Abstract
Enterotoxigenic Escherichia coli CFA/I is a protective antigen and has been overexpressed in bacterial vectors, such as Salmonella Typhimurium H683, to generate vaccines. Effects that overexpressed CFA/I may engender on the bacterial host remain largely unexplored. To investigate, we constructed a high CFA/I expression strain, H683-pC2, and compared it to a low CFA/I expression strain, H683-pC, and to a non-CFA/I expression strain, H683-pY. The results showed that H683-pC2 was less able to migrate into semisolid agar (0.35%) than either H683-pC or H683-pY. Bacteria that migrated showed motility halo sizes of H683-pC2 < H683-pC < H683-pY. In the liquid culture media, H683-pC2 cells precipitated to the bottom of the tube, while those of H683-pY did not. In situ imaging revealed that H683-pC2 bacilli tended to auto-agglutinate within the semisolid agar, while H683-pY bacilli did not. When the cfaBE fimbrial fiber encoding genes were deleted from pC2, the new plasmid, pC2(−), significantly recovered bacterial swimming capability. Our study highlights the negative impact of overexpressed CFA/I fimbriae on bacterial swimming motility.
1. Introduction
Although flagella are responsible for bacterial motility, the motion of bacteria is affected by certain other surface appendages, such as extracellular polysaccharides, lipopolysaccharides (LPS) and pili. For example, Bacillus subtilis extracellular polysaccharide matrix gene epsE was expressed to break the flagella rotor to inhibit motility in order to form a biofilm (Guttenplan et al 2010). LPS is associated with flagella-mediated motility in Pseudomonas aeruginosa through the modulation of cell-surface attachment (Lindhout et al 2009). Enteric bacteria attach to surfaces and host tissues by means of curli fibers (Epstein and Chapman 2008). When MR/P fimbriae were produced, MrpJ repressed transcription of the flagellar regulon, thus reducing flagella synthesis (Li et al 2001). These studies illustrate that outer membrane proteins and surface appendages play an important role in bacterial motility. However, there have been no previous reports on how fimbriae affect bacterial motility (Gibiansky et al 2010). Since motility is crucial to bacterial invasion (Betts and Finlay 1992), anti-phagocytosis (Amiel et al 2010) and virulence (Meng et al 2011), studying the effects of fimbriae on bacterial motility may assist in determining whether overexpressed fimbrial proteins alter bacterial pathogenesis.
Colonization factor antigen 1 (CFA/I) is a virulence factor from enterotoxigenic Escherichia coli (ETEC). It is responsible for adherence to the host intestinal epithelium, which initiates the first step of infection (Baker et al 2009, Evans et al 1975). Five genes from two genomic regions are required for the expression of CFA/I (Willshaw et al 1985). The structural genes for CFA/I fimbrial biogenesis, cfaABCE, are located in the genomic region 1, while the positive regulator gene, cfaR, is located in the genomic region 2 (Savelkoul et al 1990). When cfaABCE were installed downstream of the tetracycline resistance gene promoter (PtetA), overexpression of CFA/I fimbriae occurred in both E. coli and Salmonella (Wu et al 1995), suggesting that regulator gene cfaR is not required for fimbrial subunit expression after its promoter is replaced with PtetA. The four structural genes encode four proteins, which coordinate to form CFA/I fimbriae: cfaB encodes the ~15 kDa major subunit which forms the fimbrial rods, cfaE encodes the minor subunit that locates at the tip of the fimbria and is also responsible for nucleating fiber formation (Baker et al 2009, Li et al 2007), cfaA encodes the periplasmic chaperone that promotes subunit folding and transportation (Li et al 2007), and cfaC encodes the usher that assembles into a channel structure in the cell outer membrane (Saulino et al 2000). Although CFA/I has been overexpressed in various bacterial vectors such as Salmonella enterica serovar Typhimurium H71 (Suo et al 2009), S. Typhimurium H683 (Ochoa-Repáraz et al 2008, Yang et al 2011), E. coli (Tobias et al 2010) and Vibrio cholerae (Tobias et al 2008), its impact on the behavior of a bacterial host has not been investigated.
Previous studies of bacterial behavior have mainly focused on the relationships between genes and behavioral phenotypes. With the introduction of molecular biology in the past several decades, increasing numbers of recombinant bacteria have been generated for the overexpression of heterologous genes. For instance, by means of vaccine vector S. Typhimurium H683 (Wu et al 1995), human ETEC CFA/I fimbriae were overexpressed to prevent and treat experimental autoimmune encephalomyelitis (Ochoa-Repáraz et al 2008), the Y. pestis F1 capsule and LcrV were simultaneously overexpressed to prevent plague (Yang et al 2007), and bovine ETEC K99 fimbriae were overexpressed to immunize neonates (Ascón et al 1998). In all these studies, attention was exclusively paid to protein expression and the immune responses elicited against the proteins. The impact of these foreign proteins on the bacterial vectors remains unknown. Since all these proteins—CFA/I fimbriae, F1 capsule, LcrV and K99 fimbriae—are located on the bacterial surface, it is reasonable to speculate that they may affect bacterial motility. In this study, we chose to overexpress ETEC CFA/I in S. Typhimurium H683 to examine whether CFA/I alters the motile behavior of Salmonellae. The results suggest that bacterial swimming capability is significantly impaired by the overexpression of CFA/I.
2. Materials and methods
2.1. Bacterial strains, media, plasmids, primers and growth conditions
The bacterial strains used are shown in table 1. E. coli H681, a Δasd mutant strain, and S. Typhimurium H683, and a ΔasdΔaroA mutant strain (Wu et al 1995), were used to transform recombinant asd+ plasmids carrying cfa/I genes. All strains were cultured using the Lysogeny broth (LB) medium without antibiotics. Strains H681 and H683 were grown in the medium supplemented with diaminopimelic acid (50 μg ml−1) (Galán et al 1990). To analyze growth rates, recombinant Salmonella strains were taken from a −80 °C freezer and used to inoculate LB agar for overnight incubation at 37 °C. Bacterial organisms were harvested, and the cell optical density at 600 nm (OD600) was adjusted to ~0.05 with the LB medium. Then, the bacteria were grown in BioScreen C (Oy Growth Curves AB Ltd) at a continuous agitation of 150 rpm and 37 °C for 5.5 h. Each strain was analyzed in triplicate in each experiment, and three independent experiments were conducted.
Table 1.
