SUMMARY
Fbw7 is the substrate recognition component of the SCF (Skp1-Cullin-F-box)-type E3 ligase complex and a well-characterized tumor suppressor that targets numerous oncoproteins for destruction. Genomic deletion or mutation of FBW7 has been frequently found in various types of human cancers, however, little is known about the upstream signaling pathway(s) governing Fbw7 stability and cellular functions. Here we report that Fbw7 protein destruction and tumor suppressor function are negatively regulated by the prolyl isomerase Pin1. Pin1 interacts with Fbw7 in a phoshorylation-dependent manner and promotes Fbw7 self-ubiquitination and protein degradation by disrupting Fbw7 dimerization. Consequently, over-expressing Pin1 reduces Fbw7 abundance and suppresses Fbw7’s ability to inhibit proliferation and transformation. By contrast, depletion of Pin1 in cancer cells leads to elevated Fbw7 expression, which subsequently reduces Mcl-1 abundance, sensitizing cancer cells to Taxol. Thus, Pin1-mediated inhibition of Fbw7 contributes to oncogenesis and Pin1 may be a promising drug target for anti-cancer therapy.
INTRODUCTION
SCFFbw7 is a multi-component RING-type E3 ligase involved in the regulation of numerous cellular processes by promoting the degradation of critical regulatory proteins including cyclin E (Koepp et al., 2001; Strohmaier et al., 2001), c-Myc (Welcker et al., 2004; Yada et al., 2004), c-Jun (Nateri et al., 2004; Wei et al., 2005), NOTCH-1 (Gupta-Rossi et al., 2001; Oberg et al., 2001; Wu et al., 2001), Sterol regulatory element binding protein-1 (SREBP1) (Punga et al., 2006), and Mcl-1 (Inuzuka et al., 2011; Wertz et al., 2011). The F-box protein Fbw7 is a vital component of this complex, as it is the substrate recognition subunit whose primary function is to bind phosphorylated targets (Cardozo and Pagano, 2004; Petroski and Deshaies, 2005; Skowyra et al., 1997). As it plays such an important role in the overall function of this E3 ligase, dysregulation of Fbw7 leads to various pathological diseases notably cancer (Crusio et al., 2010; Welcker and Clurman, 2008).
Characterized substrates recognized by Fbw7, including cyclin E, c-Jun, c-Myc, Mcl-1 and NOTCH-1, are well-known oncogenes involved in a variety of human tumors (Nakayama and Nakayama, 2005; Welcker and Clurman, 2008). Thus, it has been well documented that Fbw7 is a tumor suppressor whose mutation occurs in multiple neoplasms including colon cancer, breast cancer, and leukemia (Akhoondi et al., 2007; Malyukova et al., 2007; Strohmaier et al., 2001). However, although earlier yeast studies have indicated that Cdc4, the yeast homologue of Fbw7 could undergo self-ubiquitination (Galan and Peter, 1999; Pashkova et al., 2010; Zhou and Howley, 1998), so far nothing is known of the upstream signaling pathways that govern Fbw7 stability and/or function. Recent studies suggest that Pin1 plays a critical role in regulating the stability of various phosphoproteins (Liou et al., 2011) including most Fbw7 substrates, such as Mcl-1 (Ding et al., 2008) and c-Jun (Wulf et al., 2001), but it is currently unknown whether Pin1 could directly regulate the stability and/or function of Fbw7, or any other F-box protein.
Pin1 is the only peptidyl-prolyl cis/trans isomerase (PPIase) that binds to and isomerizes specific phosphorylated Ser/Thr-Pro (pSer/Thr-Pro) motifs in a subset of proteins, resulting in conformational changes in the proteins (Lu and Zhou, 2007; Nakamura et al., 2012; Tun-Kyi et al., 2011). These Pin1-induced conformational changes have been shown to regulate various protein functions, including protein stability, catalytic activity, phosphorylation status, protein-protein interactions, and/or subcellular localization (Liou et al., 2011; Lu and Zhou, 2007; Nakamura et al., 2012; Wulf et al., 2005).
Given the important role for Pin1 in regulating proline-directed phosphorylation, Pin1 has a pivotal role in a variety of biological processes, and its deregulation contributes to various pathological conditions, most notably cancer (Liou et al., 2011; Lu, 2004; Lu and Zhou, 2007; Tun-Kyi et al., 2011). Pin1 is overexpressed and also activated due to loss of its inhibitor kinase DAPK1, a tumor suppressor, in various human cancers, which contributes to centrosome amplification, chromosome instability and tumor development in vitro and in vivo, and its overexpression correlates with poor clinical outcomes in human cancer patients (Ayala et al., 2003; Bao et al., 2004; Lee et al., 2011a; Ryo et al., 2001; Suizu et al., 2006). In contrast, inhibition of Pin1 suppresses tumorigenesis in vitro (Ryo et al., 2002) and prevents cancer development induced by overexpression of oncogenes such as Neu or Ras (Wulf et al., 2004) or by knockout of tumor suppressors such as p53 (Takahashi et al., 2007) in mice. Consistent with these oncogenic phenotypes, Pin1 could either activate a number of oncogenes such as c-Jun (Wulf et al., 2001), Mcl-1 (Ding et al., 2008), NOTCH-1 (Rustighi et al., 2009), c-Myb (Pani et al., 2008) and Steroid Receptor Coactivator 3 (SRC-3) (Yi et al., 2005); or inactivate multiple tumor suppressors including p53 (Girardini et al., 2011; Takahashi et al., 2007), Promyelocytic Leukemia Protein (PML) (Yuan et al., 2011), FOXOs (Brenkman et al., 2008) and SMRT (Stanya et al., 2008). In doing so, Pin1 amplifies various oncogenic pathways to facilitate cancer development, thus making it an attractive anti-cancer target (Lee et al., 2011b; Lu and Zhou, 2007). However, although Pin1 has been shown to regulate the stability of many proteins, nothing is known about whether Pin1 might directly act on any F-box proteins such as the Fbw7 tumor suppressor.
RESULTS
Fbw7 Abundance Inversely Correlates with Pin1 Expression in Human Cancer Tissues, and Is Regulated via the Proteasomal Degradation Pathway
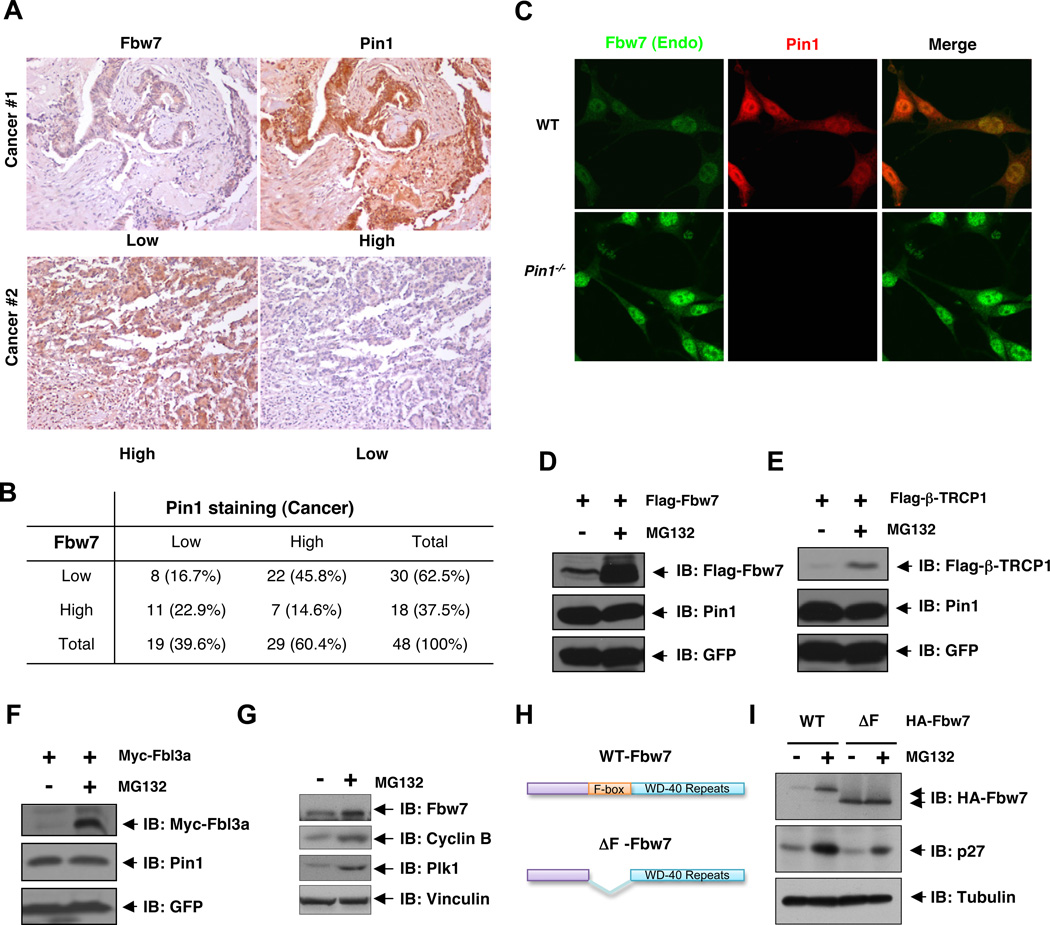
Loss of Fbw7 tumor suppressor expression is frequently observed in various human cancers, which could be attributed to the genetic deletion or mutation of the Fbw7 locus. However, these Fbw7 genetic changes are only observed in up to 6% of all primary human tumors (Akhoondi et al., 2007; Malyukova et al., 2007; Strohmaier et al., 2001; Welcker and Clurman, 2008). Therefore, it remains largely unknown what other mechanisms may contribute to reduced Fbw7 tumor suppressor functions in cancers. Accumulated evidence have demonstrated that the activities of multiple Fbw7 downstream targets are subject to regulation by Pin1 (Lee et al., 2011b; Liou et al., 2011; Lu and Zhou, 2007), suggesting that Pin1 might be involved in regulating the Fbw7 tumor suppressor pathway. Notably, we observed an obvious inverse correlation between Fbw7 and Pin1 abundance in colon cancer clinical samples (Figures 1A–B and S1A). Additionally, an increased expression of Fbw7 was also observed in Pin1−/− MEFs when compared to Pin1 WT MEFs (Figures 1C and S1C), further supporting a possible signaling link between the Pin1 oncoprotein and the Fbw7 tumor suppressor. Surprisingly, depletion of Pin1 did not lead to upregulation of Fbw7 mRNA levels (Figure S1E), arguing for a post-transcriptional regulation of Fbw7 by Pin1.
