Abstract
Mycobacterium ulcerans causes Buruli ulcer in humans, a progressive ulcerative epidermal lesion due to the mycolactone toxin produced by the bacterium. Molecular analysis of M. ulcerans reveals it is closely related to M. marinum, a pathogen of both fish and man. Molecular evidence from diagnostic PCR assays for the insertion sequence IS2404 suggests an association of M. ulcerans with fish. However, fish infections by M. ulcerans have not been well documented and IS2404 has been found in other mycobacteria. We have thus, employed two experimental approaches to test for M. ulcerans in fish. We show here for the first time that M. ulcerans with or without the toxin does not mount acute or chronic infections in Japanese Medaka “Oryzias latipes” even at high doses. Moreover, M. ulcerans-infected medaka do not exhibit any visible signs of infection nor disease and the bacteria do not appear to replicate over time. In contrast, similar high doses of the wild-type M. marinum or a mycolactone producing M. marinum “DL” strain are able to mount an acute disease with mortality in medaka. Although these results would suggest that M. ulcerans does not mount infections in fish we have evidence that CLC macrophages from goldfish are susceptible to mycolactones.
Keywords: Mycobacterium ulcerans, fish, medaka, mycolactone
1. Introduction
Mycobacterium ulcerans is an environmental pathogen that causes a severe necrotizing skin lesion in humans called Buruli ulcer. It has been identified as the third most important human mycobacterial disease by the World Health Organization [1], followed by diseases caused by M. tuberculosis and M. leprae. Buruli ulcer has been reported in over 30 countries but is becoming an important emerging infectious disease of West Africa [2, 3, 4] and Australia [5]. Molecular analysis of chromosomal genes reveals that M. ulcerans is closely related to M. marinum, which causes disease in both fish and man [6]. M. ulcerans also carries a large 174-kb plasmid that produces a cytotoxic macrolide toxin, called mycolactone [7, 8].
M. ulcerans is a slow-growing mycobacterial species characterized by temperature-restricted growth between 28–34°C and is thought to have evolved from an M. marinum-like ancestor [9]. M. ulcerans is distinguished from M. marinum by three major differences; 1) loss of approximately 1 Mb of its genome, 2) acquisition of a large virulence plasmid which encodes a toxic immunosuppressive macrolide, mycolactone and 3) accumulation of 771 pseudo-genes, some inactivated by acquisition of 213 and 91 copies of the insertion elements IS2404 and IS2606 respectively [10]. Thus the functional M. ulcerans genome (5.8 MB) is about 2 MB smaller than that of M. marinum (6.6 MB) [10]. Taken together, these studies strongly suggest that M. ulcerans has evolved from an M. marinum ancestor through reductive evolution driven by host adaptation.
The mode of infection and transmission of M. ulcerans from the environment to humans is unknown but acquisition of Buruli ulcer has been globally associated with residence close to slow moving water and the disease is not found in the arid regions of West Africa [11, 12]. Attempts to identify the potential vectors and reservoirs by culturing M. ulcerans from the environment have been largely unsuccessful, primarily due to overgrowth of faster growing bacteria on solid media. Recently the first environmental isolate was obtained from a water strider (Gerridae) after serial passage of the insect homogenate through mice [13]. In contrast, there has been abundant molecular evidence for M. ulcerans in the environment. Using IS240-based diagnostic PCR assays, potential M. ulcerans DNA has been identified in a wide range of aquatic organisms including plants, detritus, fish, snails, tadpoles and insects [14, 15, 16, 17]. Of particular interest is the fact that many IS2404-PCR positive predaceous insects such as Naucorids and Belostomatids have been collected in Cameroon, Benin and Ghana. These insects are known to attack and bite some fish [18], and may play a role in the trophic transfer of M. ulcerans through food webs [19]. However, IS2404 is also carried by several other species of mycobacteria including pathogens of fish and frogs [17, 20, 21, 22] and molecular assays using M. ulcerans specific variable nucleotide random repeat [VNTR] typing suggests that most of the fish analyzed from endemic water bodies, do not contain M. ulcerans [17].
Mycobacteriosis in fish has been well documented for decades [23, 24] with the three most important pathogenic species being M. abscessus, M. fortuitum and M. marinum. Recently, outbreaks of mycobacteriosis have been reported in various locations along the Mediterranean and Red Seas and the Chesapeake Bay attributed to another IS2404-positive mycobacterial pathogen M. pseudoshottsii, and a novel clade of M. marinum [25, 26]. One striking revelation is that these novel strains also possess plasmids that encode for a toxic macrolide, mycolactone, similar to that of M. ulcerans [27, 28].
The virulence of M. marinum in fish has been studied in a number of models including goldfish (Carassius auratus) [29], Zebrafish (Danio rerio) [30], and Japanese medaka (Oryzias latipes) [31]. In the studies reported here we have used medaka as a model for studying the virulence of M. ulcerans in fish and the role of mycolactone for virulence. Medaka are small (2–3 cm long by 0.5–1 cm wide) oviparous fresh water fish native to Asia [32]. They are widely used as a laboratory animal in various fields of biology, especially developmental biology [33, 34] and a wide range of medaka resources including extensive databases in toxicology, molecular genetics, genome project and existing inbred and transgenic lines are available. The M. marinum infection model can be reproduced to yield either acute or chronic infection in a dose dependant manner similar to that of M. tuberculosis infections in humans. In this model, infection leads to slow but progressive granuloma formation in target organs, as well as inflammation of the spleen [31].
We are presenting here, the first experimental study to assess the pathogenic potential of M. ulcerans for fish as well as the first to provide evidence regarding the toxicity of mycolactone for fish. In contrast to the severe infections that were induced by high doses of M. marinum for medaka, data reported here show that infective doses as high as 108 CFU M. ulcerans do not lead to overt disease in medaka. Although M. ulcerans DNA can be detected by PCR in all anatomical sections of infected fish up to 23 wks post infection, signs of disease are absent. Data from quantitative PCR show decreasing numbers of M. ulcerans during the infection period suggesting that the organism not only fails to replicate in medaka but may be cleared in this host. Neither molecular genetics nor biochemical studies presented in this work support a role for mycolactone in the virulence of M. ulcerans for fish.