Bacterial strains, plasmids and primers used in this study.
| Strains | Characteristics | Sources |
|---|---|---|
| E. coli H681 | Δasd | (Wu et al 1995) |
| S. Typhimurium H683 | ΔasdΔaroA | (Wu et al 1995) |
| Plasmids | Characteristics and derivation | Sources |
| pC | pJGX15C-asd+, cfaABCE under control of PtetA | (Wu et al 1995) |
| pC2 | asd+, cfaABCE under control of PtetA~PphoP, derived from pC | This study |
| pY | asd+, cfa/I−, derived from pC | (Yang et al 2012) |
| pTP2cfaB(−) | asd+, cfaA+cfaB−cfaC+cfaE+, derived from pC2 | This study |
| pC2(−) | asd+, cfaA+cfaB−cfaC+cfaE−, derived from pTP2cfaB(−) | This study |
| Primers | Oligonucleotide sequencesa | Enzyme sitesb |
| cfA-F | GAGGCGGTAAAACCAGATAGC | XbaI |
| cfB-R | GGATCCCATTGTGGTCAGAGCCATTGC | KpnI |
| cfC-F | TTATGAAGGAAGTCTGAAATGG | EcoRI |
| cfC-R | CTCGAGTTATCCTTTATCATTCTTTGCCAG | XhoI |
Note:
The sequences in bold print are the integrated restriction enzyme sites.
The restriction enzyme sites for XbaI and EcoRI are not designed in primers but contained in the template DNA sequence downstream of the primers.
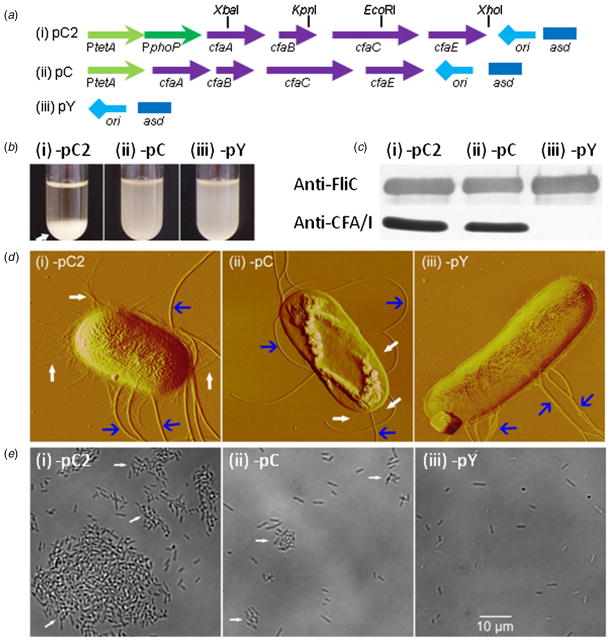
To determine whether CFA/I fimbriae impact bacterial motility, pC, the plasmid that harbors operon, cfa/I, was utilized (table 1). In pC, cfaABCE is under the control of PtetA (Wu et al 1995). To enhance cfa/I expression, the strong fusion promoter PtetA~PphoP from pV55 (Yang et al 2007) was cloned and used to replace PtetA in pC. The new plasmid was named pC2 (table 1). In pC2, cfaABCE was under the control of PtetA~PphoP. Plasmid pY was derived from pC by deleting cfaABCE from pC, and pY was used as a control plasmid (Yang et al 2012) (table 1). To determine whether CfaBE fimbrial fibers were involved in bacterial motility, the inner DNA sequences of cfaB and cfaE in plasmid pC2 were deleted, respectively, in-frame and completely. This took several steps as detailed in the following. First, using primers of cfA-F/cfB-R (table 1), with pC as a template, a DNA fragment of 456 bp containing 3′-end cfaA and 5′-end cfaB was amplified by polymerase chain reaction (PCR). After digestion with XbaI and KpnI, it was inserted between the XbaI and KpnI in pC2 (figure 1(a)). Thus, the 459-bp inner DNA fragment sequence of cfaB was deleted from pC2, and the new plasmid was termed pTP2cfaB(−) (table 1). Next, based on pTP2cfaB(−), the cfaE gene encoding the minor subunit was entirely deleted as follows: using primers of cfC-F/cfC-R with pC as a template, a DNA fragment of 1394 bp containing 3′-end cfaC was amplified by PCR. After digestion with XhoI and EcoRI (figure 1(a)), it was inserted between the EcoRI and XhoI sites in pTP2cfaB(−) to completely remove cfaE, obtaining plasmid pC2(−). Plasmids pC2, pC, pC2(−) and pY were transformed to S. Typhimurium H683 to generate H683-pC2, -pC, -pTP2cfaB(−), -pC2(−) and -pY for evaluation of the CFA/I’s involvement in the bacterial motilities.
Figure 1.
Effect of overexpression of CFA/I on bacterial phenotypes. (a) Schematic plasmid maps. The cfaABCE is regulated by PtetA~PphoP in pC2 (i) and by PtetA in pC (ii), and control pY does not harbor cfaABCE (iii). (b) Overexpression of CFA/I resulting in cell auto-precipitation. The overnight cell culture of H683-pC2 precipitated to the bottom of the tube (i), while those of -pC (ii) and -pY (iii) did not. The arrow indicates the precipitated cell pellets. (c) Western blot of anti-flagella and -CFA/I fimbriae. The anti-FliC blots showed no differences for flagellum expression among H683-pC2 (i), -pC (ii) and -pY(iii), while anti-CFA/I blots showed that H683-pC2 (i) produced more CFA/I fimbriae than -pC (ii) but -pY (iii) did not synthesize CFA/I. (d) AFM imaging of bacterial flagella and CFA/I fimbriae. The H683-pC2 (i), -pC (ii) and -pY (iii) images all revealed flagella associated with the bacterial organisms, but CFA/I fimbriae were associated with H683-pC2 and -pC, but not with -pY. The wide and narrow arrows indicate flagella and CFA/I fimbriae, respectively. (e) Light microscope images of agglutinated bacteria. Imaging revealed that a majority of H683-pC2 cells (i) were auto-agglutinated within the liquid LB medium, while a great number of H683-pC (ii) and all -pY (iii) cells existed in the planktonic form.
2.2. Evaluation of bacterial swimming ability
After being removed from the −80 °C freezer, H683-pC2, -pC, -pTP2cfaB(−), -pC2(−) and -pY were used to inoculate LB agar plates and grown overnight at 37 °C. Individual colonies were selected using tips and used to inoculate the surfaces of LB plates containing 0.35% agar (Stafford and Hughes 2007), and then incubated at 37 °C. After the inoculants entered the semisolid agar, the bacteria swam in various directions, which formed a pattern of concentric circles around the inoculation sites. We assumed that the diameters of the ‘motility halos’ correlated positively with the bacterial swimming capability. Thus, to compare the swimming capabilities of these strains, we used the halo diameter as a criterion as previously described (Tareen et al 2011). The halo diameters of 20 colonies were measured in two cross directions at 8 h post-inoculation, and their averages were used as the final values.