Figure 1. Fbw7 Abundance Is Inversely Correlated with Pin1 Expression in Human Colon Cancer Tissues, and Is Regulated via the Proteasomal Degradation Pathway.
A–B. Fbw7 abundance is inversely correlated with Pin1 expression in human colon cancer. Serial sections of tissue arrays of 48 colon cancer specimens were subjected to immunohistochemistry with anti-Pin1 and anti-Fbw7 antibodies, and visualized by DAB staining (A). In each sample, Fbw7 and Pin1 levels were semi-quantified in a double-blind manner as high or low according to the standards presented in (A) and summarized in (B). Their correlation was analyzed by Spearman rank correlation test (p < 0.01).
C. The abundance of endogenous Fbw7 is increased in Pin1−/− Mouse Embryonic Fibroblasts (MEFs). Wild type (WT) and Pin1−/− MEFs were subjected to immunofluoresence analysis with anti-Fbw7 (green) and anti-Pin1 (red) immuno-staining.
D–F. The abundance of ectopically expressed Fbw7 (D), β-TRCP1 (E), and Fbl3a (F) are regulated via proteasomal degradation. HeLa cells transfected with Flag-Fbw7, Flag-β-TRCP1, or Myc-Fbl3a together with GFP as an internal transfection control were treated with MG132 for 12 hr, followed by immunoblot analysis with the indicated antibodies.
G. The abundance of endogenous Fbw7 is also regulated by the proteasomal degradation pathway. 293T cells were treated with MG132, followed by immunoblot analysis with the indicated antibodies.
H. Schematic representation of full length Fbw7 and ΔF-Fbw7 used in I.
I. The expression of the ΔF-Fbw7 mutant is not affected by MG132. 293T cells transiently expressing wild type (WT) HA-Fbw7 or ΔF-Fbw7 were treated with MG132, followed by immunoblot analysis with the indicated antibodies.
(See also Figure S1)
In support of this idea, it has been reported that several F-box proteins including Fbw7 are unstable and regulated post-translationally through proteasomal degradation (Bashir et al., 2004; Pashkova et al., 2010; Wei et al., 2004). However, how Fbw7 protein stability is regulated in vivo remains poorly understood. We found that the abundance of ectopically expressed Flag-Fbw7 (Figure 1D), Flag-β-TRCP1 (Figure 1E), and Myc-Fbl3a (Figure 1F) were increased following treatment with the proteasome inhibitor, MG132. We also observed an increased expression in endogenous Fbw7 after MG132 treatment (Figures 1G and S1F). Interestingly, deletion of the F-box-motif created a stabilized version of Fbw7 that did not respond to MG132 treatment (Figures 1H–I), arguing that self-ubiquitination might contribute to the destabilization of Fbw7. Altogether, these results suggest that the Pin1 isomerase may be an upstream regulatory component of Fbw7, whose expression inversely correlates with the abundance of the Fbw7 tumor suppressor.
Pin1 Interacts with Fbw7 in a Phosphorylation-Dependent Manner
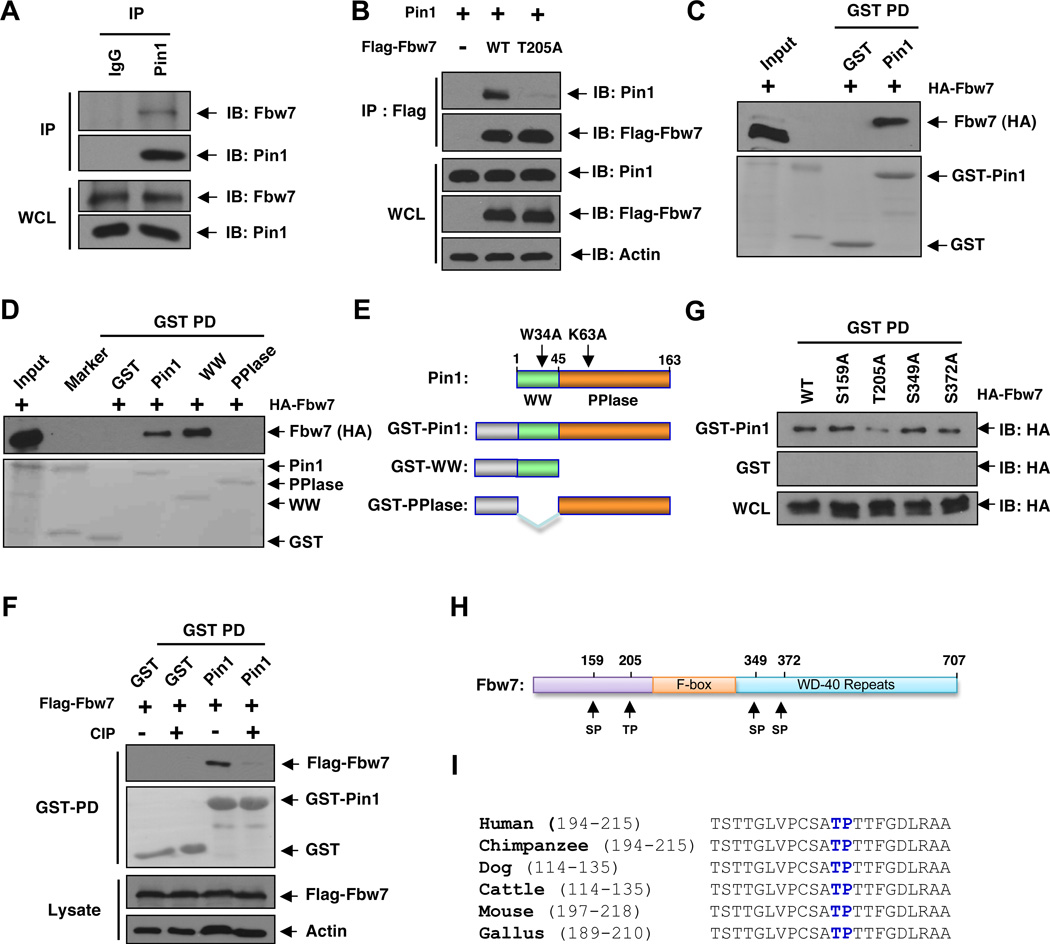
As previous studies have shown that the Pin1 prolyl isomerase is capable of modulating the protein stability of various key cell fate regulators, the fact that depletion of Pin1 does not affect Fbw7 mRNA levels (Figure S1E) would suggest a direct role for Pin1 in regulating the half-life of Fbw7. In support of Fbw7 being a possible Pin1 target, we detected the interaction between Pin1 and Fbw7 using both in vivo (Figures 2A–B and S2A) and in vitro (Figure 2C) binding assays. In addition to Fbw7, we also identified interactions between Pin1 and several other F-box proteins (Figure S2B). Consistent with other reported Pin1 substrates (Lu and Zhou, 2007), Fbw7 binds to both the full-length Pin1 and its WW domain, but not the PPIase domain (Figure 2D–E). Additionally, this interaction was abolished by the de-phosphorylation of Fbw7 with calf intestinal phosphatase (CIP) (Figure 2F), indicating that Pin1 binds to Fbw7 in a phosphorylation-dependent manner. This was further confirmed by the observations that an Alanine substitution of T205, but not other possible phosphorylation sites specifically diminished the interaction between Pin1 and Fbw7 in vitro (Figures 2G–I). Importantly, we found that the presence of T205 correlates with its sensitivity to MG132 treatment (Figures S2C–D), indicating that the Thr205-Pro motif of Fbw7 is important for binding to Pin1 as well as regulating Fbw7 stability.
Figure 2. Pin1 Interacts with Fbw7 in a Phosphorylation-Dependent Manner in vitro and in vivo.