2. Materials and methods
2.1. Bacterial strains and growth conditions
The M. ulcerans (MU) 1615 (ATCC 35840) is a well-characterized Malaysian human isolate with physical and biochemical properties similar to the Ghanaian strain Agy99 for which the complete genome sequence is available [3]. Transposon mutagenesis [8] was used to generate the mycolactone negative mutant MU1615::TN118 which contains an insertion in the FABH-like gene, mup045. This strain does not produce either the core or the side chain of mycolactone and has been well characterized by mass spectrometry and cytopathicity assays [7]. Both M. ulcerans strains used in this study were intrinsically labeled with a green fluorescent protein [GFP] which was inserted via an integrating vector psm5 [19]. The GFP gene is inserted into the chromosome of MU1615 GFP and MU1615::TN118 GFP in the phage attachment site (att) and has no effect on the virulence of the bacterium. The M. marinum (MM) 1218 strain (ATCC 972) is a saltwater fish isolate. M. marinum DL (MMDL) strain is a sea bass isolate which makes mycolactone F [27]. All strains were grown to mid-log phase in Middlebrook 7H9 (M7H9) media supplemented with 10% oleic acid-albumin-dextrose enrichment (OADC) {DIFCO}. M. ulcerans and M. marinum 1218 strains were incubated at 32°C and M. marinum DL was incubated at the permissive room temperature.
2.2. Medaka aquaculture
Japanese medaka used in this study were obtained from Don Ennis (University of Louisiana, Lafayette, LA, USA). Medaka were maintained in the laboratory in aquaria at 28°C, as described [31]. Fish were infected with mycobacteria and maintained post infection in a BSL-2 laboratory at 28°C. All infections and maintenance of infected animals were held in BSL-2 facilities; they were conducted as prescribed by NIH BMBL guidelines and approved by both the Institutional Biosafety Committee and by the IACUC at the University of Louisiana, Lafayette.
2.3. Experimental design
Bacterial inocula was prepared by serial passage through a 25-gauge needle to break up the mycobacterial clumps and diluted in PBS to obtain the desired concentration range of 102–108 initially based on OD600 and confirmed by colony forming units (CFU) obtained by plating the inocula on M7H10 agar plates supplemented with 10% OADC. Medaka were anaesthetized with tricaine methanesulphonate (MS-222) [0.0175%] and 30μl of bacterial suspension at respective doses was administered to each fish by intraperitoneal injection. Sham infections were also performed where medaka were inoculated with 30 μl of sterile PBS. An initial study was conducted to determine optimal infective dose (102–108). In a subsequent study 10 fish each were inoculated with 104 and 108 CFU of MU1615 GFP, MU 1615::TN118 GFP, MM1218 or MMDL and 5 PBS sham infections based on earlier studies [31]. At 1 and 8 wks post infection, 4 fish from each group were sacrificed, dissected and assayed for presence of bacteria and 1 fish per group was preserved in 10% formal in for sectioning and histopathology. This experiment was repeated and similar results were obtained. In a follow up study to induce chronic infection, 30 fish each were injected with 102 CFU of MU1615 GFP or MU1615::TN118 GFP and 10 fish were subjected to sham infections with PBS. At 1, 8 and 23 wks post infection 8 fish per bacterial strain were sacrificed and assayed for presence of M. ulcerans while 2 fish per bacterial strain were preserved in 10% formal in for sectioning and histopathology. All fish were maintained separately under similar environmental conditions and monitored for survival, mortality, gross behavioral changes and gross morphological pathology.
2.4. Histopathology
At the set time points noted above, 1–2 infected fish were euthanized using an overdose of tricaine methanesulphonate (MS-222) [0.1%]. Fishes were processed whole in 10% neutral-buffered formal in followed by embedding in paraffin wax. Thin sections of the paraffin embedded fish were made and stained with hematoxylin and eosin (H&E) and Ziehl-Neelsen acid fast stain. The histopathology and presence of acid fast bacilli (AFB) in the fish tissues were examined. “Sham” control infected fish were sacrificed and subjected to similar treatments for comparison.
2.5. Microscopic evidence of microbial colonization
Four fish per group from the initial study and 8 fish from the subsequent chronic study respectively were euthanized as described in order to determine infectivity at respective time points. Each fish was dissected by making a single anterior to posterior incision along the abdomen followed by removal of the kidney, liver, spleen, gut and heart. Whole organs from fish infected with fluorescently labeled bacteria were inspected microscopically as described in [31]. Briefly, fish organs were placed in an empty Petri dish and observed for fluorescence using a Nikon SMZ800 (Nikon, Tokyo, Japan) stereomicroscope equipped with X-cite TM 120 for epifluorescence system. The isolated organs were kept separately and homogenized in 500μl of M7H9 broth media supplemented with 100μg/ul cyloheximide, 20μg/ul chloramphenicol and 25μg/ul ampicillin. For detection of acid fast bacilli [AFB] in the dissected organs, smears were made from the homogenized suspensions and stained using the Zeihl-Neelsen technique. AFBs were viewed by light microscopy using an Olympus BX51 microscope [USA]. Wet mounts of representative homogenized organs were also viewed using a Nikon ECLIPSE E400 fluorescent microscope for the detection of the fluorescently labeled bacteria.
2.6. PCR analysis of infected Tissue
DNA was extracted from organ homogenates using a protocol adapted from Lamour and Finely [35]. Amplification of the enoyl reductase domain of mlsA, which encodes the lactone core of mycolactone, was used for identification of MU1615 GFP and MU1615::Tn118 GFP DNA in fish tissues. The early secreted antigen protein gene esxA, was used for identification of MMDL and MM1218 in fish tissues. Two and a half microlitres of each respective DNA sample was amplified with the mlsA primer pair; 5′– GAGATCGGTCCCGACGTCTAC-3′ and 5′-GGCTTGACTCATGTCACGTAAG-3′ or the esxA primer pair; 5′ – GACAGCAGCAGTGGAATTTCG – 3′ and 5′ – CTTCTGCTGCACACCCTGGTA – 3′ in 25μl polymerase chain reaction mixtures using the GoTaq polymerase-buffer system (Promega). Each reaction contained 18.3μl double-distilled water, 2.5μl GoTaq green master mix [400μl of each deoxynucleoside triphosphate, 3mM MgCl2, blue and yellow dyes], 0.5μM of forward and reverse primers, 0.75U GoTaq polymerase each and 5μl of DNA template. Cycling was performed in a Matercycler gradient thermal cycler (Eppendorf) as follows: 95°C for 5 min; 35 cycles of for 95°C for 1 min, 55°C for 1 min 72°C for 1 min; and 72°C for 10 min. Seven microlitres of each reaction mixture was analyzed on 1.5% agarose gels in 1X Tris-acetate-EDTA stained with 1μg/ml ethidium bromide for visualization of amplicons.