To compare the agar migration rates of the recombinant Salmonella strains, the inoculants were counted for agar migration and non-migration at 8 h post-inoculation. The percentage of the inoculants that accomplished agar migration was calculated for each strain, which was used for assessment of statistical difference. The agar concentrations used in the migration assay were 0.45%, 0.35% and 0.25%. Twenty inoculants were monitored for each strain per experiment. This experiment was repeated six times.
2.3. Imaging of bacterial cells using atomic force microscopy and light microscopy
The bacteria were observed using an atomic force microscope (AFM) as previously described (Suo et al 2008, 2009). In short, H683-pC2, -pC and -pY were grown in a liquid LB medium, then rolled at 60 rpm at 37 °C until their OD600 reached ~0.6. Cell culture suspensions of 100 μl were deposited directly onto a freshly cleaved mica disk. The suspension-covered mica was incubated for 15 min at room temperature, then rinsed with Nanopure water and dried with nitrogen gas flow. All AFM images were acquired in air in tapping mode using a Multimode V system from Veeco (Suo et al 2007). At least 20 cells were observed for each strain, and images with an approximately average density of fimbriae were presented.
Supernatants of cell cultures of strains H683-pC2, -pC and -pY were imaged optically using an Olympus BX61 system equipped with a DP71 camera in phase contrast mode. To image bacteria contained within semisolid agar, cells of H683-pC2 and -pY were used to inoculate surfaces of the LB medium containing 0.35% agar with a medium thickness of ~1.0 mm. At 8 h post-inoculation, a layer of swimming halos was imaged using phase contrast mode. The video clips of bacteria swimming were converted using Movie Maker, designed by Microsoft, from 140 time-lapse images per 10 s.
2.4. Western blot analysis
H683-pC2, -pC and -pY cells were collected from LB medium cultures during the logarithmic (log) growth phase. Cell pellets were rigorously vortexed for shearing flagella and CFA/I fimbriae as previously described (Bahrani et al 1991). Comparative sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analyses of CFA/I and flagellin from H683-pC2, -pC and -pY were performed (Yang et al 2007). For anti-flagellin blotting, 1.35 × 108 colony forming units (CFU) of H683-pC2, -pC and -pY were used per well, and for anti-CFA/I blotting 1.35 × 108 CFU of H683-pC2 and 2.7 × 109 CFU of H683-pC and -pY were used, respectively. After electrophoresis, protein transfer and immunoblot development were performed according to a method previously described (Yang et al 2007). The mouse monoclonal FliC antibody (Biocompare) and the rabbit polyclonal CFA/I antiserum (laboratory preparation) were used for detecting flagellin and CFA/I fimbria expression, respectively.
2.5. Statistical analysis
The Student’s t-test was used to evaluate the statistical differences in each experiment. The P values of <0.05, <0.01 and <0.001 were used to differentiate significant differences among different parameters.
3. Results and discussion
3.1. Overexpression of CFA/I causes auto-agglutination of Salmonellae
The plasmids pC2, pC and pY (figure 1(a)) were transformed to S. Typhimurium H683, and three new recombinant strains of H683-pC2, -pC and -pY were generated. After overnight growth at 37 °C in the liquid LB medium, the cell cultures were allowed to stand without agitation for 1 h at ambient temperature. As shown in figure 1(b), H683-pC2 cells precipitated to the bottom of the tube and left a clear supernatant, implying the auto-agglutination of H683-pC2, while -pC and -pY remained an even turbid mixture of cells and medium. Agglutination is typically observed for bacterial organisms expressing CFA/I in the presence of anti-CFA/I serum (Alves et al 2000). The auto-agglutination of H683-pC2 in the absence of anti-CFA/I serum suggests that -pC2 may produce an enormous amount of CFA/I fimbriae on cell surfaces, endowing it with this new phenotype. This auto-agglutination phenomenon was also previously observed in our laboratories for another CFA/I expression strain, S. Typhimurium Δasd::kanR H71-pHC (Suo et al 2008). The auto-agglutination of H683-pC2 is unlikely to result from flagella, since no differences were observed for flagella production using either Western blot or AFM imaging (figures 1(c) and (d)). As the CFA/I expression level of H683-pC is fairly low, we utilized a bacterial CFU 20-fold higher than that of -pC2 to emphasize the protein band in Western blotting analysis. Despite the low concentration of H683-pC2 compared to -pC, -pC2 still generated a band ~1.5-fold denser than that of -pC (figure 1(c)), suggesting that -pC2 yields ~30 times more CFA/I fimbriae than -pC. The results from AFM imaging confirmed the Western blot assay, as H683-pC2 produced a massive amount of fimbriae that densely covered the cell surface (figure 1(d)). The fimbriae were observed to decorate the bacterial surface only sparsely for H683-pC, while no fimbriae were imaged for -pY (figure 1(d)). Our previous study showed that fusion promoter PtetA~PphoP is stronger than PtetA (Yang et al 2007). Hence, when PtetA~PphoP was applied to cfaABCE, it elevated the expression of CFA/I fimbriae as compared to PtetA. The massive CFA/I fimbriae may be the cause of the subsequent auto-agglutination of H683-pC2. The auto-agglutination phenomenon was further observed using a light microscope (figure 1(e)). Even in the cell culture supernatant, a majority of H683-pC2 cells remained in agglutinated form, with only a minority of cells maintained in planktonic form. For H683-pC, a few cells remained in the agglutinated state while many cells resided in the planktonic pattern. As for H683-pY, all cells resided in the planktonic pattern (figure 1(e)). A previous study showed that purified CFA/I fimbrial molecules tend to aggregate to form bundles (Evans et al 1979), so the auto-agglutination of H683-pC2 and -pC is very likely due to the fimbrial adhesive feature.
3.2. Overexpression of CFA/I impedes bacterial swimming
Flagella contribute to bacterial adhesion and motility, while fimbriae are involved in attachment (Finlay and Falkow 1997, Zolghadr et al 2010). To investigate whether overexpression of CFA/I fimbriae interferes with flagella-driven bacterial motion, a swimming experiment was conducted. The use of semisolid agar to analyze bacterial motility can be traced back to approximately one century (Tittsler and Sandholzer 1936). Within semisolid agar, motility is manifested macroscopically by a diffusion zone of bacterial growth spreading from the inoculation spots (Tittsler and Sandholzer 1936). Therefore, in addition to the microscopic swimming ability of individual bacteria, the macroscopic swimming ability of massive bacteria can also be determined using the conventional method, which measures the bacterial capability of movement within semisolid agar.