A. Pin1 binds to endogenous Fbw7 in vivo. Mouse embryonic fibroblasts (MEFs) were subjected to immunoprecipitation with anti-Pin1 antibody or control pre-serum, followed by immunoblotting with the anti-Fbw7 or anti-Pin1 antibodies.
B. Co-immunoprecipitation of ectopically expressed WT-Fbw7 or T205A-Fbw7 and Pin1 in vivo. 293T cells were co-transfected with Flag-Fbw7 and HA-Pin1 constructs and then subjected to immunoprecipitation with anti-Flag, followed by immunoblotting with anti-Pin1 or anti-Flag antibodies.
C. Detection of Fbw7 and Pin1 interaction in vitro. Glutathione-agarose beads containing GST or GST-Pin1 were incubated with whole cell extracts derived from 293T cells expressing HA-Fbw7. After washing, proteins pulled down by GST beads were subjected to immunoblot analysis with anti-HA antibody.
D–E. Fbw7 interacts with the WW domain, but not the PPIase domain of Pin1. Whole cell lysates derived from 293T cells expressing HA-Fbw7 were incubated with glutathione-agarose beads containing GST, GST-Pin1 or GST-Pin1 truncation mutants (E). After washing, bound proteins were subjected to immunoblot analysis with anti-HA antibody (D).
E. Schematic representation of full length Pin1 and the various truncation mutants used in D.
F. Pin1 binds to Fbw7 in a phosphorylation-dependent manner. Lysates from 293T cells transiently expressing Flag-Fbw7 were treated with calf intestinal phosphatase (CIP) and subjected to GST-Pin1 pulldown, followed by immunoblot analysis with the indicated antibodies.
G. Pin1 interacts with wild type Fbw7, but not the T205A-Fbw7 mutant. Whole cell lysates derived from 293T cells expressing wild type Fbw7 or the indicated mutants were subjected to GST-Pin1 pulldown, followed by immunoblot analysis with anti-HA antibody.
H. Schematic representation of the various SP or TP sites that were replaced by alanine to generate the mutants used in G.
I. Sequence alignment of Fbw7 with the T205P206 site recognized by Pin1 in a variety of species.
(See also Figure S2)
Pin1 Negatively Regulates Fbw7 Stability via Promoting Its Ubiquitination
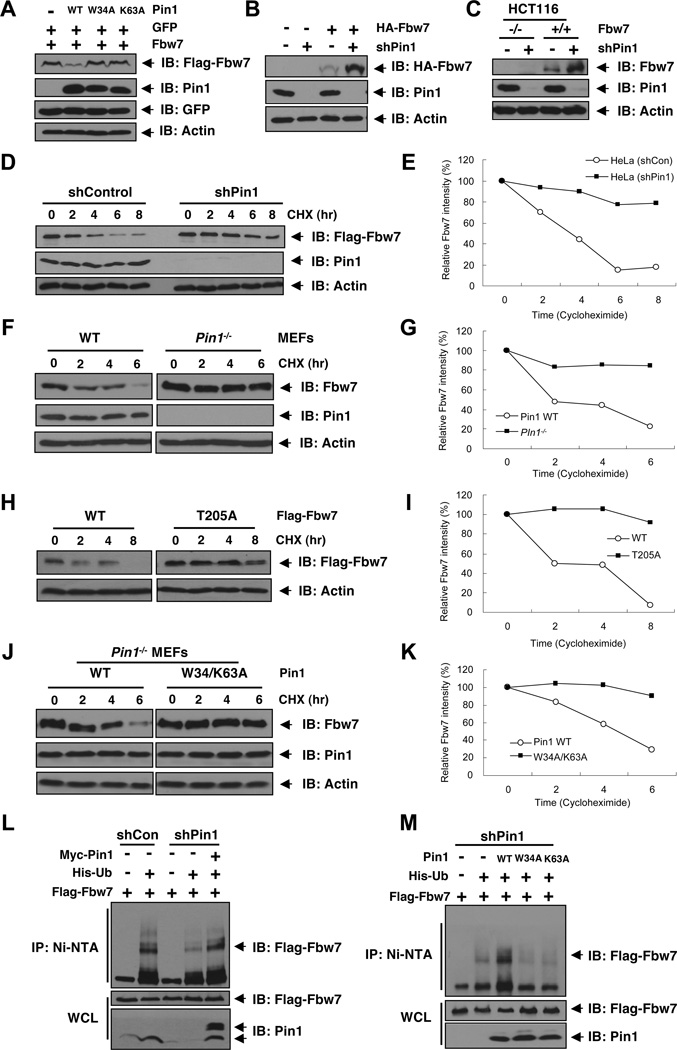
Given the inverse correlation between Pin1 and Fbw7 abundance in human clinical cancer samples and the phosphorylation-dependent interaction between Pin1 and Fbw7, we next examined whether Pin1 could directly regulate Fbw7 stability. We found that while WT-Pin1 reduced Fbw7 expression (Figures 3A and S3A), neither the W34A nor the K63A mutant had any effect (Figure 3A), suggesting that both the WW and PPIase domains are required for Pin1 to regulate Fbw7 abundance, as shown for all other known Pin1 substrates (Lu and Zhou, 2007). Furthermore, T205A-Fbw7 that could not interact with Pin1 (Figure 2B), displayed resistance to Pin1-mediated downregulation of Fbw7 (Figure S3B). Reciprocally, depleting endogenous Pin1 in HCT116 cells led to elevated expression of both endogenous and exogenous Fbw7 (Figures 3B–C).
Figure 3. Pin1 Reduces Fbw7 Protein Stability By Promoting Its Ubiquitination.
A. Fbw7 abundance is inhibited by wild-type Pin1, but not the W34A or the K63A dominant negative mutant. 293T cells were transfected with plasmids expressing Flag-Fbw7 and GFP in the presence or absence of WT, W34A (substrate binding domain mutant), or K63A (PPIase inactive form) Pin1, followed by immunoblot analysis with the indicated antibodies.
B. Depletion of Pin1 leads to elevated Fbw7 expression. HCT116 cells stably expressing HA-Fbw7 were infected with shPin1 lentivirus (or shGFP as a negative control), followed by selection with 2 µg/ml puromycin, and then subjected to immunoblot analysis with the indicated antibodies.
C. Depletion of Pin1 stabilizes endogenous Fbw7. WT or Fbw7−/− HCT116 cells were infected with shPin1 lentivirus (or shGFP as a negative control), followed by selection with 2 µg/ml puromycin, and then subjected to immunoblot analysis with the indicated antibodies.
D–E. Depletion of Pin1 increases the half-life of ectopically expressed Fbw7. HeLa cells stably expressing Pin1 shRNA or shControl were transfected with Flag-Fbw7 and treated with cyclohexamide for the indicated times, followed by immunoblot analysis with the indicated antibodies (D) and a semi-quantification with actin as a loading control and relative Fbw7 levels at time 0 set as 100% (E).
F–G. Depletion of Pin1 increases endogenous Fbw7 stability. MEF cells derived from wild type or Pin1 knockout mice were treated with cyclohexamide for the indicated times, followed by immunoblot analysis with the indicated antibodies (F) and a semi-quantification with actin as a loading control and relative Fbw7 levels at time 0 set as 100% (G).
H–I. Pin1 does not affect of the stability of the T205A mutant of Fbw7. HeLa cells were transfected with wild type or T205A Flag-Fbw7 and Fbw7 stability was then determined using cycloheximide chase (H). A semi-quantification was performed using actin as a loading control and relative Fbw7 levels at time 0 set as 100% (I).
J–K. A dominant negative mutant of Pin1 fails to regulate Fbw7 stability. Pin1−/− MEFs were stably transfected with either WT or the W34/K63A dominant negative mutant of Pin1. Fbw7 stability was then determined by using cycloheximide chase (J). A semi-quantification was performed using actin as a loading control and relative Fbw7 levels at time 0 set as 100% (K).
L. Pin1 promotes in vivo Fbw7 ubiquitination. HeLa cells stably expressing control shRNA or Pin1 shRNA were transfected with Flag-Fbw7, Myc-Pin1, and/or His-tagged ubiquitin or vector control as indicated, followed by lysis in a buffer containing 6M urea. Ubiquitin-conjugated proteins were captured with nickel-agarose beads and subjected to immunoblot analysis with anti-Flag antibody.
M. Dominant negative mutants of Pin1 do not affect in vivo Fbw7 ubiquitination. Stable Pin1 shRNA infected HeLa cells were transfected with Flag-Fbw7, Pin1 (WT, W34A, or K63A), and/or His-tagged ubiquitin or vector control as indicated, followed by immunoprecipitation with nickel-agarose beads and subjected to immunoblot analysis with anti-Flag antibody.
(See also Figure S3)
We next investigated whether the elevated Fbw7 levels were due to changes in Fbw7 half-life using the cycloheximide chase (Figure 3D–K), as previously described (Inuzuka et al., 2011; Inuzuka et al., 2010). In contrast to vector control cells where Fbw7 was unstable, Fbw7 was much more stable in Pin1-depleted cells (Figures 3D–E). A similar increase in the half-life of endogenous Fbw7 was also observed in Pin1−/− MEFs when compared to wild-type MEFs (Figures 3F–G). In confirmation of a regulatory role for Pin1 in Fbw7 stability, Fbw7 displayed increased half-life in Pin1−/− MEFs expressing W34A/K63A-Pin1 compared to those expressing WT-Pin1 (Figures 3J–K). Additionally, in support of a critical role for T205 in regulating Fbw7 stability (Figures S2C–D), T205A-Fbw7 was more stable (Figures 3H–I). Thus, Pin1 appears to be a regulator of Fbw7 by presumably interacting with Fbw7 at T205 to induce a conformational change.