Quantitative PCR was used to determine genome equivalent units (GU) of MU1615 GFP and MU1615: :TN118 GFP in fish kidney. An internal probe was constructed the enoyl reductase domain using a Taqman probe with a TET dye. “No template controls” that lacked ER positive DNA were included. Positive control standards were included using known concentrations of DNA from purified plasmid template. The presence of inhibitors was excluded by PCR using an internal positive control (Applied biosystems). Five microlitres of representative DNA was amplified with the primer pair; 5′-CGCCTACATCGCTTTGG -3′ and 5′-TTGAATCGCAGCCATACC -3′ and an internal probe; 5′ –TET CTGATCCATGCCGGCA MGBNFQ -3′ in 25μl polymerase chain reaction mixtures using the fluorescent Taqman PCR system. Each reaction mixture contained 3μl double-distilled water, 12.5μl environmental mastermix, 1μl each of forward and reverse primers and 2.5μl probe. Cycling and detection of the Taqman fluorophore was performed in a BioRad thermocycler and sequence detector with parameters as follows: 95°C for 10 minutes, and 40 cycles of 95°C 15 seconds, 56°C for 30 seconds. Results were only considered if the standard curve correlation coefficient (R2) exceeded or was equal to .99, and if the log linear slope fell within the range of − 2.9 and −3.6. DNA in duplicate was rerun in instances where duplicate reactions did not yield similar results, or if above criteria were not met. Extrapolations were made for determination of M. ulcerans GU/sample.
2.7. Tissue culture
The adherent carp monocyte cell line CLC (European Collection of Cell Cultures no. 95070628) was used. Cells were maintained at 28°C and 5% CO2 using high glucose MEM (Gibco) supplemented with 10% essential amino acids (Gibco), 10% heat-inactivated fetal bovine serum (Gibco) and 2mM L-Glutamine as previously described in [36, 37]. Cytopathicity assays were performed in 24-well tissue culture plates.
2.8. Cytopathicity assays
Synthetically synthesized pure mycolactone AB and F from MU1615 and MMDL respectively were obtained from Yoshito Kishi (Harvard University) and used for cytopathicity assays according to previous published methods [27,38]. Cytopathicity was defined as the minimal concentration of mycolactone necessary to produce greater than 80% cell rounding in 24 h and loss of the monolayer by 48 h [38]. Mycolactone-treated cells were further analyzed to determine the extent of necrotic versus apoptotic cell death. Necrosis was measured using a colorimetric assay for release of lactate dehydrogenase (Promega) as previously described [38]. Briefly, cells were suspended in culture media and seeded in a 96 – well tissue culture plate. The release of cytoplasmic lactate dehydrogenase from mycolactone treated and a permeabilized cell was measured at 24 h post infection using the colorimetric kit following the manufacturer’s instructions. Background release of LDH was determined from lysis of ethanol treated cells according to the manufacturer’s protocol. The percentage of LDH released was then computed using the following calculation: [(release of LDH from mycolactone treated cells – background release from ethanol-treated cells)/ (maximum release of LDH by cell lysis – background release)] × 100.
Apoptosis of mycolactone treated CLC cells was measured at 24 h post infection by using the Cell Death Detection Plus enzyme-linked immunosorbent assay (Roche, Indianapolis, IN) as described previously in [38]. Apoptosis was determined as fold enrichment of nucleosomes [(measurement of DNA-histone complex from treated cells)/ (background measurement of untreated cells)].
2.9. Statistical analysis
Statistics were calculated using SPSS version 17 and GraphPad Prism version 4 software. For the analysis of cytotoxicity via apoptosis and LDH release, the Students t-test was used to determine significant differences between the congeners of mycolactone used. For the analysis of percent survival of medaka post infection with different mycobacteria, standard deviations were computed to determine significance. For analysis of numbers of infected organs that were AFB and ER-PCR positive within and between strains, the Mann-Whitney test for comparison of two groups was used to determine significance.
3. Results
3.1. M. ulcerans establishes initial systemic infection medaka
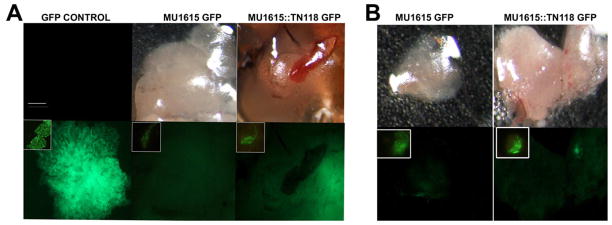
To determine the ability of M. ulcerans to establish an initial infection in medaka, 4 fish each infected with 108 CFU MU1615 GFP and MU1615::TN118 GFP were sacrificed 1 wk post infection, dissected and analyzed by fluorescent microscopy for GFP. Fluorescent M. ulcerans were detected in two target organs, the liver and kidney of the infected fish (Fig. 1A and B). To further substantiate our findings, the dissected organs were homogenized and sections were analyzed by microscopy. Wet mounts of the homogenized organs viewed under an epifluorescent microscope revealed the presence of GFP expressing bacteria (Fig. 1A and B inset). Smears made from dissected organs at each time point were stained using Ziehl – Neelsen stain and also revealed the presence of acid-fast bacilli (AFB) (Table 1) after infection. This observation was also comparable to the detection of bacterial DNA within the organs via ER-PCR. MM1218 and MMDL bacilli as well as DNA were also detected in organs following infection with 100% infectivity in MM1218 (Table 1).
Figure 1. M. ulcerans establishes initial infection in medaka.
Whole organ of dissected liver [A] and kidney [B] from 108 CFU MU1615 GFP and MU1615::TN118 GFP infected medaka expressing fluorescent bacteria 1 wk post-infection [inset of wet mount of respective homogenized organ showing fluorescent bacteria]. First two panels show PBS negative control and bacteria-only control slide of colony and wet mount [inset]. Scale bar = 50μm
Table 1.
Infected fish sections positive for acid fast bacilli (AFB) and bacterial DNA (PCR) at 7 and 60 days p.i. respectively.