Strains H683-pC2, -pC and -pY were used to inoculate semisolid LB agar (0.35%) surfaces. At 8 h post-inoculation, although a majority of the inoculants had started swimming and formed motility halos (figure 2(a-i, -iii, -v)), inoculants that had been unable to start swimming were observed for all three tested strains (figure 2(a-ii, iv, vi). To facilitate description, we used ‘migrating inoculants’ to describe the inoculants able to migrate into semisolid agar to start swimming. Swimming capability was determined by two parameters, the percentage of the migrating inoculants and the diameters of the motility halos the migrating inoculants formed. To determine whether the swimming capabilities were inherent to the strains tested, this experiment was performed six times. H683-pC2 displayed the lowest percentage of migrating inoculants (70.8%), significantly lower than those of -pC (99.2%) and -pY (97.5%) (P < 0.05; P < 0.05) (figure 2(b-i)). H683-pC2 also yielded the smallest motility halo size (8.5 mm), very significantly smaller than those of -pC (10.4 mm) and -pY (13.7 mm) (P < 0.001; P < 0.001) (figure 2(b-ii)). It is noteworthy that the motility halo size of H683-pC was smaller than that of -pY (P < 0.001) but larger than that of -pC2 (P < 0.05) (figure 2(b-ii)). These results suggest that overexpression of CFA/I prevents Salmonella from migrating into semisolid agar and that the swimming ability of those Salmonella with overexpressed CFA/I which did migrate into agar is decreased. The results also suggest a negative correlation between the level of CFA/I expression and the bacterial swimming ability: the more fimbriae the cell carries, the less swimming capability the bacterium retains.
Figure 2.
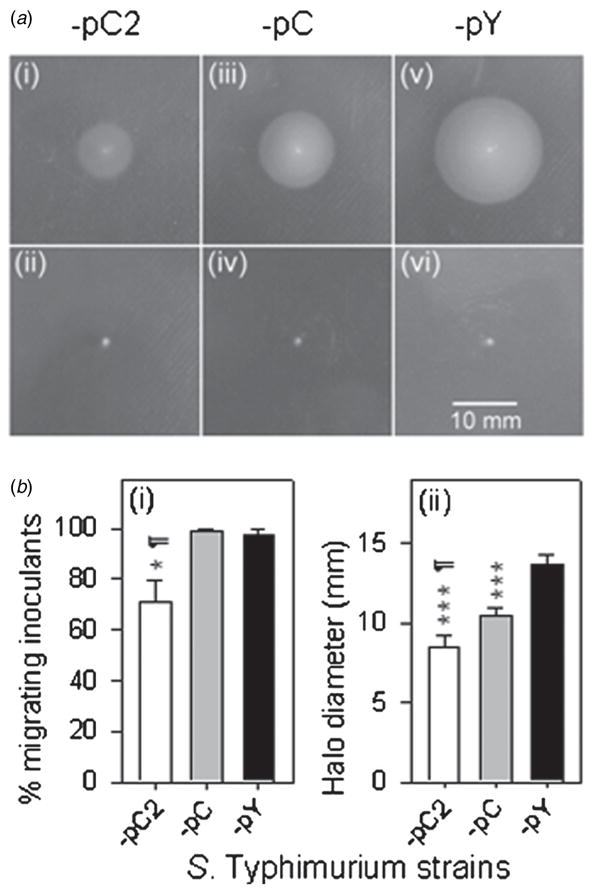
Effects of overexpression of CFA/I fimbriae on bacterial swimming. (a) Not all inoculants were able to start swimming in 0.35% agar at 8 h post-inoculation. Images of H683-pC2 (i), -pC (iii) and -pY (v) inoculants which started swimming versus those unable to start swimming, (ii), (iv) and (vi), respectively. The scale bar, 10 mm, is applicable to all the images (i–vi). (b) CFA/I fimbriae counteract bacterial swimming capability. (i) The migrating inoculants rates were determined for H683-pC2, -pC and -pY, and the statistical differences were evaluated and are indicated as * P < 0.05 for -pC2 versus -pY and ¶ P < 0.05 for -pC2 versus -pC. Values are the mean ± SEM (n = 6). (ii) The motility halo diameters at 8 h post-inoculation were recorded, and the statistical differences were evaluated and are indicated as *** P < 0.001 for H683-pC2 or -pC versus -pY, and ¶ P < 0.05 for -pC2 versus -pC. Values are the mean ± SEM (n = 3).
The decreased capability of H683-pC2 to enter semisolid agar can be understood based on the following. Overexpression of CFA/I caused the aggregation of bacterial cells, and more severe aggregation was observed for more fimbriated strains. This is evidenced by the auto-agglutination of H683-pC2 grown in a liquid medium (figure 1(b)) and the microscopic observations (figure 1(e)). In comparison to non-aggregated cells, the aggregates had more difficulty entering semisolid agar. Cells contained in aggregates must overcome the agglutination forces to enter semisolid agar. Strain H683-pC2 showed the lowest percentage of migrating inoculants because it had fewer non-aggregated cells than the other two strains. Although smaller aggregates were also identified for H683-pC (figure 1(e-ii)), a large number of non-aggregated cells was observed for this strain, which then displayed a migrating inoculants percentage close to 100%, similar to that of -pY (figure 2(b-i)).
3.3. Factors that contribute to impeded bacterial swimming capability
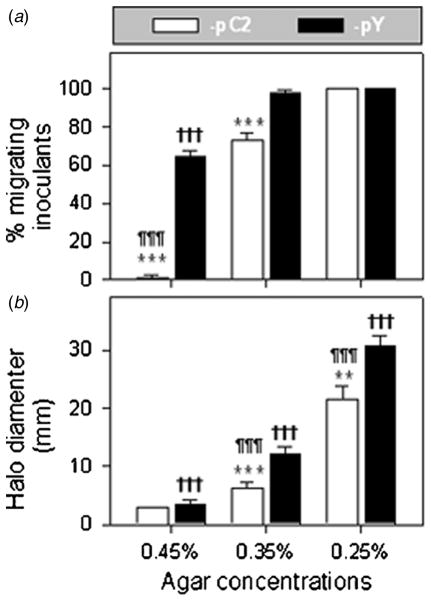
In general, two aspects determine how fast bacteria swim: environmental conditions and the innate swimming ability of the bacteria. The impact of environmental factors, such as chemotaxis (Pandey and Jain 2002), pH (Celli et al 2009), temperature (Magariyama et al 1995) and viscosity (Magariyama and Kudo 2002) on bacterial swimming have been addressed. The impact of agar concentration on bacterial swimming was analyzed in this study. Agar with a lower concentration has a larger pore size (Maaloum et al 1998). Since bacterial cells enter the semisolid agar through these pores (Ames et al 1996), the pore size largely determines whether the cells are able to enter the agar. Indeed, even for strain H683-pY, more than one-third of the cells were unable to enter the agar when the agar concentration reached 0.45% (figure 3(a)). The AFM imaging of 0.35% agar indicated an average pore size around 1 μm (figure S1 (available from stacks.iop.org/PhysBio/9/036005/mmedia)). H683-pC2 cells were surrounded by a dense layer of fimbriae (figure 1(d)), which rendered the bacterial cell surface rougher and gave the cell a size larger than that of -pY. We speculate that this effect introduces additional resistance to H683-pC2 cells when they migrate into agar micropores. This is supported by the observation that when the agar concentration was decreased to 0.25%, all the H683-pC2 inoculants migrated into the agar (figure 3(a)), because a pore size of 0.25% agar is sufficient to accommodate the rough H683-pC2 cells. After successful agar migration, as the hydrodynamic resistances encountered by these constructs are in the order of H683-pC2 > -pC > -pY, unsurprisingly, their swimming abilities are in the order of -pC2 < -pC < -pY (figure 3(b)). Moreover, the fimbriae may interact strongly with the gel environment, causing entanglement and stickiness, which may also be a reason that the fimbriated cells possess a reduced swimming capability. Thus, in addition to the previously observed external environmental conditions, agar concentrations may constitute an external factor impacting the densely fimbriated H683-pC2.
