotype similar to Sepp1−/− mice. A subtle learning deficit is observed when Scly−/− animals are fed a low-Se diet, and these animals also have reduced expression of selenoproteins in brain. We further report a male bias in the neurological motor phenotype of Sepp1−/− mice. The disparity of neurological problems in Scly−/− and Sepp1−/− mice suggests that Sepp1 is more critical than Scly for maintenance of brain Se, but that recycling Se from Sec via Scly is physiologically important during dietary Se deficiency. Altogether these findings highlight that Se is critically important for the nervous system, and that Se metabolism through Sepp1 and Scly impacts spatial learning.
Acknowledgments
The authors thank Dr. Robert A. Nichols for critical review of the manuscript. This research was supported by National Institutes of Health grants to the University of Hawaii for the Behavioral-Electrophysiology Core (G12-RR003061), to MJB (DK047320), and FPB (RR016467 and DA027318).
Footnotes
The authors report no financial disclosures or potential conflicts of interest.
References
- Agamy O, Ben Zeev B, Lev D, Marcus B, Fine D, Su D, Narkis G, Ofir R, Hoffmann C, Leshinsky-Silver E, Flusser H, Sivan S, Soll D, Lerman-Sagie T, Birk OS. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am J Hum Genet. 2010;87:538–544. doi: 10.1016/j.ajhg.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Weeber EJ, Durudas A, Qiu S, Masiulis I, Sweatt JD, Li WP, Adelmann G, Frotscher M, Hammer RE, Herz J. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Behne D, Weiler H, Kyriakopoulos A. Effects of selenium deficiency on testicular morphology and function in rats. J Reprod Fertil. 1996;106:291–297. doi: 10.1530/jrf.0.1060291. [DOI] [PubMed] [Google Scholar]
- Bellinger FP, He QP, Bellinger MT, Lin Y, Raman AV, White LR, Berry MJ. Association of selenoprotein p with Alzheimer’s pathology in human cortex. J Alzheimers Dis. 2008;15:465–472. doi: 10.3233/jad-2008-15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger FP, Raman AV, Reeves MA, Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J. 2009;422:11–22. doi: 10.1042/BJ20090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosschaerts T, Guilliams M, Noel W, Herin M, Burk RF, Hill KE, Brys L, Raes G, Ghassabeh GH, De Baetselier P, Beschin A. Alternatively activated myeloid cells limit pathogenicity associated with African trypanosomiasis through the IL-10 inducible gene selenoprotein P. J Immunol. 2008;180:6168–6175. doi: 10.4049/jimmunol.180.9.6168. [DOI] [PubMed] [Google Scholar]
- Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr. 2005;25:215–235. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- Burk RF, Hill KE. Selenoprotein P-expression, functions, and roles in mammals. Biochim Biophys Acta. 2009;1790:1441–1447. doi: 10.1016/j.bbagen.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk RF, Hill KE, Olson GE, Weeber EJ, Motley AK, Winfrey VP, Austin LM. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J Neurosci. 2007;27:6207–6211. doi: 10.1523/JNEUROSCI.1153-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caito SW, Milatovic D, Hill KE, Aschner M, Burk RF, Valentine WM. Progression of neurodegeneration and morphologic changes in the brains of juvenile mice with selenoprotein P deleted. Brain Res. 2011;1398:1–12. doi: 10.1016/j.brainres.2011.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu-Ugalde J, Theilig F, Behrends T, Drebes J, Sieland C, Subbarayal P, Kohrle J, Hammes A, Schomburg L, Schweizer U. Mutation of megalin leads to urinary loss of selenoprotein P and selenium deficiency in serum, liver, kidneys and brain. Biochem J. 2010;431:103–111. doi: 10.1042/BJ20100779. [DOI] [PubMed] [Google Scholar]
- Combs GF, Jackson MI, Watts JC, Johnson LK, Zeng H, Idso J, Schomburg L, Hoeg A, Hoefig CS, Chiang EC, Waters DJ, Davis CD, Milner JA. Differential responses to selenomethionine supplementation by sex and genotype in healthy adults. Br J Nutr. 2011:1–12. doi: 10.1017/S0007114511004715. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res. 1999;835:18–26. doi: 10.1016/s0006-8993(98)01258-x. [DOI] [PubMed] [Google Scholar]
- Deagen JT, Butler JA, Beilstein MA, Whanger PD. Effects of dietary selenite, selenocystine and selenomethionine on selenocysteine lyase and glutathione peroxidase activities and on selenium levels in rat tissues. J Nutr. 1987;117:91–98. doi: 10.1093/jn/117.1.91. [DOI] [PubMed] [Google Scholar]