| MU1615 GFP | MU1615:TN118 GFP | MMDL | MM1218 | PBS sham | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Days p.i. | 7 | 60 | 7 | 60 | 7 | 60 | 7 | 60 | 7 | 60 | |
| GUT | Microscopy | 6/8 | 7/8 | 7/8 | 8/8 | 8/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 |
| PCR | 4/8 | 6/8 | 6/8 | 6/8 | 7/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 | |
| HEART | Microscopy | 1/8 | 3/8 | 2/8 | 6/8 | 4/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 |
| PCR | 5/8 | 5/8 | 3/8 | 5/8 | 2/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 | |
| KIDNEY | Microscopy | 6/8 | 5/8 | 5/8 | 7/8 | 3/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 |
| PCR | 6/8 | 5/8 | 6/8 | 6/8 | 3/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 | |
| LIVER | Microscopy | 3/8 | 6/8 | 2/8 | 8/8 | 7/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 |
| PCR | 4/8 | 6/8 | 5/8 | 5/8 | 6/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 | |
| SKIN | Microscopy | 6/8 | 7/8 | 7/8 | 8/8 | 8/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 |
| PCR | 7/8 | 7/8 | 7/8 | 6/8 | 8/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 | |
| SPLEEN | Microscopy | 3/8 | 5/8 | 3/8 | 8/8 | 3/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 |
| PCR | 4/8 | 5/8 | 4/8 | 8/8 | 2/8 | 8/8 | 8/8 | 8/8 | 0/8 | 0/8 | |
3.2. M. ulcerans is avirulent in medaka
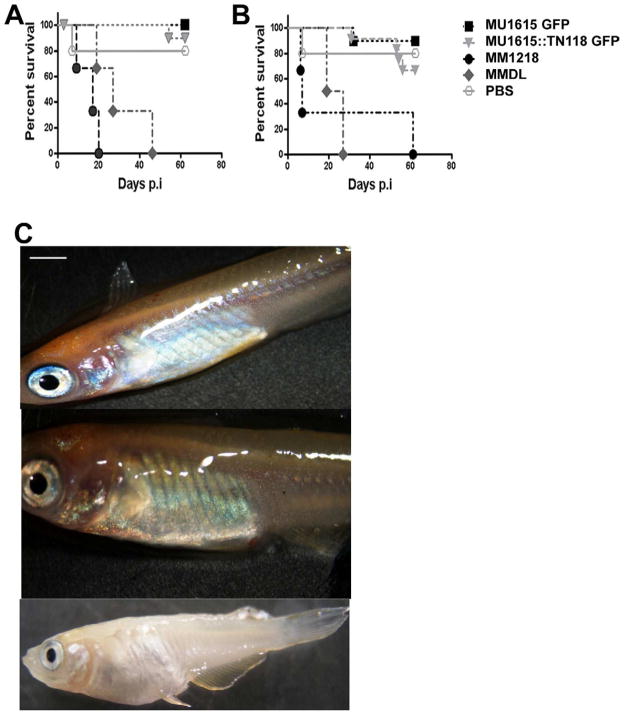
In zebrafish, medaka and goldfish, an infective dose of >105 CFU M. marinum will cause an acute lethal infection whereas injection of < 103 CFU produces chronic granulomatous disease at concentrations below [30, 31, 39]. In a previous experiment we infected medaka with 102–108 CFU MU1615 GFP to establish the appropriate dose for later experiments. To determine whether M. ulcerans is pathogenic to medaka and whether presence of mycolactone affects virulence, 10 fish per strain per dose were inoculated with 104 and 108 CFU of MU1615 GFP, MU1615::TN118 GFP, MMDL and MM1218. Infected fish were monitored for the development of disease and/or death. At low (104) and high (108) doses, 75% of MU1615 GFP and MU1615::TN118 GFP infected fish survived up to 8 wks post infection (Fig. 2A and 2B) when they were sacrificed. In stark contrast to the above, 50% of medaka infected with 104 and 108 MM1218 CFU were dead by 2 wks pi and 1wk post infection respectively (Fig. 2A and 2B). The death rate for medaka infected with MMDL was similar to that of MM1218 although the time to death was somewhat retarded. Gross inspection of medaka infected with 108 CFU of MU1615 GFP and MU1615::TN118 GFP at 60 days p.i. appeared comparable to PBS control fish where no external lesions or bloating were seen (Fig. 2C). In contrast, medaka infected with 108 CFU of MM1218 and MMDL were slightly bloated prior to death and enlarged organs were seen post dissection. External lesions were not present (Fig. 2C). This experiment was repeated with similar results.
Figure 2. M. ulcerans is avirulent in medaka.
Percent survival of medaka infected with 104 [A] and 108 [B] CFU of MU1615 GFP, MU1615::TN118 GFP, MM1218 and MMDL. [C] Gross morphology of medaka 8wks post infection with PBS [top panel], 108 CFU MU1615 GFP[middle panel] and 108 CFU MM1218 [bottom panel]. Scale bar = 10mm.
3.3. Survival of M. ulcerans in medaka is not Mycolactone-dependent
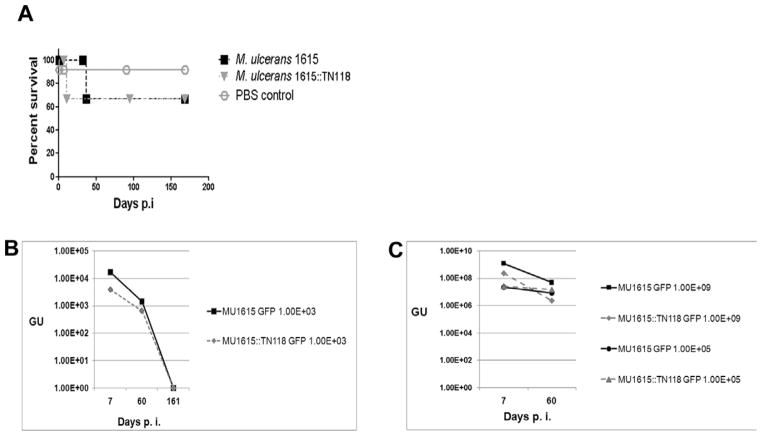
M. marinum causes lethal infections in natural populations of fish as well as in laboratory infections using both medaka and zebrafish [23, 40]. The lack of mortality in M. ulcerans infected fish led us to investigate the ability of M. ulcerans to replicate in fish, as well as to determine whether mycolactone plays a role in this phenotype. We established a chronic infection of both strains by administering 102 CFU of MU1615 GFP and MU1615::TN118 GFP to 30 fish per strain and compared observations over a 23 week infection period. The bacteria load at 1, 8 and 23 wks for each fish was monitored by microscopy, conventional PCR and qPCR. Over 70% (21 out of 30) of fish infected with either strain survived up to 23 wks post infection when they were sacrificed (Fig. 3A). During the 23 week infection period both MU1615 GFP and MU1615::TN118 GFP GU declined and by 23 weeks neither strain could be detected in the kidney by qPCR (Fig 3B). There was no significant difference in the numbers of fish that were positive for either bacterium at all the time points assessed in this study by either microscopy or PCR. We also determined the representative GU in the kidney of medaka infected with 104 CFU of MU1615 GFP and MU1615::TN118 GFP from the previous study and our data shows a slight but insignificant decline in the GU by 8 wks post infection (Fig. 3C). These results suggest that neither strain actively replicates within medaka. Furthermore the presence of mycolactone does not seem to confer a survival advantage to M. ulcerans within medaka.