Figure 3.
CFA/I-inhibited bacterial swimming depends on agar concentration. (a) The migrating inoculant rates of the fimbriated bacteria were elevated with the decrease in agar concentration. The migrating inoculants rates of H683-pC2 and -pY were statistically compared for 0.45%, 0.35% and 0.25% agar, and the differences are indicated as *** P < 0.001 for H683-pC2 versus -pY, ¶¶¶ P < 0.001 for H683-pC2 at 0.35% versus 0.45% agar, and ††† P < 0.001 for H683-pY at 0.35% versus 0.45% agar. Values are the mean ± SEM (n = 6). (b) The motility halo sizes of the fimbriated bacteria increase with the decrease in agar concentration. H683-pC2 was compared with -pY for halo diameters at 8 h post-inoculation, and the statistical differences between H683-pC2 and -pY were evaluated and are indicated as *** P < 0.001 at each of the three agar concentrations; ¶¶¶ P < 0.001 for -pC2 at 0.35% versus 0.45% agar, and 0.25% versus 0.35% agar; and ††† P < 0.001 for -pY at 0.35% versus 0.45% agar, and 0.25% versus 0.35% agar. Values are the mean ± SEM (n = 3).
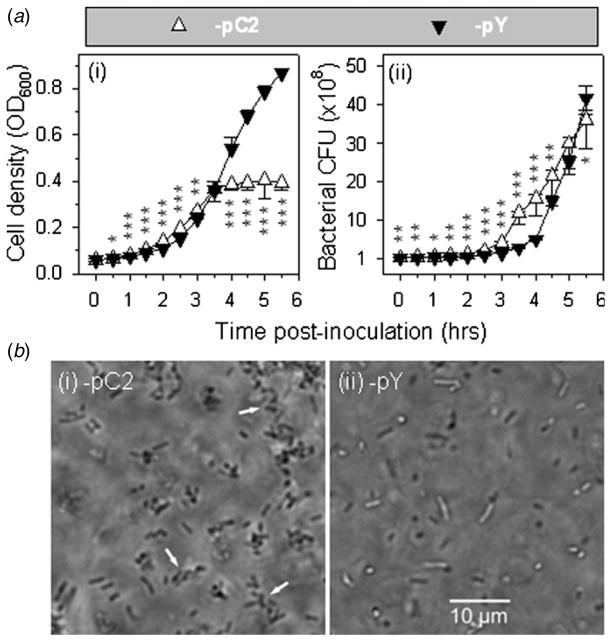
Three factors innate to the bacteria are anticipated to contribute to the small motility halo size of H683-pC2: rate of cell division, bacterial aggregation and the swimming capability of individual cells. Halo size is determined by bacterial swimming capability and cell division rate. For two strains that have identical swimming capabilities, the one with the higher division rate is expected to show the larger motility halo size. H683-pC2 and -pY were grown in the liquid LB medium with the initial inoculation cell density of 0.05 at OD600. Between 0.5 and 3.0 h post-inoculation, the OD600 values of H683-pC2 were significantly higher than those of -pY (figure 4(a-i)). After 3.5 h, no difference in OD600 was observed between H683-pC2 and -pY. However, between 4.0 and 5.5 h, the OD600 values of H683-pC2 were significantly lower than those of -pY. Actually, between 3.5 and 5.5 h post-inoculation, the OD600 values of H683-pC2 remained nearly unchanged (its growth curve has a sharp turnover from a rapid log growth phase to a slow stationary growth phase) (figure 4(a-i)). The reason why the OD600 values of H683-pC2 were higher than those of -pY in the lag and early log growth phases is not readily evident, but its rapid shift from the log growth phase to the ‘stationary growth phase’ may be due to auto-agglutination. Since the bacterial pellets of auto-agglutinated H683-pC2 cells grew larger with time, they presumably became too heavy to suspend in the medium and finally precipitated from the medium. This left the culture supernatant of H683-pC2 clearer than that of -pY, which resulted in the light absorption of -pC2 being weaker than that of -pY at 600 nm. Since the growth rate of H683-pC2 could not be measured accurately, we analyzed the bacterial CFU every half hour by taking cell culture samples out of the BioScreen C and rigorously vortexing the samples for 1 min at the maximum speed since microscopic observation showed that 1 min of vortexing is sufficient for completely separating the agglutinated cells into individual cells. Next, the samples were serially diluted and plated onto LB agar for CFU enumeration after overnight growth at 37 °C. H683-pC2 had significantly more CFU than -pY at the time of the inoculation (t = 0 h) (P < 0.01) (figure 4(a-ii)). The reason for this difference is not currently clear, but the higher initial CFU of H683-pC2 led to a ‘faster’ growth rate than that of -pY in terms of both OD600 (figure 4(a-i)) and CFU (figure 4(a-ii)) in the lag and early log phases. At 5 h post-inoculation, no difference in CFU was observed between H683-pC2 and -pY. At 5.5 h post-inoculation, the growth rate of H683-pC2 was surpassed by that of -pY (figure 4(a-ii)). Although the CFU values of the H683-pC2 and -pY strains accurately index their cell growth rates, these data still cannot be used to clearly address which strain grows faster because of the different initial inoculants and the abnormal CFU profile of H683-pC2. Therefore, we calculated the maximum growth rates of these two strains using the minimum doubling time of the bacterial population. Through calculation, we found that the rapidest growth for both strains occurred between 3.0 and 3.5 h post inoculation. Correspondingly, the minimum doubling time of the populations of H683-pC2 and -pY are 20.2 ± 2.5 versus 18.9 ± 2.6 min, respectively, and the difference did not reach a significant level (P > 0.05), suggesting these two strains show no differences in growth rate. Therefore, we may infer that growth rate does not contribute to the impaired swimming capability of H683-pC2. Possible reasons for the impaired swimming capability of H683-pC2 are bacterial aggregation and the slow swimming capability of individual cells.