- Esaki N, Nakamura T, Tanaka H, Soda K. Selenocysteine lyase, a novel enzyme that specifically acts on selenocysteine. Mammalian distribution and purification and properties of pig liver enzyme. J Biol Chem. 1982;257:4386–4391. [PubMed] [Google Scholar]
- Fan QW, Iosbe I, Asou H, Yanagisawa K, Michikawa M. Expression and regulation of apolipoprotein E receptors in the cells of the central nervous system in culture: A review. Age (Dordr) 2001;24:1–10. doi: 10.1007/s11357-001-0001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiou MD, Bisle B, Reckow S, Teplytska L, Maccarrone G, Turck CW. Profiling of mouse synaptosome proteome and phosphoproteome by IEF. Electrophoresis. 2010;31:1294–1301. doi: 10.1002/elps.200900647. [DOI] [PubMed] [Google Scholar]
- Fleming SM, Salcedo J, Fernagut PO, Rockenstein E, Masliah E, Levine MS, Chesselet MF. Early and progressive sensorimotor anomalies in mice overexpressing wild-type human alpha-synuclein. J Neurosci. 2004;24:9434–9440. doi: 10.1523/JNEUROSCI.3080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan P, Viteri FE, Bertrais S, Czernichow S, Faure H, Arnaud J, Ruffieux D, Chenal S, Arnault N, Favier A, Roussel AM, Hercberg S. Serum concentrations of beta-carotene, vitamins C and E, zinc and selenium are influenced by sex, age, diet, smoking status, alcohol consumption and corpulence in a general French adult population. Eur J Clin Nutr. 2005;59:1181–1190. doi: 10.1038/sj.ejcn.1602230. [DOI] [PubMed] [Google Scholar]
- Hill KE, Zhou J, Austin LM, Motley AK, Ham AJ, Olson GE, Atkins JF, Gesteland RF, Burk RF. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J Biol Chem. 2007;282:10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- Hill KE, Zhou J, McMahan WJ, Motley AK, Burk RF. Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J Nutr. 2004;134:157–161. doi: 10.1093/jn/134.1.157. [DOI] [PubMed] [Google Scholar]
- Hoffmann FW, Hashimoto AC, Shafer LA, Dow S, Berry MJ, Hoffmann PR. Dietary selenium modulates activation and differentiation of CD4+ T cells in mice through a mechanism involving cellular free thiols. J Nutr. 2010;140:1155–1161. doi: 10.3945/jn.109.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann PR, Hoge SC, Li PA, Hoffmann FW, Hashimoto AC, Berry MJ. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 2007;35:3963–3973. doi: 10.1093/nar/gkm355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston MV, Hagberg H. Sex and the pathogenesis of cerebral palsy. Dev Med Child Neurol. 2007;49:74–78. doi: 10.1017/s0012162207000199.x. [DOI] [PubMed] [Google Scholar]
- Korschineck I, Ziegler S, Breuss J, Lang I, Lorenz M, Kaun C, Ambros PF, Binder BR. Identification of a novel exon in apolipoprotein E receptor 2 leading to alternatively spliced mRNAs found in cells of the vascular wall but not in neuronal tissue. J Biol Chem. 2001;276:13192–13197. doi: 10.1074/jbc.M011795200. [DOI] [PubMed] [Google Scholar]
- Kurokawa S, Takehashi M, Tanaka H, Mihara H, Kurihara T, Tanaka S, Hill K, Burk R, Esaki N. Mammalian selenocysteine lyase is involved in selenoprotein biosynthesis. J Nutr Sci Vitaminol (Tokyo) 2011;57:298–305. doi: 10.3177/jnsv.57.298. [DOI] [PubMed] [Google Scholar]
- Lacourciere GM, Mihara H, Kurihara T, Esaki N, Stadtman TC. Escherichia coli NifS-like proteins provide selenium in the pathway for the biosynthesis of selenophosphate. J Biol Chem. 2000;275:23769–23773. doi: 10.1074/jbc.M000926200. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Mayoral SR, Omar G, Penn AA. Sex differences in a hypoxia model of preterm brain damage. Pediatr Res. 2009;66:248–253. doi: 10.1203/PDR.0b013e3181b1bc34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meplan C, Crosley LK, Nicol F, Beckett GJ, Howie AF, Hill KE, Horgan G, Mathers JC, Arthur JR, Hesketh JE. Genetic polymorphisms in the human selenoprotein P gene determine the response of selenoprotein markers to selenium supplementation in a gender-specific manner (the SELGEN study) FASEB J. 2007;21:3063–3074. doi: 10.1096/fj.07-8166com. [DOI] [PubMed] [Google Scholar]