Figure 3. Survival of M. ulcerans in medaka is not mycolactone dependant.
[A] Percent survival of medaka infected with 102 CFU of MU1615 GFP and MU1615::TN118 GFP. [B] Genome equivalent of MU1615 GFP and MU1615::TN118GFP in medaka infected with 102 CFU bacteria. [C] Genome equivalent of MU1615 GFP and MU1615::TN118GFP in medaka infected with 104 and 108 CFU bacteria. Data are mean log genome forming units [GFU] of MU1615GFP and MU1615::TN118 GFP infected medaka at 1, 8 and 23 wks p.i. R2=0.99
3.4. M. ulcerans causes minimal histopathology in medaka
Results from analysis of H&E stained sections generally reflected bacterial load and corresponding inflammatory response. Whereas massive numbers of AFBs were detected in stained sections prepared from MM1218 and MMDL infected fish, very few AFBs were detected in the organs of MU1615 GFP and MU1615::TN118 GFP infected fish AT 8 weeks p.i. (Fig. 4). PBS sham infected fish revealed no AFB (Fig. 4A), influx of inflammatory cells or granuloma (Fig. 4B). Very few AFB were detected in the kidney (Fig. 4C) and peritoneum of MU1615 infected medaka and little inflammatory response was present (Fig. 4D). Interestingly, MU1615::TN118 infected medaka showed small clusters of intracellular and extracellular AFB (Fig. 4E, arrows) and a greater inflammatory response than MU1615 (Fig 4F). Both intracellular and extracellular bacteria (Fig. 4G and 4I arrows) that were associated with granulomatous lesions (Fig. 4H, 4J, arrows) were seen in the kidneys of MM1218 and MMDL infected fish. Small clusters of extracellular bacteria were detected scattered in the entire gastrointestinal tract of MU1615 GFP and MU1615::TN118 GFP infected fish; a finding not observed in infections with other strains. These observations further confirmed our findings that M. ulcerans does not cause disease or replicate in medaka.
Figure 4. Histopathology of medaka kidney with 104 CFU of MU1615 GFP, MU1615::TN118 GFP, MM1218 and MMDL.
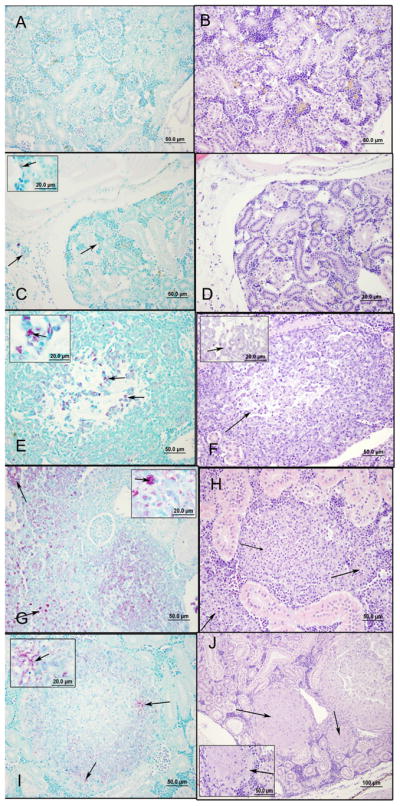
All sections were fixed and stained with Ziehl – Neelsen stain [left panels] and hematoxylin and eosin stain [right panels]. [A and B] PBS negative control; [C and D] MU1615 GFP infected medaka showing few intracellular bacteria [C- arrow and inset];little inflammatory response [D]; [E and F] MU1615::TN118 GFP infected medaka showing scattered pockets of intracellular and extracellular bacteria [E– arrow and inset] and diffuse inflammatory response [F]; [G and H] MM1218 infected medaka showing intracellular and extracellular bacteria [G – arrow and inset] and associated granuloma [H]; [I and J] MMDL infected medaka showing AFB bacteria [I] and associated well organized granuloma with necrotic centers [J – arrow]. Data represents fish sacrificed at 8 wks post infection. Scale bars = 50 μm for all panels and 20μm for insets
3.5. Effects of mycolactone on cultured fish cells
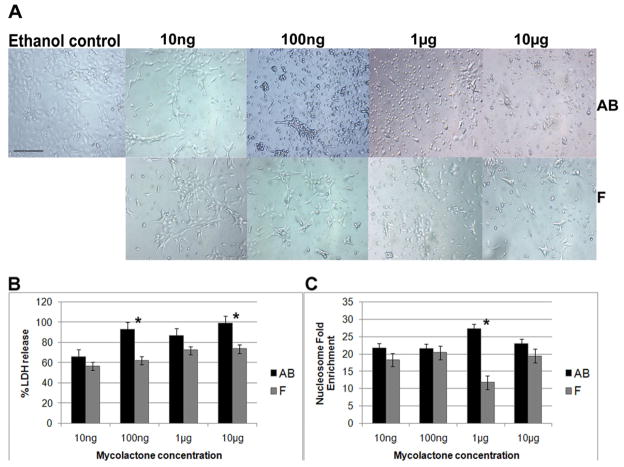
One explanation for the lack of M. ulcerans virulence for medaka could be that fish in general are not susceptible to the toxic effects of mycolactone. The cellular effects of mycolactone AB (M. ulcerans) and mycolactone F (MMDL and M. pseudoshottsii) on cultured murine and human cell lines and human neutrophils have been well characterized [31]. Mycolactone cytotoxicity is characterized by morphological changes and rapid necrosis at concentrations above 1 μg/ml and delayed apoptosis at concentrations as low as 1ng/ml within 24 h. To determine the effect of mycolactone on cultured CLC fish macrophages, a semi-confluent layer of cells was treated with 10 fold dilutions of mycolactone AB or mycolactone F between 10ng and 10μg and evaluated for morphological and biochemical evidence of toxicity. At 10 ng, cells treated with both mycolactones showed cell rounding with a non confluent monolayer (Fig. 5A). At concentrations above 1μg/ml, mycolactone AB treated cells appeared rounded and swollen and the density of the monolayer was very sparse compared to the control monolayer. By 48h post treatment, the cell monolayer had completely detached. Cytotoxic effects were also observed with mycolactone F treated cells; however, the phenotype was not as profound as for mycolactone AB treated cells (Fig. 5A).
Figure 5. Analysis of Mycolactone – mediated cytopathicity on the fish macrophages.