Figure 4.
Evaluation of growth rate and observation of in situ auto-agglutination of H683-pC2. (a) Assay of the growth rates of H683-pC2 and -pY via OD600. The statistical differences in growth rate between H683-pC2(−) and -pY were evaluated and are indicated as * P < 0.05, ** P < 0.01 and *** P < 0.001 at each time point. Values are the mean ± SEM (n = 3). (b) Assay of the growth rates of H683-pC2 and -pY via CFU. The statistical differences in CFU between H683-pC2(−) and -pY were evaluated and are indicated as * P < 0.05, ** P < 0.01 and *** P < 0.001 at each time point. Values are the mean ± SEM (n = 3). (c) Observation of H683-pC2 auto-agglutination inside agar. The motility halos formed within 0.35% agar by strains H683-pC2 (i) and -pY (ii) were observed for individual cell behavior using an optical microscope in phase contrast mode. The arrows indicate cell auto-agglutination. No aggregates were detected for control H683-pY.
CFA/I-mediated aggregation may adversely affect bacterial swimming capability in two ways. First, the large volume of the aggregates may bring additional hydrodynamic resistance when they swim collectively. Second, the overall propelling force of the aggregates is a combination of the flagella-driven force of all the individual cells, which is much smaller than their arithmetic sum because the cells inside the aggregates do not orient themselves in the same direction. Thus, the propelling forces of the aggregates in different directions counteract each other. Our results show that the H683-pC2 cells did form aggregates both in the liquid medium and in semisolid agar (figures 1(e-i) and 4(b-i)), and this aggregation may also have contributed to the small motility halo size of H683-pC2, since the aggregates stopped the cells from swimming toward the edges of the halos. No aggregates were discerned for H683-pY in either liquid medium or semisolid agar (figures 1(e-iii) and 4(b-ii)), and it maintained normal swimming in these media.
We also observed different swimming abilities for the non-aggregated cells of all three strains, as shown in the video clips in the supporting materials (videos S1–S4 (available from stacks.iop.org/PhysBio/9/036005/mmedia)). Although we are not able to give a quantitative description of the swimming speed at this stage, a significant difference can be seen in the video clips: in the liquid LB medium the H683-pY cells swam the most rapidly while most of the -pC2 cells moved slowly. The detailed mechanism of the impeded swimming capability of H683-pC2 remains unclear, but potential reasons include increased hydrodynamic resistance associated with the fimbriae and the tangling of fimbriae with flagella. To examine this hypothesis, we removed the CFA/I fimbrial fibers from H683-pC2 and investigated whether the non-fimbriated cells recovered their swimming capability compared to the control -pY, as follows.
3.4. Removal of CFA/I fimbrial shaft restores bacterial swimming capability
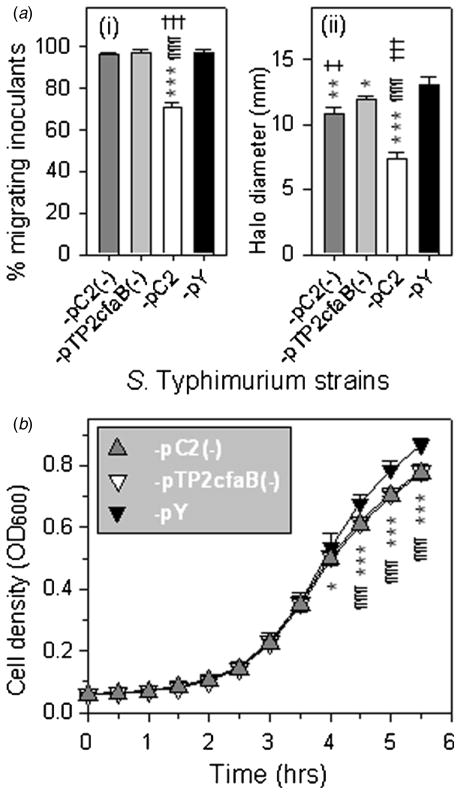
To further investigate the association between the impeded swimming of H683-pC2 and the CFA/I fimbrial fibers, we constructed a strain that does not express the shaft portion of CFA/I fimbriae, naming it H683-pC2(−). Among the four genes harbored in the cfa/I operon, cfaB and cfaE encode the fimbrial shaft (Baker et al 2009), so cfaB was in-frame deleted from pC2 to obtain pTP2cfaB(−). Then, pC2(−) was generated by complete removal of cfaE from pTP2cfaB(−). Similar to H683-pY, -pC2(−) and -pTP2cfaB(−) did not auto-agglutinate. H683-pC2(−) and -pTP2cfaB(−) were evaluated for swimming capability in 0.35% agar, with -pC2 and -pY as controls. Similar to what we observed in figure 2(a), two swimming phenotypes were observed 8 h post-inoculation: the inoculants able to enter agar to swim and those unable to migrate into agar. The percentages of migrating inoculants of H683-pC2(−) and -pTP2cfaB(−) showed no differences from that of -pY but were significantly higher than that of -pC2 (figure 5(a-i)), suggesting that fimbrial fibers block bacterial agar migration. The motility halo sizes of H683-pC2(−) and -pTP2cfaB(−) were between those of -pC2 and -pY (figure 5(a-ii)), suggesting that the fimbrial fibers adversely affect swimming ability. It is probable that these projected, rigid fibers confer extra hydrodynamic resistance to the swimming cells, thus slowing them down. On one hand, this supports our hypothesis that fimbrial fibers affect bacterial agar migration and swimming ability. On the other hand, it may imply that other unidentified factors interfere with bacterial swimming. Of note is that both H683-pC2(−) and -pTP2cfaB(−) do not possess fimbrial fibers, but -pTP2cfaB(−) gave a slightly larger average swimming halo size than that of -pC2(−) (P < 0.05) (figure 5(a-ii)), implying CfaE is subtly involved in the bacterial motility in some way irrelevant to the fimbrial fibers. To test whether cell division contributed to the low swimming ability of H683-pC2(−), we compared its growth rate with -pTP2cfaB(−) and -pY. The result showed that the growth rates of H683-pC2(−) and -pTP2cfaB(−) showed no difference, but significantly slower than that of -pY at 4 and 4.5 h post-inoculation (P < 0.05 or 0.001) (figure 5(b)). Although the mechanisms of the retarded growth of H683-pC2(−) and -pTP2cfaB(−) require further investigation, this result supports our assumption that the slow swimming capabilities of H683-pC2(−) and -pTP2cfaB(−) correlates to its slow growth rate. Overall, these experiments indicate that CFA/I fimbrial fibers fundamentally affect bacterial motility by impeding swimming.