- Mihara H, Kurihara T, Watanabe T, Yoshimura T, Esaki N. cDNA cloning, purification, and characterization of mouse liver selenocysteine lyase. Candidate for selenium delivery protein in selenoprotein synthesis. J Biol Chem. 2000;275:6195–6200. doi: 10.1074/jbc.275.9.6195. [DOI] [PubMed] [Google Scholar]
- Misu H, Takamura T, Takayama H, Hayashi H, Matsuzawa-Nagata N, Kurita S, Ishikura K, Ando H, Takeshita Y, Ota T, Sakurai M, Yamashita T, Mizukoshi E, Honda M, Miyamoto K, Kubota T, Kubota N, Kadowaki T, Kim HJ, Lee IK, Minokoshi Y, Saito Y, Takahashi K, Yamada Y, Takakura N, Kaneko S. A liver-derived secretory protein, selenoprotein P, causes insulin resistance. Cell Metab. 2010;12:483–495. doi: 10.1016/j.cmet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Morris R. Developments of a water-maze procedure for studying spatial learning in the rat. J Neurosci Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Hill KE, Austin LM, Motley AK, Burk RF. All regions of mouse brain are dependent on selenoprotein P for maintenance of selenium. J Nutr. 2007;137:690–693. doi: 10.1093/jn/137.3.690. [DOI] [PubMed] [Google Scholar]
- Peters MM, Hill KE, Burk RF, Weeber EJ. Altered hippocampus synaptic function in selenoprotein P deficient mice. Mol Neurodegener. 2006;1:12. doi: 10.1186/1750-1326-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- Renko K, Werner M, Renner-Muller I, Cooper TG, Yeung CH, Hollenbach B, Scharpf M, Kohrle J, Schomburg L, Schweizer U. Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Sepp-knockout mice. Biochem J. 2008;409:741–749. doi: 10.1042/BJ20071172. [DOI] [PubMed] [Google Scholar]
- Riese C, Michaelis M, Mentrup B, Gotz F, Kohrle J, Schweizer U, Schomburg L. Selenium-dependent pre- and posttranscriptional mechanisms are responsible for sexual dimorphic expression of selenoproteins in murine tissues. Endocrinology. 2006;147:5883–5892. doi: 10.1210/en.2006-0689. [DOI] [PubMed] [Google Scholar]
- Scharpf M, Schweizer U, Arzberger T, Roggendorf W, Schomburg L, Kohrle J. Neuronal and ependymal expression of selenoprotein P in the human brain. J Neural Transm. 2007;114:877–884. doi: 10.1007/s00702-006-0617-0. [DOI] [PubMed] [Google Scholar]
- Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer U, Michaelis M, Kohrle J, Schomburg L. Efficient selenium transfer from mother to offspring in selenoprotein-P-deficient mice enables dose-dependent rescue of phenotypes associated with selenium deficiency. Biochem J. 2004;378:21–26. doi: 10.1042/BJ20031795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer U, Streckfuss F, Pelt P, Carlson BA, Hatfield DL, Kohrle J, Schomburg L. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem J. 2005;386:221–226. doi: 10.1042/BJ20041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert P, Bachner D, Flohe L. Analysis of the mouse selenoprotein P gene. Biol Chem. 1998;379:683–691. doi: 10.1515/bchm.1998.379.6.683. [DOI] [PubMed] [Google Scholar]
- Stoedter M, Renko K, Hog A, Schomburg L. Selenium controls the sex-specific immune response and selenoprotein expression during the acute-phase response in mice. Biochem J. 2010;429:43–51. doi: 10.1042/BJ20091868. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Hursting SD, Perkins SN, Wang TC, Wang TT. Genistein affects androgen-responsive genes through both androgen- and estrogen-induced signaling pathways. Mol Carcinog. 2006;45:18–25. doi: 10.1002/mc.20153. [DOI] [PubMed] [Google Scholar]
- Tobe R, Mihara H, Kurihara T, Esaki N. Identification of proteins interacting with selenocysteine lyase. Biosci Biotechnol Biochem. 2009;73:1230–1232. doi: 10.1271/bbb.90065. [DOI] [PubMed] [Google Scholar]
- Valentine WM, Abel TW, Hill KE, Austin LM, Burk RF. Neurodegeneration in mice resulting from loss of functional selenoprotein P or its receptor apolipoprotein E receptor 2. J Neuropathol Exp Neurol. 2008;67:68–77. doi: 10.1097/NEN.0b013e318160f347. [DOI] [PubMed] [Google Scholar]
- Valentine WM, Hill KE, Austin LM, Valentine HL, Goldowitz D, Burk RF. Brainstem axonal degeneration in mice with deletion of selenoprotein p. Toxicol Pathol. 2005;33:570–576. doi: 10.1080/01926230500243045. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou Y, Schweizer U, Savaskan NE, Hua D, Kipnis J, Hatfield DL, Gladyshev VN. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J Biol Chem. 2008;283:2427–2438. doi: 10.1074/jbc.M707951200. [DOI] [PubMed] [Google Scholar]