Consistent with the tight regulation of Fbw7 turnover by Pin1, we observed reduced levels of Fbw7 ubiquitination in Pin1-depleted cells compared to control vector infected cells (Figure 3L). Moreover, re-expression of Pin1 in Pin1-depleted cells rescued Fbw7 ubiquitination (Figure 3L). In contrast, overexpression of inactive Pin1 mutants, including W34A or K63A, in Pin1-depleted cells did not cause any significant changes in Fbw7 ubiquitination (Figure 3M). Moreover, ubiquitination of Fbw7 was significantly decreased in cells expressing T205A-Fbw7, a mutant that fails to interact with Pin1 (Figures S3C–D) and was more stable (Figures 3H–I). These results indicate that Pin1 regulates Fbw7 ubiquitination and stability through its direct interaction with Fbw7. Moreover, depletion of endogenous Pin1 reduced the sensitivity of transfected Fbw7 to trypsin digestion in vitro, suggesting that Pin1 might affect Fbw7 conformation by regulating the isomerization state of Fbw7 (Figure S3E), as shown for other Pin1 substrates (Stukenberg and Kirschner, 2001; Zita et al., 2007).
Pin1 Promotes Fbw7 Self-Ubiquitination By Disrupting Fbw7 Dimerization
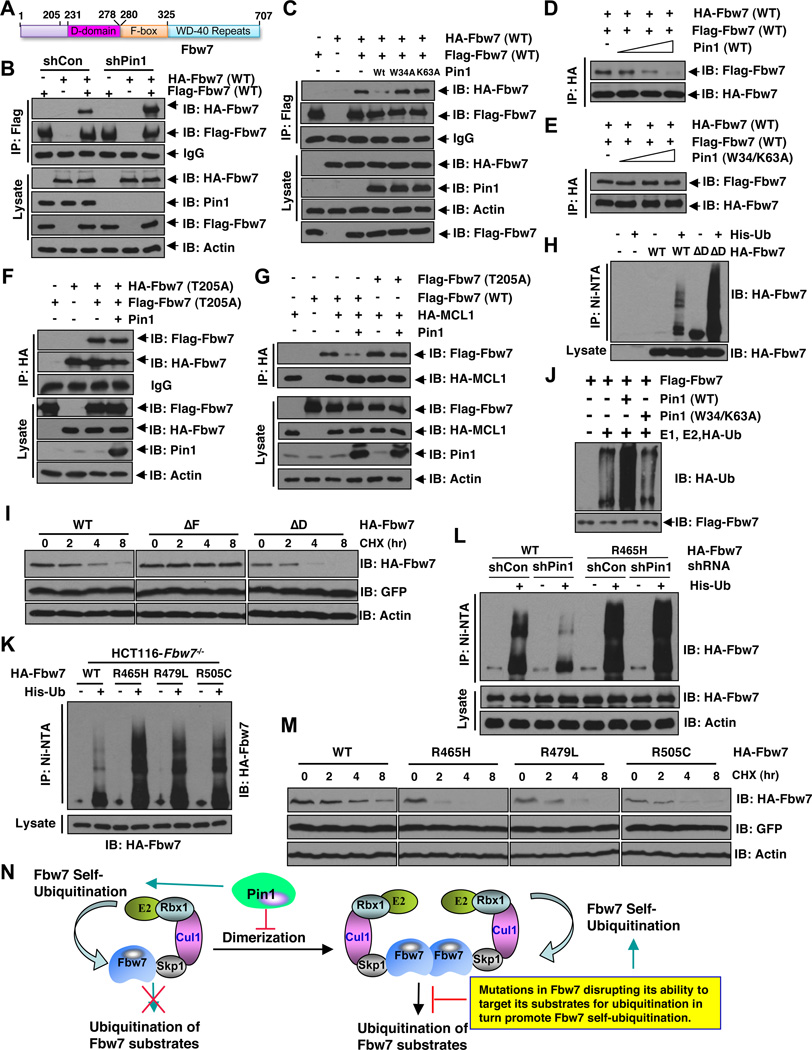
Recently, emerging evidences have suggested that F-box protein dimerization is critical for various SCF E3 ligase complexes to exert its E3 ubiquitin ligase activity towards downstream substrates (Barbash et al., 2008; Tang et al., 2007; Welcker and Clurman, 2007). More importantly, an evolutionarily conserved D-domain (also known as a dimerization domain) has been identified in Fbw7, just next to the F-box motif (Figure 4A) (Bai et al., 1996). Interestingly, the Pin1 interacting T205 site is in close proximity to the D-domain (Figure 4A), which prompted us to further investigate whether Pin1 binding could affect Fbw7 dimerization.
Figure 4. Pin1 Promotes Fbw7 Self-Ubiquitination By Dirupting Fbw7 Dimerization.
A. Schematic representation of the identified D-domain and F-box motif in Fbw7.
B. Stable Pin1 shRNA infected HCT116 Fbw7−/− cells (or shGFP cells as a negative control) were transfected with Flag-Fbw7 and HA-Fbw7 constructs or vector control as indicated, followed by immunoprecipitation with Flag-agarose beads and subjected to immunoblot analysis with the indicated antibodies.
C. Stable Pin1 shRNA infected HCT116 Fbw7−/− cells were transfected with Flag-Fbw7 and HA-Fbw7 constructs in the presence or absence of Pin1 (with empty vector or the indicated Pin1 mutants as negative controls), followed by immunoprecipitation with Flag-agarose beads and subjected to immunoblot analysis with the indicated antibodies.
D–E. HCT116 Fbw7−/− cells were transfected with either a HA-Fbw7 or a Flag-Fbw7 construct. 40 hours post-transfection, anti-Flag immunoprecipitation was performed to affinity-purify Fbw7 protein. The Flag-Fbw7/HA-Fbw7 complex was eluted out with 3xFlag peptide and 0.3 µM of eluted Fbw7 was incubated with increasing doses (0, 0.3, 1 or 3 µM) of recombinant WT-Pin1 (D) or W34/K63A-mutant Pin1 (E) before performing the anti-HA immunoprecipitation. The recovered Fbw7 dimer was monitored by immunoblot analysis.
F. Stable Pin1 shRNA infected HCT116 Fbw7−/− cells were transfected with Flag-Fbw7-T205A and HA-Fbw7-T205A constructs in the presence or absence of Pin1 (with empty vector as a negative control as indicated), followed by immunoprecipitation with HA-agarose beads and subjected to immunoblot analysis with the indicated antibodies.
G. Expression of Pin1 leads to reduced in vivo co-immunoprecipitation between Mcl-1 and ectopically expressed WT-Fbw7, but not T205A-Fbw7. HCT116-Fbw7−/− cells were co-transfected with the indicated Flag-Fbw7, HA-Mcl-1 and Pin1 constructs and then subjected to immunoprecipitation with anti-HA, followed by immunoblotting with the indicated antibodies.
H. Deletion of the D-domain results in increased in vivo Fbw7 ubiquitination. HCT116 cells were transfected with the indicated HA-Fbw7 and/or His-tagged ubiquitin or vector control as indicated, followed by immunoprecipitation with nickel-agarose beads and subjected to immunoblot analysis with anti-HA antibody.
I. Deletion of the F-box motif extended Fbw7 half-life while deletion of the D-domain shortened Fbw7 half-life. HCT116 cells were transfected with wild type, ΔF, or ΔD HA-Fbw7 and Fbw7 stability was then determined using cycloheximide chase.
J. HCT116 Fbw7−/− cells were transfected with a Flag-Fbw7 construct. 40 hours post-transfection, anti-Flag immunoprecipitation was performed to affinity-purify the SCFFbw7 complex followed by elution with 3×Flag peptides. 0.3 µM of affinity purified and eluted SCFFbw7 complex was then incubated with recombinant E1, E2, HA-Ubiquitin protein in the presence of 1 µM of purified recombinant WT-Pin1 or the W34/K63A-mutant Pin1. Fbw7 self-ubiquitination was monitored by immunoblot analysis with the indicated antibodies.
K. T-ALL derived hot-spot Fbw7 mutants displayed increased in vivo Fbw7 ubiquitination. HCT116-Fbw7−/− cells were transfected with the indicated HA-Fbw7 constructs and/or His-tagged ubiquitin or vector control as indicated, followed by immunoprecipitation with nickel-agarose beads and subjected to immunoblot analysis with anti-HA antibody.
L. Depletion of endogenous Pin1 leads to reduced self-ubiquitination of WT-Fbw7, but not R465H-Fbw7. HCT116 Fbw7−/− cells were infected with the indicated shRNA lentiviral vectors and selected in 1 µg/ml puromycin for 72 hours to eliminate the non-infected cells. Afterwards, the generated cells were transfected with the indicated HA-Fbw7 and/or His-tagged ubiquitin or vector control as indicated, followed by immunoprecipitation with nickel-agarose beads and subjected to immunoblot analysis with anti-HA antibody.