Figure 5.
Impact of the CFA/I fimbrial fibers on bacterial swimming. (a) CFA/I fimbrial fibers hinder bacterial swimming. (i) The migration inoculants rates at 8 h post-inoculation were determined for H683-pC2(−), -pTP2cfaB(−), -pC2 and -pY, and the statistical differences were evaluated and are indicated as *** P < 0.001 for H683-pC2 versus -pY, ¶¶¶ P < 0.001 for -pC2 versus -pC2(−) and ††† P < 0.001 for H683 -pC2 versus -pTP2cfaB(−). Values are the mean ± SEM (n = 6). (ii) The motility halo sizes at 8 h post-inoculation were recorded, and the statistical differences were evaluated and are indicated as *** P < 0.001 for H683-pC2 versus -pY, ** P < 0.01 for H683-pC2(−) versus -pY, * P < 0.05 for H683-pTP2cfaB(−) versus -pY, ¶¶¶ P < 0.001 for H683-pC2 versus -pC2(−), ††† P < 0.001 for H683-pC2 versus -pTP2cfaB(−) and ‡ P < 0.05 for -pC2(−) versus -pTP2cfaB(−). Values are the mean ± SEM (n = 3). (b) Growth rates of the non-fimbriated strains versus control. The OD600 was recorded and the statistical differences were evaluated and are indicated as * P < 0.05, ** P < 0.01 and *** P < 0.001 for H683-pC2(−) versus -pY, ¶ P < 0.05, ¶¶ P < 0.01 and ¶¶¶ P < 0.001 for -pTP2cfaB(−) versus -pY. Values are the mean ± SEM (n = 3).
This study has focused on the population-scale behavior of fimbriated Salmonella. Future studies may focus on the behavior of single cells, which may help us understand how fimbriae affect the dynamics of individual bacteria. This may be accomplished using the three-dimensional (3D) defocused particle tracking (DPT) method, which tracks multiple swimming cells in 3D at a single-cell resolution (Wu et al 2006). This technique will allow us to measure how bacterial motor power and motility are affected by overexpressed fimbriae. It may also enable us to analyze the fimbrial impacts on the cell diffusion coefficient, collective dynamics and quorum sensing behavior. We predict that these DPT experiments will deepen our understanding of the impact of the fimbriae on swimming behavior, as well as bacterial behavior at both the collective and individual levels. Knowledge acquired in these areas may assist in the development of highly efficient biosensors that immobilize fimbriated pathogenic bacteria to expedite the process of clinical diagnosis (Suo et al 2012).
4. Conclusion
Our work indicates that overexpressed ETEC CFA/I adversely affects bacterial swimming through two mechanisms. First, CFA/I causes cells to form aggregates, which is deleterious to their swimming capability. Second, CFA/I fimbrial fibers impede the swimming of non-aggregated cells. These two effects correlate negatively to the CFA/I expression levels, i.e. the more CFA/I fimbriae are expressed, the less swimming capability the bacteria have.
Supplementary Material
Acknowledgments
We appreciate Dr Jerod Skyberg for his critical reviews of this manuscript. This study was supported by National Institutes of Health Grant R21 AI080960 and P20 RR020185, an equipment grant from the M J Murdock Charitable Trust, and the Montana State University Agricultural Experiment Station. It is also supported in part by Office of Navy Research Award N00014-10-1-0946. The authors declare that they have no conflict of interest.
Nomenclature
- ETEC
enterotoxigenic E. coli
- CFA/I
colonization factor antigen I
- LB
lysogeny broth
- OD600
optical density at 600 nm
- AFM
atomic force microscope
- PCR
polymerase chain reaction
- log
logarithmic
- CFU
colony forming units
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PtetA
tetracycline resistance gene promoter
- 3D
three-dimensional
- DPT
defocused particle tracking
Footnotes
S Online supplementary data available from stacks.iop.org/PhysBio/9/036005/mmedia
References
- Alves AM, Lasaro MO, Almeida DF, Ferreira LC. DNA immunization against the CFA/I fimbriae of enterotoxigenic Escherichia coli (ETEC) Vaccine. 2000;19:788–95. doi: 10.1016/s0264-410x(00)00244-9. [DOI] [PubMed] [Google Scholar]
- Ames P, Yu YA, Parkinson JS. Methylation segments are not required for chemotactic signalling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol Microbiol. 1996;19:737–46. doi: 10.1046/j.1365-2958.1996.408930.x. [DOI] [PubMed] [Google Scholar]
- Amiel E, Lovewell RR, O’Toole GA, Hogan DA, Berwin B. Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression. Infect Immun. 2010;78:2937–45. doi: 10.1128/IAI.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascón MA, Hone DM, Walters N, Pascual DW. Oral immunization with a Salmonella typhimurium vaccine vector expressing recombinant enterotoxigenic Escherichia coli K99 fimbriae elicits elevated antibody titers for protective immunity. Infect Immun. 1998;66:5470–6. doi: 10.1128/iai.66.11.5470-5476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani FK, Johnson DE, Robbins D, Mobley HL. Proteus mirabilis flagella and MR/P fimbriae: isolation, purification, N-terminal analysis, and serum antibody response following experimental urinary tract infection. Infect Immun. 1991;59:3574–80. doi: 10.1128/iai.59.10.3574-3580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KK, Levine MM, Morison J, Phillips A, Barry EM. CfaE tip mutations in ETECCFA/I fimbriae define critical human intestinal binding sites. Cell Microbiol. 2009;11:742–54. doi: 10.1111/j.1462-5822.2009.01287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts J, Finlay BB. Identification of Salmonella typhimurium invasiveness loci. Can J Microbiol. 1992;38:852–7. doi: 10.1139/m92-138. [DOI] [PubMed] [Google Scholar]
- Celli JP, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci USA. 2009;106:14321–6. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein EA, Chapman MR. Polymerizing the fibre between bacteria and host cells: the biogenesis of functional amyloid fibres. Cell Microbiol. 2008;10:1413–20. doi: 10.1111/j.1462-5822.2008.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Evans DJ, Jr, Clegg S, Pauley JA. Purification and characterization of the CFA/I antigen of enterotoxigenicEscherichia coli. Infect Immun. 1979;25:738–48. doi: 10.1128/iai.25.2.738-748.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Silver RP, Evans DJ, Jr, Chase DG, Gorbach SL. Plasmid-controlled colonization factor associated with virulence in Esherichia coli enterotoxigenic for humans. Infect Immun. 1975;12:656–67. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–69. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE, Nakayama K, Curtiss R., 3rd Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene. 1990;94:29–35. doi: 10.1016/0378-1119(90)90464-3. [DOI] [PubMed] [Google Scholar]