[A] Cytopathicity on CLC cells, showing ethanol control treated cells [first left]; mycolactone AB treated cells [top panel] and mycolactone F treated cells [bottom panel]. The final concentration of mycolactone added to treated cells was 10μg. Scale bar = 50 μm [B and C] Cytotoxicity measured by LDH release and nucleosome enrichment. Culture supernatants were collected from wells containing CLC cells 24 h after treatment with mycolactone. [B] The amount of LDH released by necrosis was measured using a Cytotox 96 assay kit [Promega]. Data are means and standard deviations of the values obtained from triplicate samples; P=0.7, 0.03, 0.06 and 0.04 for 10ng, 100ng, 1μg and 10μg respectively [Student’s t test]. [C] Apoptosis was assessed at 24 h with the cell death detection enzyme-linked immunosorbent assay kit [Roche] and expressed as fold enrichment of nucleosomes. Data are means and standard deviations of the values obtained from triplicate samples; P=0.06, 0.9, 0.02 and 0.06 for 10ng, 100ng, 1μg and 10μg respectively [Student’s t test].
To determine the mechanism of mycolactone – mediated toxicity, either mycolactone AB or F was added to a semi-confluent layer of CLC cells and cells were assayed for necrosis and apoptosis at 24h (Fig. 5B, 5C). Mycolactone AB produced more necrosis on CLC cells at all concentrations tested compared to necrosis produced by mycolactone F and this difference was significant for the 100ng [p=0.03] and 10 μg [p=0.04] doses (Fig. 5B). Although mycolactone AB treated CLC cells showed a slightly higher level of apoptosis compared to mycolactone F treated cells at all concentrations (Fig. 5C), this difference was only significant at 1 μg [p=0.02]. There was a significant dose response in death by necrosis in cells treated with 10ng and 100ng mycolactone AB as measured by LDH release with 100ng of the toxin being sufficient to cause 90% necrosis in the cell monolayer. Due to the significant depletion of the monolayer after 100ng of mycolactone AB treatment, death by apotosis did not follow the standard dose-response trend as is observed in human cells [39]. Mycolactone F appeared to induce less cell death by necrosis in CLC cells compared to mycolactone AB there was a slight dose-response in the concentration of toxin used. There was no dose response cell death due to apoptosis in cells treated with mycolactone F which cannot be explained. All experiments were performed more than once and in triplicate. These results show that CLC cells are sensitive to both mycolactone AB and F; however mycolactone AB appears to be somewhat more toxic to CLC cells than mycolactone F.
4. Discussion
M. ulcerans and M. marinum share 99% identity in 16sRNA gene sequence. A large body of molecular and whole-genome analyses suggests that M. ulcerans has evolved from an M. marinum- like ancestor [10] through reductive evolution. M. marinum is a well characterized fish pathogen [23, 30, 32, 40] which can also cause a granulomatous skin disease in humans and may occasionally invade deeper tissue [41]. In a review of over 190 cases with known exposure over 75% of the cases could be linked to contact with aquaria, fish or shellfish [41]. In contrast, although M. ulcerans DNA has been tentatively identified in many aquatic samples including fish, the primary reservoirs in the environment have not been identified. The transmission of M. ulcerans from the environment to humans is one of the central mysteries of Buruli ulcer.
In this study we have used a well-established medaka fish-infection model to assess the pathogenic potential of M. ulcerans in a fish host. Our results show that at an infective dose of about 104 M. marinum cells caused both disease and mortality in medaka, whereas M. ulcerans was avirulent even at the much higher infective dose of 108 organisms. Of particular interest is the fact that although M. ulcerans DNA is widely distributed in fish organs by 1 week post infection, few bacteria can be observed in organs by microscopy at 8 weeks and after 23 weeks M. ulcerans DNA cannot be detected in infected fish. Consistent with these findings is the fact that little histopathology is associated with M. ulcerans infection. In medaka, M. marinum was found to be highly lethal and massive numbers of M. marinum were detected at 8 weeks in all organs of the surviving moribund fish. Histopathology of M. marinum infected organs shows characteristic granuloma formation with many intra-and extracellular bacteria present. Although it is possible that M. ulcerans may cause infection in other fish species we think this unlikely for the following reason: neither M. marinum nor M. ulcerans are characterized by a high degree of host specificity [11, 42]. M. marinum causes disease in a wide spectrum of fish and frog species [25, 26] and M. ulcerans is pathogenic for many vertebrates including several species of marsupials, horses and cats [43]. Although initial studies reported the presence of M. ulcerans DNA in fish, this identification of M. ulcerans was based on a PCR detection assay of IS2404 sequence [14]. The presence of M. ulcerans DNA in fish has not been confirmed using more specific DNA probes although large scale studies of fish have not yet been conducted. Results from this study are consistent with epidemiologic studies showing that fishermen working in endemic waters are actually at low risk for acquiring M. ulcerans infection [12].
This work does not support the role of mycolactone as a virulence determinant for mycobacterial infection in fish either in M. ulcerans or M. marinum. In fact, a greater inflammatory response was found following infection with a mycolactone-negative M. ulcerans mutant compared to wild type M. ulcerans although neither were virulent. Even though mycolactone-producing M. marinum have been identified as a cause of disease in over 30 species of fish from the Red Sea [26], there could be numerous reasons for the severity of disease in this habitat including degradation of water quality. Alternatively other pathogens may have initiated these infections in the Red Sea and M. marinum may have subsequently colonized these animals as an opportunist. The preliminary studies presented here suggests the following; first, the pathogenesis M. marinum for fish is much greater than that of M. ulcerans, and second, that expressing mycolactones did not significantly increase virulence for either of these two mycobacteria.
The genetic coding capacity for mycolactone is considerable, requiring 100 kb of DNA and considerable reducing power. The metabolic cost of producing the molecule is high. A key question then is, “Why do mycobacteria make mycolactone?” Evidence suggests that mycolactone does not play an obvious role in the virulence of M. ulcerans for fish. The role of the molecule in M. marinum infection especially requires further study since our studies compared the virulence of two different isolates of M. marinum rather than the virulence of isogenic strains with and without the mycolactone. Conclusive evidence for the role of mycolactone in M. marinum fish disease must await availability of an isogenic mycolactone-negative M. marinum DL mutant. Our biochemical data shed little light on the question since both mycolactone AB and F show high potency for cultured CLC fish phagocytes. Further studies to determine in vivo transcription of mycolactone genes in fish could be very informative.
The apparent inability of M. ulcerans to productively colonize medaka is intriguing from the evolutionary standpoint and adds to the evidence that novel niche adaptation may play a role in the evolution of M. ulcerans from an M. marinum-like ancestor. In this regards it is particularly interesting that mycolactone-producing M. marinum strains appear to have a chromosomal gene repertoire more similar to that of M. marinum than to M. ulcerans [44].