M. T-ALL derived hot-spot Fbw7 mutants displayed shortened Fbw7 half-life. HCT116 cells were transfected with the indicated HA-Fbw7 constructs and Fbw7 stability was then determined using cycloheximide chase.
N. A proposed model for Pin1 regulation of Fbw7 self-ubiquitination and stability via inhibiting Fbw7 dimer formation. In addition, we also propose that mutations in Fbw7 that disrupt its ability to ubiquitinate its downstream substrates will phenocopy Pin1 overexpression in promoting Fbw7 self-ubiquitination and subsequent degradation.
(See also Figure S4)
Consistent with a potential negative role for Pin1 in the Fbw7 dimerization process, depletion of endogenous Pin1 increased Fbw7 dimerization (Figure 4B). On the other hand, re-expression of WT-Pin1, but not the inactive mutant, W34A or K63A Pin1, in Pin1-knockdown cells reduced Fbw7 dimerization in vivo (Figure 4C). More importantly, purified recombinant Pin1, but not its inactive W34A/K63A mutant, could disrupt Fbw7 dimerization in a dose-dependent manner in vitro at concentrations as used previously (Lu et al., 1999; Nakamura et al., 2012; Zhou et al., 2000) (Figures 4D–E, and S4A–B). In contrast, the dimerization of the T205A-Fbw7 mutant, which failed to interact with Pin1 (Figure 2G), was not affected by Pin1 expression (Figure 4F), suggesting that a physical interaction with Fbw7 may be required for Pin1 to disrupt Fbw7 dimerization. Additionally, depletion of endogenous Pin1 did not affect the ability of Fbw7 to complex with Cullin-1 (Figure S4C). Furthermore, depleting the F-box motif, which prevents Fbw7 from forming a functional SCF complex, did not affect Fbw7 interaction with Pin1 (Figure S4D). Moreover, the T205A-Fbw7 mutant, which fails to interact with Pin1, had comparable ability as WT-Fbw7 to interact with Cullin1 (Figure S4E) and Skp1 (Figure S4F). These results suggest that Pin1 does not inhibit Fbw7 from forming an active SCF-type of E3 ligase so that the ability of Pin1 to regulate Fbw7 stability might stem from its ability to interfere with Fbw7 dimerization.
Several groups have clearly demonstrated that deletion of the D-domain significantly reduced the ability of Fbw7 to ubiquitinate downstream targets (Figure 4N) (Tang et al., 2007; Welcker and Clurman, 2007). Consistent with their findings, ectopic expression of Pin1, which affects the dimerization of WT, but not the T205A mutant form of Fbw7 (Fig. 4C–F), severely reduced the ability of WT-Fbw7, but not T205A-Fbw7 to interact with Mcl-1 (Figure 4G). Next, we went on to explore mechanistically how manipulation of Fbw7 dimerization might affect Fbw7 stability. We found that ΔD-Fbw7, a mutant that fails to form dimers (Figure S4G), still interacted with Cullin-1 as efficiently as WT-Fbw7 (Figure S4H). These results suggest that although Fbw7 monomers could not efficiently ubiquitinate downstream substrates, there may still be an intact and functional SCFFbw7 complex. Thus, we further reasoned that the blockage of SCFFbw7 activity towards its downstream substrates might re-channel the E3 ligase activity of Fbw7 towards itself, leading to increased self-ubiquitination (Figure 4N). In support of this hypothesis, we found a significant increase in ΔD-Fbw7 ubiquitination compared to WT-Fbw7 (Figure 4H). Furthermore, the F-box-deleted form of Fbw7 (ΔF-Fbw7) had a much more reduced ubiquitination than WT-Fbw7 (Figure S4I), suggesting that self-ubiquitination of Fbw7 may be the primary means of regulating Fbw7 stability in this experimental setting. Consistent with this model, we further demonstrated that compared to WT-Fbw7, ΔF-Fbw7 displayed a longer half-life while ΔD-Fbw7 exhibited a much shorter half-life (Figures 4I and S4J).
In keeping with the notion that Pin1 regulates Fbw7-stability and E3 ligase activity by disrupting Fbw7 dimerization (Figures 4B–F), purified recombinant Pin1, but not its inactive point mutant, efficiently increased Fbw7 self-ubiquitination in vitro (Figures 4J and S4K). Conversely, depletion of endogenous Pin1 led to reduced Fbw7 self-ubiquitination in vivo (Figure S4L). More importantly, consistent with Pin1-mediated disruption of Fbw7-dimerization being critical for Fbw7 to ubiquitinate its downstream substrates, ectopic expression of Pin1 severely reduced the ability of WT-Fbw7, but not T205A-Fbw7 to promote Mcl-1 ubiquitination in vivo (Figure S4M). Furthermore, purified recombinant Pin1, but not its inactive point mutant, also reduced Fbw7-mediated ubiquitination of Mcl-1 in vitro (Figure S4N). These results together indicate that Pin1 can promote Fbw7 self-ubiquitination, leading to destabilization of Fbw7, in part by disrupting Fbw7 dimerization, which subsequently also leads to reduced SCFFbw7 E3 ligase activity towards Fbw7 substrates.
However, neither F-box nor D-domain deletions have been observed in any pathological condition. Instead, mutations in the WD-40 substrate interaction motif of Fbw7 have frequently been observed in many types of human cancers. These mutations typically result in the abolishment of Fbw7 interaction with its downstream substrates. While not mechanistically identical to Pin1 overexpression that is associated with tumorigenesis, we reasoned that these types of Fbw7 mutations should functionally resemble Pin1 overexpression, as they ultimately lead to reduced ubiquitination of Fbw7 substrates. Therefore, we examined whether these Fbw7 genetic mutations also promoted Fbw7 self-ubiquitination. Indeed, we observed a significant increase in ubiquitination of the three hot-spot Fbw7 mutants derived from T cell acute lymphoblastic leukemia (T-ALL) (Inuzuka et al., 2011; Maser et al., 2007) (Figure 4K). More importantly, depletion of endogenous Pin1 led to a significant reduction in the self-ubiquitination of WT-Fbw7, but not R465H-Fbw7, a mutant that cannot interact with its downstream targets (Figure 4L). Our findings that both mutation of the substrate binding motif of Fbw7 and overexpression of Pin1 leads to reduced Fbw7 interaction with its substrates suggest that they may utilize a similar mechanism to re-channel the E3 ligase activity of Fbw7 towards self-ubiquitination, thereby excluding a possible additive effect. Furthermore, compared with WT-Fbw7, these three mutants displayed much shorter half-life (Figure 4M), although their binding to Cullin-1 was not affected by the mutations (Figure S4O). These results suggest that in addition to inactivating SCFFbw7 E3 ligase activity, the mutations in the Fbw7 substrate recognition motif might also result in accelerated self-destruction of the Fbw7 tumor suppressor, facilitating tumor formation. In keeping with this hypothesis, we observed that T-ALL cell lines harboring mutations in the WD40-repeat domain displayed reduced expression of Fbw7 (Figure S4P), which is largely due to a shortened Fbw7 half-life (Figure S4Q).
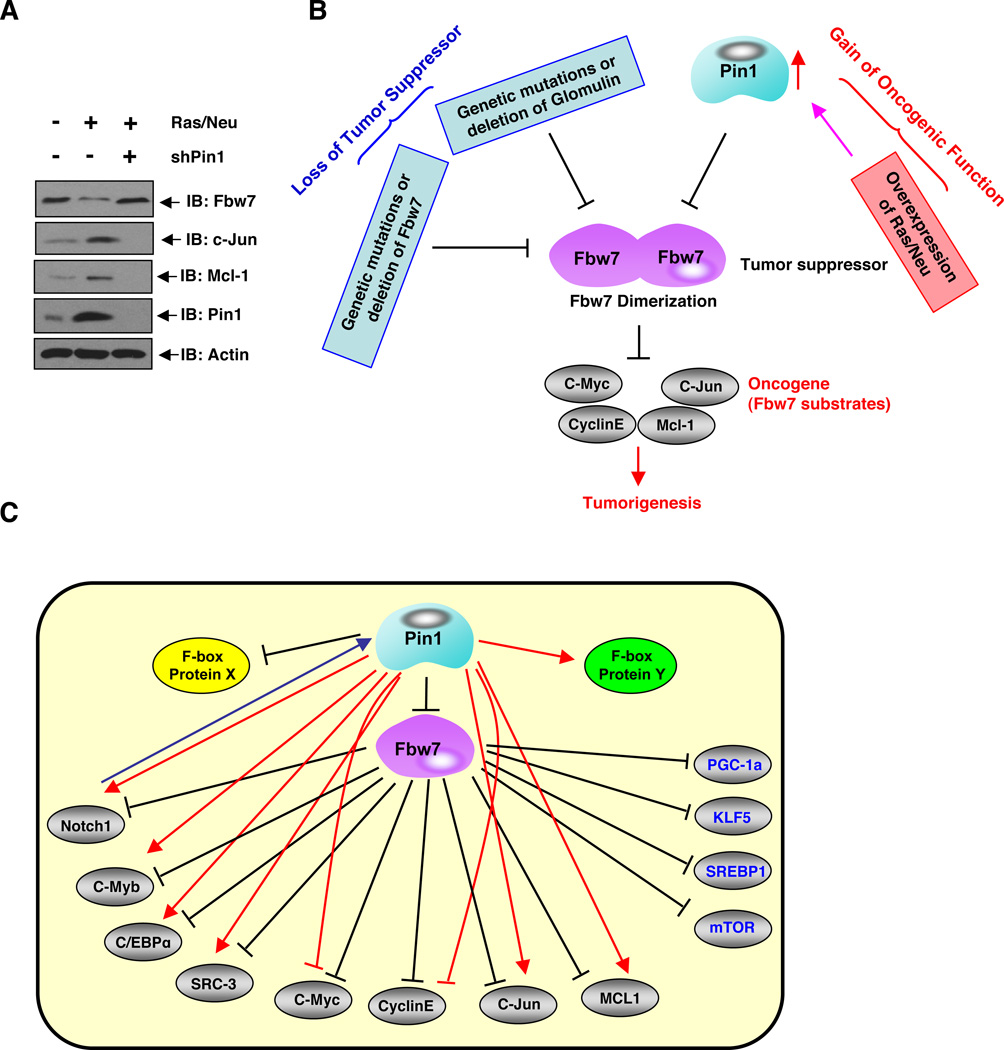
In addition, Glomulin, a putative tumor suppressor protein mutated in the vascular disorder glomuvenous malformation (GVM), has been recently identified to be a negative regulator of Fbw7 stability (Tron et al., 2012). However, the underlying molecular mechanism for this regulation is not fully understood. Interestingly, we found that ectopic expression of Glomulin promoted Fbw7 dimerization in vivo (Figure S4R), whereas depletion of endogenous Glomulin impaired Fbw7 dimerization in vivo (Figure S4S). Taken together, the above results indicate that Fbw7 dimerization plays an important role in regulating Fbw7 stability under various pathological conditions (Figure 6B).
Figure 6. Fbw7 dimerization and E3 ligase activity can be impaired by multiple mechanisms including gain of oncogenic function and/or loss of tumor suppressor function in human cancers.
A. Immunoblot analysis of whole cell lysates derived from the MCF10A cells that were engineered to express the oncogenic Ras and Neu proteins. Where indicated, shPin1 lentiviral vector was used to deplete endogenous Pin1 before harvesting for immunoblot analysis.
B. Schematic representation of a model by which physiological upstream signals, such as acquired activation of oncogenic Ras/Neu, overexpression of Pin1, genetic mutations in Fbw7, or loss of Glomulin activity can regulate Fbw7 tumor suppressor function in part through disrupting Fbw7 dimerization, leading to destabilization of Fbw7.
C. A Proposed model for the potential role of Pin1 in regulating Fbw7 protein stability and its downstream ubiquitination targets.
Pin1-Mediated Downregulation of Fbw7 Contributes to Tumorigenesis
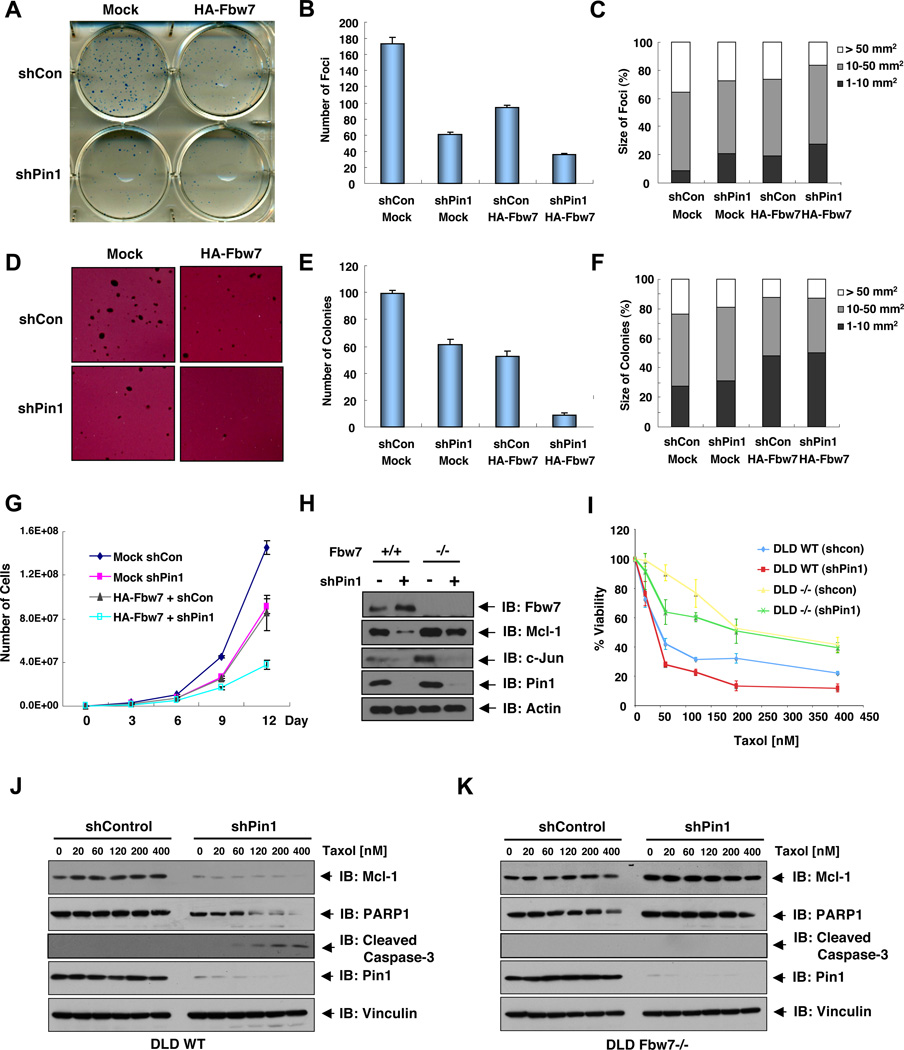
Given the important role of Pin1 in regulating Fbw7 stability, a key question is whether Pin1 affects the cellular functions of Fbw7. Consistent with the tumor suppressor function of Fbw7, cells expressing HA-Fbw7 showed a significantly lower number of foci (Figures 5A–C) and colonies in soft agar (Figures 5D–F). Moreover, depletion of Pin1 also significantly reduced foci number and anchorage independent colony growth, which displayed a more dramatic suppression effect in the HA-Fbw7-expressing cells (Figures 5A–F), a phenotype that might be partly due to elevated levels of Fbw7 after Pin1 depletion (Figure S5A). Consistent with the regulatory function of Pin1 on Fbw7, depletion of Pin1 impaired cell growth similarly to ectopic expression of HA-Fbw7, while expression of Fbw7 in Pin1-depleted cells delivered a synergistic inhibitory effect on cellular proliferation (Figure 5G). More importantly, ectopic expression of WT, but not the enzyme-dead version (W34/K63A) of Pin1, led to a significant increase in foci formation (Figure S5H) and anchorage-independent growth (Figure S5I–J), arguing that the prolyl isomerase activity of Pin1 is required for its oncogenic activity. In keeping with this finding, cells expressing T205A-Fbw7 displayed reduced foci formation (Figure S5H) and anchorage-independent growth (Figure S5I–J) in response to exogenous Pin1 expression. These results together demonstrate that Pin1 may exert its oncogenic function by suppressing the ability of Fbw7 to reduce cell transformation and cell proliferation.
Figure 5. Pin1-Mediated Downregulation of Fbw7 Contributes to Tumorigenesis.
A–F. Pin1 depletion increases HA-Fbw7 expression and its function to suppress foci formation on plastic plates (A–C) and anchorage-dependent cell growth in soft agar (D–F). The various generated HCT116 Fbw7−/− cells were seeded on plastic plates (A) or in soft agar (D) for 2 or 3 weeks respectively, followed by crystal violet staining (A) or P-iodonitrotetrazolium violet staining (D). The number of colonies formed per 500 cells was scored (B, E). Colony numbers are the mean ±SD of three independent experiments. The percentage of size of colonies formed per 500 cells was scored (C, F).
G. Depletion of Pin1 increases the ability of Fbw7 to suppress cell proliferation. 1 × 104 of various indicated HCT116 Fbw7−/− cells were seeded in 6 well plates and the number of cells was counted every 3 days to generate the growth curve. The error bars represent mean ± SD; n=3.
H. Depletion of Pin1 leads to the reduction of Fbw7 substrates Mcl-1 and c-Jun in WT-DLD1 cells, presumably due to the upregulation of endogenous Fbw7 levels. However, in contrast to c-Jun, Mcl-1 is not significantly reduced after depletion of Pin1 in Fbw7−/− DLD1 cells. The indicated DLD1 cells were infected with shPin1 lentivirus (or shGFP as a negative control), followed by selection with 1 µg/ml puromycin, and then subjected to immunoblot analysis with the indicated antibodies.