In conclusion, our data suggest that M. ulcerans is not capable of mounting a productive infection in fish and does not support the hypothesis based on epidemiological evidence shown by IS2404-PCR that fish may be a reservoir for M. ulcerans. Whether the acquisition of the mycolactone plasmid by M. marinum results in altered virulence requires further study with genetically well characterized strains.
Acknowledgments
We are grateful to Dr. Jeffery Cirillo for the kind gift of the gold fish CLC macrophages (Texas A&M Health Science Center). We thank Amrita Mallick and Jesse Soileau for assistance with maintenance of the medaka. We thank Lalita Ramakrishnan for the kind gift of the plasmid for the construction of the strain MU1615 GFP and the mycolactone-negative strain MU161::TN118 GFP. Finally we are grateful to the Afrique One consortium for the support of Buruli ulcer research through its postdoctoral fellowship.
This research was supported in part by the NIH National Institute of Allergy and Infectious Diseases Grant Award 1 R03 AI026719-01A1, the UBS Optimus Foundation - Stop Buruli Consortium, Louisiana Board of Regents Research and Development Grant (RD01-A-38) and NIH grant (5R21AI055964-01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. Buruli ulcer: diagnosis of Mycobacterium ulcerans disease. World Health Organization; Geneva, Switzerland: 2004. [Google Scholar]
- 2.Bayley AC. Buruli ulcer in Ghana. Br Med J. 1971;2:401–402. doi: 10.1136/bmj.2.5758.401-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pettit JH, Marchette NJ, Rees RJ. Mycobacterium ulcerans infection. Clinical and bacteriological study of the first cases recognized in South East Asia. Br J Dermatol. 1966;78:187–97. doi: 10.1111/j.1365-2133.1966.tb12204.x. [DOI] [PubMed] [Google Scholar]
- 4.Smith JH. Epidemiologic observations on cases of Buruli ulcer seen in a hospital in the Lower Congo. Am J Trop Med Hyg. 1970;19:657–63. doi: 10.4269/ajtmh.1970.19.657. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell PJ, Jerrett IV, Slee KJ. Skin ulcers caused by Mycobacterium ulcerans in koalas near Bairnsdale, Australia. Pathology. 1984;16:256–60. doi: 10.3109/00313028409068533. [DOI] [PubMed] [Google Scholar]
- 6.Kaser M, Rondini S, Naegeli M, Stinear T, Portaels F, Certa U, Pluschke G. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evol Biol. 2007;7:177. doi: 10.1186/1471-2148-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George KM, Chatterjee D, Gunawardana G, Welty D, Hayman J, Lee R, Small PL. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 1999;283:854–7. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- 8.Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PD, Davies JK, Lee RE, Adusumilli S, Garnier T, Haydock SF, Leadlay PF, Cole ST. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A. 2004;101:1345–9. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Käser M, Hauser J, Small P, Pluschke G. Large sequence polymorphisms unveil the phylogenetic relationship of environmental and pathogenic mycobacteria related to Mycobacterium ulcerans. Appl Environ Microbiol. 2009;75:5667–75. doi: 10.1128/AEM.00446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, Meurice G, Simon D, Bouchier C, Ma L, Tichit M, Porter JL, Ryan J, Johnson PD, Davies JK, Jenkin GA, Small PL, Jones LM, Tekaia F, Laval F, Daffé M, Parkhill J, Cole ST. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;17(20):192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson PD, Stinear T, Small PL, Pluschke G, Merritt RW, Portaels F, Huygen K, Hayman JA, Asiedu K. Buruli ulcer “M. ulcerans infection”: new insights, new hope for disease control. PLoS Med. 2005;2:e108. doi: 10.1371/journal.pmed.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stienstra Y, van der Graaf WT, te Meerman GJ, The TH, de Leij LF, van der Werf TS. Susceptibility to development of Mycobacterium ulcerans disease: review of possible risk factors. Trop Med Int Health. 2001;6:554–62. doi: 10.1046/j.1365-3156.2001.00746.x. [DOI] [PubMed] [Google Scholar]
- 13.Portaels F, Meyers WM, Ablordey A, Castro AG, Chemlal K, de Rijk P, Elsen P, Fissette K, Fraga AG, Lee R, Mahrous E, Small PL, Stragier P, Torrado E, Van Aerde A, Silva MT, Pedrosa J. First Cultivation and Characterization of Mycobacterium ulcerans from the Environment. PLoS Negl Trop Dis. 2008;2:e178. doi: 10.1371/journal.pntd.0000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eddyani M, Ofori-Adjei D, Teugels G, De Weirdt D, Boakye D, Meyers WM, Portaels F. Potential role for fish in transmission of Mycobacterium ulcerans disease “Buruli ulcer”: an environmental study. Appl Environ Microbiol. 2004;70:5679–81. doi: 10.1128/AEM.70.9.5679-5681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsollier L, Sévérin T, Aubry J, Merritt RW, Saint André JP, Legras P, Manceau AL, Chauty A, Carbonnelle B, Cole ST. Aquatic snails, passive hosts of Mycobacterium ulcerans. Appl Environ Microbiol. 2004;70:6296–8. doi: 10.1128/AEM.70.10.6296-6298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portaels F, Elsen P, Guimaraes-Peres A, Fonteyne PA, Meyers WM. Insects in the transmission of Mycobacterium ulcerans infection. Lancet. 1999;353:986. doi: 10.1016/S0140-6736(98)05177-0. [DOI] [PubMed] [Google Scholar]