I–K. Depletion of Pin1 leads to downregulation of Mcl-1 in WT, but not Fbw7−/− DLD1 cells, sensitizing WT-DLD1 cells to Taxol. DLD1 cell lines (WT or Fbw7−/−) expressing Pin1 shRNA or Control shRNA were treated with Taxol for 3 days and the cell viability was determined by cell counts (I), the error bars represent mean ± SD; n=3. The cell lysates derived from WT-DLD1 (J) or Fbw7−/− DLD1 (K) cells were subjected to immunoblot analysis with the indicated antibodies.
(See also Figure S5)
Recent studies have indicated an important role for Fbw7 in regulating apoptosis by modulating the stability of Mcl-1 (Inuzuka et al., 2011), and in regulating the resistance of colon cancer cells to Taxol (Wertz et al., 2011). Consistent with a previous report (Wertz et al., 2011), Fbw7−/− DLD1 cells were much more resistant to Taxol compared to Fbw7-WT DLD1 cells (Figure 5I). Interestingly, depletion of endogenous Pin1 led to a sharp decrease of Mcl-1 (Figure 5H), presumably due to increased Fbw7 (Figures 3C, 5H and S5A). In keeping with the reduction of the pro-survival factor Mcl-1 (Figure 5J), Pin1-depleted DLD1-WT cells became more sensitive to Taxol treatment, compared with control shRNA-infected DLD1-WT cells (Figure 5I). On the other hand, depletion of endogenous Pin1 did not significantly affect the Mcl-1 abundance in Fbw7−/− DLD1 cells (Figure 5K). As a result, there was no significant change of Taxol sensitivity after depletion of Pin1 (Figure 5I), especially at high levels of Taxol treatment. These results indicate that Pin1 mainly regulates the abundance of Mcl-1 by modulating the stability of Fbw7, leading to changes in cellular sensitivities to chemotherapeutic agents.
Given the critical role of Pin1 in regulating the tumor suppressor function of Fbw7, next we were interested in further understanding whether in pathological conditions such as cancer, disregulation of upstream signaling pathways could lead to the inactivation of the tumor suppressor function of Fbw7. To this end, our previous studies have shown that activation of many oncogenic pathways, such as the Ras/Neu signaling pathway, can lead to elevated expression of Pin1 (Ryo et al., 2002). In keeping with this finding, we demonstrated that there was an obvious reduction of endogenous Fbw7 protein levels coupled with a significant induction of Fbw7 substrates, including c-Jun and Mcl-1, in Ras/Neu-expressing cells (Figure 6A). Importantly, depletion of Pin1 in Ras/Neu-expressing cells led to restoration of Fbw7 expression levels and a subsequent decrease in various Fbw7 substrates. These results indicate that Pin1 may be a possible route through which oncogenic Ras/Neu might suppress Fbw7 activity to promote tumorigenesis (Figure 6B). Therefore, our work demonstrates that aberrant gain of oncogenic functions, including overexpression of Ras/Neu or Pin1, in tumor cells can possibly lead to impaired Fbw7 dimerization (Figures 4B–C), resulting in reduced E3 ligase activity of Fbw7 that facilitates tumorigenesis. Moreover, our work also indicates that genetic mutations in Fbw7 (Figures 4K–L), and loss of Glomulin activity (Figures S4R–S), can disrupt Fbw7 dimerization leading to reduced E3 ligase activity, which would also facilitate tumorigenesis. These findings uncover the physiological significance of Fbw7 dimerization in controlling the E3 ligase and tumor suppressor function of Fbw7, which can be impaired in tumor cells by multiple means including gain of oncogenic mutations and/or loss of certain tumor suppressor functions (Figure 6B).
DISCUSSION
Fbw7 is a well-characterized major tumor suppressor, which is frequently inactivated by mutation or genetic deletion in various types of human cancers (Akhoondi et al., 2007; Malyukova et al., 2007; Strohmaier et al., 2001). However, little is known about the upstream regulation of Fbw7. Here we report that Fbw7 can be regulated by proteasomal degradation, a process that is promoted by Pin1. Specifically, we found that Pin1 directly interacts with Fbw7 to disrupt Fbw7 dimerization, a process that has been recently identified as a key regulatory mechanism for the function of many well-characterized F-box proteins (Barbash et al., 2008; Tang et al., 2007; Welcker and Clurman, 2007). As a result, Pin1 blocks the ability of Fbw7 to mediate substrate degradation, but instead promotes Fbw7 self-ubiqutination. Interestingly, compared to WT-Fbw7, T-ALL derived Fbw7 mutants with impaired ability to ubiquitinate substrates also displayed increased self-ubiquitination and shorter half-life. Therefore, our result support a model in which blockage of the normal E3 ligase activity of Fbw7 towards its physiological substrates may re-channel this destruction machinery to self-degradation (Figure 4N).
In addition to providing the molecular mechanisms for Pin1-mediated regulation of Fbw7 stability, we demonstrated that overexpression of Pin1 suppresses the ability of Fbw7 to inhibit cell transformation and proliferation. Moreover, depletion of Pin1 sensitized cells to Taxol, a phenotype that correlates with the reduction of the pro-survival factor Mcl-1. These results implicate Pin1 as an upstream regulatory protein for the Fbw7 signaling pathway, and further suggest that overexpression of Pin1 may contribute to the frequent loss of Fbw7 tumor suppressor function found in human cancers. Consistent with this hypothesis, we found that in a panel of breast cancer cell lines, there is a positive correlation between Pin1 overexpression and the elevated expression of Fbw7 substrates in cells bearing wild-type Fbw7 (Figure S1B).
In contrast to loss of Fbw7, Pin1 overexpression is frequently observed in human cancers, which promotes cell proliferation and transformation and correlates with poor clinical outcome (Lee et al., 2011b; Lu and Zhou, 2007). The oncogenic potential of Pin1 is mostly attributed to its ability to regulate a wide range of signaling pathways (Lee et al., 2011b). Interestingly, most Fbw7 downstream substrates, including, c-Jun (Wulf et al., 2001), NOTCH-1 (Rustighi et al., 2009), c-Myb (Pani et al., 2008), SRC-3 (Yi et al., 2005), CCAAT/Enhancer Binding Protein α (C/EBPα) (Jeong et al., 2009) and Mcl-1 (Ding et al., 2008), have been identified to be Pin1 substrates as well. We found that the expression of the Fbw7 substrates SREBP1, Mcl-1 and c-Jun were also regulated by Pin1 (Figure S5C–F). This presents an intriguing and unanswered question regarding the exact molecular mechanism by which Pin1 modulates these key cell regulators.
Our data clearly demonstrate that this could be achieved by two complementary means (Figure 6C). For some substrates such as Mcl-1, Pin1 primarily regulates its abundance through modulating Fbw7 stability. In Fbw7−/− cells, Pin1 failed to influence Mcl-1 abundance (Figures 5H, 5K and S5B). However, other Fbw7 substrates such as c-Jun still responds to depletion of Pin1 in the Fbw7−/− background, indicating that Pin1 may directly regulate c-Jun stability through an additional E3 ligase other than Fbw7 (Figure 5H). We recognize that it requires additional investigation to fully understand the exact physiological role for Pin1 in governing the stability of multiple Fbw7 substrates. However, it is clear that either route, direct regulation of the substrate or indirectly through Fbw7 stability, leads to elevated expression of the substrate. Since most Fbw7 substrates are well-characterized oncoproteins (Welcker and Clurman, 2008), this may offer a molecular understanding for the oncogenic role for Pin1.
The findings that Pin1 inhibits Fbw7 offer an exciting mechanism to manipulate Fbw7 activity in human diseases. To this end, it has been shown that reducing Pin1 expression or restoring Fbw7 function in cancer cells effectively suppresses tumorigenesis (Inbal et al., 1997; Wulf et al., 2004). Our results indicate that oncogenic Pin1 may be an important enzyme, which can physiologically suppress Fbw7 functions. In addition to Fbw7, we also identified the interaction between Pin1 and many other F-box proteins (Figure S2B). This indicates that Pin1 might potentially regulate the stability of a wide spectrum of F-box proteins, allowing Pin1 to influence a greater range of signaling pathways. However, much more studies are warranted to fully understand the exact role of Pin1 in the stability control of other F-box proteins. Nonetheless, these works together make Pin1 an attractive therapeutic target, inhibition of which could potentially upregulate the expression of Fbw7 and other F-box proteins to retard the growth of human tumor cells.
EXPERIMENTAL PROCEDURES
Immunohistochemistry, Immunostaining and Immunoblotting analyses
Formalin-fixed and paraffin-embedded tissue microarrays of human colon cancer tissues were purchased from Imgenex (IMH-359; San Diego, CA). Immunohistochemical staining for Pin1 and Fbw7 was performed as described previously (Lee et al., 2011a; Ryo et al., 2003). Fbw7 antibody for IHC was purchased from Bethyl.
In vitro Fbw7 dimerization Assay
The in vitro Fbw7 dimerization assays were performed similarly as described before (Ryo et al., 2003; Zhou et al., 2000). Briefly, Flag-Fbw7 and HA-Fbw7 constructs were transfected in HCT116 Fbw7−/− shPin1 cells. 40 hours post-transfection, cells were treated with nocodazole (50 ng/ml) for 12 hours and anti-Flag immunoprecipitation was performed to affinity-purify Fbw7 protein. Flag-Fbw7 was eluted out using 3xFlag peptide (from Sigma), and 0.3 µM of eluted proteins were incubated with increasing doses (0, 0.3, 1 or 3 µM) of recombinant WT-Pin1 or W34/K63A-mutant Pin1 as described previously (Lu et al., 1999; Nakamura et al., 2012; Zhou et al., 2000) before HA-immunoprecipitation was performed. The recombinant Pin1 (0, 0.3, 1 or 3 µM) used in the assay is in the stoichoimetric range of concentrations relative to eluted Fbw7, as described for other Pin1 substrates (Lu et al., 1999; Nakamura et al., 2012; Zhou et al., 2000). The recovered proteins were resolved by SDS-PAGE and probed with the indicated antibodies to monitor Fbw7 dimer formation.
Foci Formation and Soft Agar Colony Formation Assays
Foci formation assays were performed by seeding 500 cells in 6 well tissue culture dishes and soft agar assays were done by seeding 500 cells in 6 well tissue culture dishes containing 0.3% top low-melt agarose-0.5% bottom low-melt agarose as previously described (Ryo et al., 2002). Cells were fed every 3 days and colonies were counted and measured after 2–3 weeks.
HIGHLIGHTS.
Pin1 levels inversely correlate with Fbw7 levels in human cancer tissues
Pin1 interacts with Fbw7 and inhibits Fbw7 dimerization
Pin1 decreases Fbw7 protein stability via promoting its self-ubiquitination
Pin1 negatively regulates the tumor suppressor function of Fbw7
Supplementary Material
ACKNOWLEDGMENTS
A.W.L. is a NRSA T32 Fellow (CA081156-08), T.H.L is the recipient of the NIH Pathway to Independence (PI) Award (K99AG033104), D.Y.L. is a Human Frontier Science Program Fellow (LT000802) and C.H.C. is a Fellow of the Department of Defense Breast Cancer Research Program (W81XWH09-1-0481). W.W. is an American Cancer Society Research Scholar. The work was supported by NIH grants R01GM089763 to W.W., Susan G. Komen for the Cure grant KG100958 to X.Z.Z and R01CA167677 and R01AG017870 to K.P.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes six figures with their corresponding figure legends and Supplemental Experimental Procedures, which can be found with this article online.
REFERENCES
- Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer research. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- Ayala G, Wang D, Wulf G, Frolov A, Le R, Wheeler T, Sowadski JM, Lu KP, Bao L. Pin1 is a novel prognostic marker in prostate cancer. Cancer research. 2003;63:6244–6251. [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bao L, Sauter G, Sowadski J, Lu KP, Wang D. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727–1737. doi: 10.1016/S0002-9440(10)63731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, Lu F, Rustgi AK, Diehl JA. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- Brenkman AB, de Keizer PL, van den Broek NJ, van der Groep P, van Diest PJ, van der Horst A, Smits AM, Burgering BM. The peptidyl-isomerase Pin1 regulates p27kip1 expression through inhibition of Forkhead box O tumor suppressors. Cancer research. 2008;68:7597–7605. doi: 10.1158/0008-5472.CAN-08-1059. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Huo L, Yang JY, Xia W, Wei Y, Liao Y, Chang CJ, Yang Y, Lai CC, Lee DF, et al. Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer research. 2008;68:6109–6117. doi: 10.1158/0008-5472.CAN-08-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson A, et al. A Pin1/Mutant p53 Axis Promotes Aggressiveness in Breast Cancer. Cancer cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. The Journal of biological chemistry. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Inbal B, Cohen O, Polak-Charcon S, Kopolovic J, Vadai E, Eisenbach L, Kimchi A. DAP kinase links the control of apoptosis to metastasis. Nature. 1997;390:180–184. doi: 10.1038/36599. [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. SCFFBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HG, Pokharel YR, Lim SC, Hwang YP, Han EH, Yoon JH, Ahn SG, Lee KY, Kang KW. Novel role of Pin1 induction in type II collagen-mediated rheumatoid arthritis. J Immunol. 2009;183:6689–6697. doi: 10.4049/jimmunol.0901431. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science (New York, NY. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, Zhang YJ, Goate A, Chen RH, Zhou XZ, et al. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Molecular cell. 2011a;42:147–159. doi: 10.1016/j.molcel.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Pastorino L, Lu KP. Peptidyl-prolyl cis-trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert reviews in molecular medicine. 2011b;13:e21. doi: 10.1017/S1462399411001906. [DOI] [PubMed] [Google Scholar]
- Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36:501–514. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP. Pinning down cell signaling, cancer and Alzheimer's disease. Trends Biochem Sci. 2004;29:200–209. doi: 10.1016/j.tibs.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, Spruck C, Grander D, Lendahl U, Sangfelt O. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer research. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, Zhou XZ, Lu KP. Proline isomer-specific antibodies reveal the early pathogenic Tau conformation in Alzheimer's disease. Cell. 2012;149:232–244. doi: 10.1016/j.cell.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Seminars in cell & developmental biology. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science (New York, NY. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. The Journal of biological chemistry. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- Pani E, Menigatti M, Schubert S, Hess D, Gerrits B, Klempnauer KH, Ferrari S. Pin1 interacts with c-Myb in a phosphorylation-dependent manner and regulates its transactivation activity. Biochim Biophys Acta. 2008;1783:1121–1128. doi: 10.1016/j.bbamcr.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Molecular cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Punga T, Bengoechea-Alonso MT, Ericsson J. Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. The Journal of biological chemistry. 2006;281:25278–25286. doi: 10.1074/jbc.M604983200. [DOI] [PubMed] [Google Scholar]
- Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, Kaplan F, Capobianco A, Pece S, Di Fiore PP, et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nature cell biology. 2009;11:133–142. doi: 10.1038/ncb1822. [DOI] [PubMed] [Google Scholar]
- Ryo A, Liou YC, Wulf G, Nakamura N, Lee SW, Lu KP. Pin1 is an E2F target gene essential for the Neu/Ras-induced transformation of mammary epithelial cells. Molecular and cellular biology. 2002;22:5281–5295. doi: 10.1128/MCB.22.15.5281-5295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A, Nakamura N, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nature Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Molecular cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Stanya KJ, Liu Y, Means AR, Kao HY. Cdk2 and Pin1 negatively regulate the transcriptional corepressor SMRT. The Journal of cell biology. 2008;183:49–61. doi: 10.1083/jcb.200806172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Stukenberg PT, Kirschner MW. Pin1 acts catalytically to promote a conformational change in Cdc25. Molecular cell. 2001;7:1071–1083. doi: 10.1016/s1097-2765(01)00245-3. [DOI] [PubMed] [Google Scholar]
- Suizu F, Ryo A, Wulf G, Lim J, Lu KP. Pin1 regulates centrosome duplication and its overexpression induces centrosome amplification, chromosome instability and oncogenesis. Molecular and cellular biology. 2006;26:1463–1479. doi: 10.1128/MCB.26.4.1463-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Akiyama H, Shimazaki K, Uchida C, Akiyama-Okunuki H, Tomita M, Fukumoto M, Uchida T. Ablation of a peptidyl prolyl isomerase Pin1 from p53-null mice accelerated thymic hyperplasia by increasing the level of the intracellular form of Notch1. Oncogene. 2007;26:3835–3845. doi: 10.1038/sj.onc.1210153. [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Tron A, Arai T, Duda DM, Kuwabara H, Olszewski JL, Fujiwara Y, Bahamon BN, Sigaoretti S, Schulman BA, DeCaprio JA. Glomulin binds Rbx1 and regulates Cullin RING ligase-mediated turnover of Fbw7. Molecular cell. 2012;46:1–12. doi: 10.1016/j.molcel.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun-Kyi A, Finn G, Greenwood A, Nowak M, Lee TH, Asara JM, Tsokos GC, Fitzgerald K, Israel E, Li X, et al. Essential role for the prolyl isomerase Pin1 in Toll-like receptor signaling and type I interferon-mediated immunity. Nat Immunol. 2011;12:733–741. doi: 10.1038/ni.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell division. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature reviews. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Molecular and cellular biology. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: Is there an underlying theme? Nature Cell Biol. 2005;7:435–441. doi: 10.1038/ncb0505-435. [DOI] [PubMed] [Google Scholar]
- Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 2004;23:3397–3407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Lu KP. Pin1 is overexpressed in breast cancer and potentiates the transcriptional activity of phosphorylated c-Jun towards the cyclin D1 gene. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. The EMBO journal. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wu RC, Sandquist J, Wong J, Tsai SY, Tsai MJ, Means AR, O'Malley BW. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1) Molecular and cellular biology. 2005;25:9687–9699. doi: 10.1128/MCB.25.21.9687-9699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC, Tsai CH, Chen HY, Chiang CT, Lai CK, et al. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer cell. 2011;20:214–228. doi: 10.1016/j.ccr.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Molecular cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Küllertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Molecular cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
- Zita MM, Marchionni I, Bottos E, Righi M, Del Sal G, Cherubini E, Zacchi P. Post-phosphorylation prolyl isomerisation of gephyrin represents a mechanism to modulate glycine receptors function. The EMBO journal. 2007;26:1761–1771. doi: 10.1038/sj.emboj.7601625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.