- 17.Williamson HR, Benbow ME, Nguyen KD, Beachboard DC, Kimbirauskas RK, McIntosh MD, Quaye C, Ampadu EO, Boakye D, Merritt RW, Small PL. Distribution of Mycobacterium ulcerans in Buruli Ulcer Endemic and Non-Endemic Aquatic Sites in Ghana. PLoS Negl Trop Dis. 2008;2:e205. doi: 10.1371/journal.pntd.0000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swart CC, Deaton LE, Felgenhauer BE. The salivary gland and salivary enzymes of the giant waterbugs (Heteroptera; Belostomatidae) Comp Biochem Physiol A Mol Integr Physiol. 2006;145:114–22. doi: 10.1016/j.cbpa.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Mosi L, Williamson H, Wallace JR, Merritt RW, Small PL. Persistent association of Mycobacterium ulcerans with West African predaceous insects of the family belostomatidae. Appl Environ Microbiol. 2008;74:7036–7042. doi: 10.1128/AEM.01234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heckert RA, Elankumaran S, Milani A, Baya A. Detection of a new Mycobacterium species in wild striped bass in the Chesapeake Bay. J Clin Microbiol. 2001;39:710–5. doi: 10.1128/JCM.39.2.710-715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes MW, Kator H, Kotob S, van Berkum P, Kaattari I, Vogelbein W, Floyd MM, Butler WR, Quinn FD, Ottinger C, Shotts E. A unique Mycobacterium species isolated from an epizootic of striped bass “Morone saxatilis”. Emerg Infect Dis. 2001;7:896–9. doi: 10.3201/eid0705.017523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes MW, Kator H, McNabb A, Deshayes C, Reyrat JM, Brown-Elliott BA, Wallace R, Trott KA, Parker JM, Lifland B, Osterhout G, Kaattari I, Reece K, Vogelbein W, Ottinger CA. Mycobacterium pseudoshottsii sp. nov., a slowly growing chromogenic species isolated from Chesapeake Bay striped bass “Morone saxatilis”. Int J Syst Evol Microbiol. 2005;55:1139–47. doi: 10.1099/ijs.0.63343-0. [DOI] [PubMed] [Google Scholar]
- 23.Decostere A, Hermans K, Haesebrouck F. Piscine mycobacteriosis: a literature review covering the agent and the disease it causes in fish and humans. Vet Microbiol. 2004;99:159–66. doi: 10.1016/j.vetmic.2003.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Kent ML, Whipps CM, Matthews JL, Florio D, Watral V, Bishop-Stewart JK, Poort M, Bermudez L. Mycobacteriosis in zebrafish “Danio rerio” research facilities. Comp Biochem Physiol C Toxicol Pharmacol. 2004;138:383–90. doi: 10.1016/j.cca.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Colorni A. A systemic mycobacteriosis in the European sea bass Dicentrarchus labrax cultured in Eilat “Red Sea”. Isr J Aquacult Bamidgeh. 1992;44:75–81. [Google Scholar]
- 26.Ucko M, Colorni A. Mycobacterium marinum infections in fish and humans in Israel. J Clin Microbiol. 2005;43:892–5. doi: 10.1128/JCM.43.2.892-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranger BS, Mahrous EA, Mosi L, Adusumilli S, Lee RE, Colorni A, Rhodes M, Small PL. Globally distributed mycobacterial fish pathogens produce a novel plasmid-encoded toxic macrolide, mycolactone F. Infect Immun. 2006;74:6037–45. doi: 10.1128/IAI.00970-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mve-Obiang A, Lee RE, Portaels F, Small PL. Heterogeneity of mycolactones produced by clinical isolates of Mycobacterium ulcerans: implications for virulence. Infect Immun. 2003;71:774–783. doi: 10.1128/IAI.71.2.774-783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talaat AM, Trucksis M, Kane AS, Reimschuessel R. Pathogenicity of Mycobacterium fortuitum and Mycobacterium smegmatis to goldfish, Carassius auratus. Vet Microbiol. 1999;66:151–164. doi: 10.1016/s0378-1135(99)00002-4. [DOI] [PubMed] [Google Scholar]
- 30.Prouty MG, Correa NE, Barker LP, Jagadeeswaran P, Klose KE. Zebrafish-Mycobacterium marinum model for mycobacterial pathogenesis. FEMS Microbiol Lett. 2003;225:177–82. doi: 10.1016/S0378-1097(03)00446-4. [DOI] [PubMed] [Google Scholar]
- 31.Broussard GW, Ennis DG. Mycobacterium marinum produces long-term chronic infections in Medaka: a new animal model for studying human tuberculosis. Comp Biochem Physiol C Toxicol Pharmacol. 2007;145:45–54. doi: 10.1016/j.cbpc.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shima A, Mitani H. Medaka as a research organism: past, present and future. Mech Dev. 2004;121:599–604. doi: 10.1016/j.mod.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Goodrich HB. The Development of Mendelian Characters in Aplocheilus Latipes. Proc Natl Acad Sci U S A. 1926;12:649–52. doi: 10.1073/pnas.12.11.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naruse K, Tanaka M, Mita K, Shima A, Postlethwait J, Mitani H. A Medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 2004;14:820–8. doi: 10.1101/gr.2004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamour K, Finley L. A strategy for recovering high quality genomic DNA from a large number of Phytophthora isolates. Mycologia. 2006;98:514–7. doi: 10.3852/mycologia.98.3.514. [DOI] [PubMed] [Google Scholar]
- 36.El-Etr SH, Yan L, Cirillo JD. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect Immun. 2001;69:7310–7. doi: 10.1128/IAI.69.12.7310-7317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faisal M, Ahne W. A cell line “CLC” of adherent peripheral blood mononuclear leucocytes of normal common carp Cyprinus carpio. Dev Comp Immunol. 1990;14:255–60. doi: 10.1016/0145-305x(90)90097-x. [DOI] [PubMed] [Google Scholar]
- 38.Adusumilli S, Mve-Obiang A, Sparer T, Meyers W, Hayman J, Small PL. Mycobacterium ulcerans toxic macrolide, mycolactone modulates the host immune response and cellular location of M. ulcerans in vitro and in vivo. Cell Microbiol. 2005;7:1295–304. doi: 10.1111/j.1462-5822.2005.00557.x. [DOI] [PubMed] [Google Scholar]
- 39.Talaat AM, Reimschuessel R, Wasserman SS, Trucksis M, Goldfish Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect Immun. 1998;66:2938–42. doi: 10.1128/iai.66.6.2938-2942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aslam F, Ng B. You never asked Doc. I do fish. Clin Rheumatol. 2010;29:691–3. doi: 10.1007/s10067-009-1270-4. [DOI] [PubMed] [Google Scholar]
- 41.Jernigan JA, Farr BM. Incubation period and sources of exposure for cutaneous Mycobacterium marinum infection: case report and review of the literature. Clin Infect Dis. 2000;2:439–43. doi: 10.1086/313972. [DOI] [PubMed] [Google Scholar]
- 42.Marcus LC, Stottmeier KD, Morrow RH. Experimental infection of anole lizards “Anolis carolinensis” with Mycobacterium ulcerans by the subcutaneous route. Am J Trop Med Hyg. 1975;24:649–55. doi: 10.4269/ajtmh.1975.24.649. [DOI] [PubMed] [Google Scholar]
- 43.van Zyl A, Daniel J, Wayne J, McCowan C, Malik R, Jelfs P, Lavender CJ, Fyfe JA. Mycobacterium ulcerans infections in two horses in southeastern Australia. Aust Vet J Mar. 2010;88(3):101–6. doi: 10.1111/j.1751-0813.2009.00544.x. Review. [DOI] [PubMed] [Google Scholar]
- 44.Yip MJ, Porter JL, Fyfe JA, Lavender CJ, Portaels F, Rhodes M, Kator H, Colorni A, Jenkin GA, Stinear T. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J Bacteriol. 2007;189:2021–29. doi: 10.1128/JB.01442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]