- Gibiansky ML, Conrad JC, Jin F, Gordon VD, Motto DA, Mathewson MA, Stopka WG, Zelasko DC, Shrout JD, Wong GC. Bacteria use type IV pili to walk upright and detach from surfaces. Science. 2010;330:197. doi: 10.1126/science.1194238. [DOI] [PubMed] [Google Scholar]
- Guttenplan SB, Blair KM, Kearns DB. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010;6:e1001243. doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rasko DA, Lockatell CV, Johnson DE, Mobley HL. Repression of bacterial motility by a novel fimbrial gene product. Eur Mol Biol Organ J. 2001;20:4854–62. doi: 10.1093/emboj/20.17.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Poole S, Rasulova F, McVeigh AL, Savarino SJ, Xia D. A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli. J Biol Chem. 2007;282:23970–80. doi: 10.1074/jbc.M700921200. [DOI] [PubMed] [Google Scholar]
- Lindhout T, Lau PC, Brewer D, Lam JS. Truncation in the core oligosaccharide of lipopolysaccharide affects flagella-mediated motility in Pseudomonas aeruginosa PAO1 via modulation of cell surface attachment. Microbiology. 2009;155:3449–60. doi: 10.1099/mic.0.030510-0. [DOI] [PubMed] [Google Scholar]
- Maaloum M, Pernodet N, Tinland B. Agarose gel structure using atomic force microscopy: gel concentration and ionic strength effects. Electrophoresis. 1998;19:1606–10. doi: 10.1002/elps.1150191015. [DOI] [PubMed] [Google Scholar]
- Magariyama Y, Kudo S. A mathematical explanation of an increase in bacterial swimming speed with viscosity in linear-polymer solutions. Biophys J. 2002;83:733–9. doi: 10.1016/S0006-3495(02)75204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magariyama Y, Sugiyama S, Muramoto K, Kawagishi I, Imae Y, Kudo S. Simultaneous measurement of bacterial flagellar rotation rate and swimming speed. Biophys J. 1995;69:2154–62. doi: 10.1016/S0006-3495(95)80089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Yao J, Allen C. A motN mutant of Ralstonia solanacearum is hypermotile and has reduced virulence. J Bacteriol. 2011;193:2477–86. doi: 10.1128/JB.01360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Repáraz J, Rynda A, Ascón MA, Yang X, Kochetkova I, Riccardi C, Callis G, Trunkle T, Pascual DW. IL-13 production by regulatory T cells protects against experimental autoimmune encephalomyelitis independently of autoantigen. J Immunol. 2008;181:954–68. doi: 10.4049/jimmunol.181.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G, Jain RK. Bacterial chemotaxis toward environmental pollutants: role in bioremediation. Appl Environ Microbiol. 2002;68:5789–95. doi: 10.1128/AEM.68.12.5789-5795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulino ET, Bullitt E, Hultgren SJ. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc Natl Acad Sci USA. 2000;97:9240–5. doi: 10.1073/pnas.160070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelkoul PH, Willshaw GA, McConnell MM, Smith HR, Hamers AM, van der Zeijst BA, Gaastra W. Expression of CFA/I fimbriae is positively regulated. Microb Pathog. 1990;8:91–9. doi: 10.1016/0882-4010(90)90073-y. [DOI] [PubMed] [Google Scholar]
- Stafford GP, Hughes C. Salmonella typhimurium flhE, a conserved flagellar regulon gene required for swarming. Microbiology. 2007;153:541–7. doi: 10.1099/mic.0.2006/002576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo Z, Avci R, Yang X, Pascual DW. Efficient immobilization and patterning of live bacterial cells. Langmuir. 2008;24:4161–7. doi: 10.1021/la7038653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo Z, Yang X, Avci R, Deliorman M, Rugheimer P, Pascual DW, Idzerda Y. Antibody selection for immobilizing living bacteria. Anal Chem. 2009;81:7571–8. doi: 10.1021/ac9014484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo Z, Yang X, Avci R, Kellerman L, Pascual DW, Fries M, Steele A. HEPES-stabilized encapsulation of Salmonella typhimurium. Langmuir. 2007;23:1365–74. doi: 10.1021/la0621721. [DOI] [PubMed] [Google Scholar]
- Suo Z, Yang X, Deliorman M, Cao L, Avci R. Capture efficiency of Escherichia coli in fimbriae-mediated immunoimmobilization. Langmuir. 2012;28:1351–9. doi: 10.1021/la203348j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareen AM, Dashti JI, Zautner AE, Gross U, Lugert R. Sulphite: cytochrome c oxidoreductase deficiency in Campylobacter jejuni reduces motility, host cell adherence and invasion. Microbiology. 2011;157:1776–85. doi: 10.1099/mic.0.045567-0. [DOI] [PubMed] [Google Scholar]
- Tittsler RP, Sandholzer LA. The use of semi-solid agar for the detection of bacterial motility. J Bacteriol. 1936;31:575–80. doi: 10.1128/jb.31.6.575-580.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias J, Holmgren J, Hellman M, Nygren E, Lebens M, Svennerholm AM. Over-expression of major colonization factors of enterotoxigenic Escherichia coli, alone or together, on non-toxigenic E. coli bacteria. Vaccine. 2010;28:6977–84. doi: 10.1016/j.vaccine.2010.08.047. [DOI] [PubMed] [Google Scholar]
- Tobias J, Lebens M, Bolin I, Wiklund G, Svennerholm AM. Construction of non-toxic Escherichia coli and Vibrio cholerae strains expressing high and immunogenic levels of enterotoxigenic E. coli colonization factor I fimbriae. Vaccine. 2008;26:743–52. doi: 10.1016/j.vaccine.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Willshaw GA, Smith HR, McConnell MM, Rowe B. Expression of cloned plasmid regions encoding colonization factor antigen I (CFA/I) in Escherichia coli. Plasmid. 1985;13:8–16. doi: 10.1016/0147-619x(85)90051-4. [DOI] [PubMed] [Google Scholar]
- Wu MM, Roberts JW, Kim S, Koch DL, DeLisa MP. Collective bacterial dynamics revealed using a three-dimensional population-scale defocused particle tracking technique. Appl Environ Microbiol. 2006;72:4987–94. doi: 10.1128/AEM.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Pascual DW, VanCott JL, McGhee JR, Maneval DR, Jr, Levine MM, Hone DM. Immune responses to novel Escherichia coli and Salmonella typhimurium vectors that express colonization factor antigen I (CFA/I) of enterotoxigenic E. coli in the absence of the CFA/I positive regulator. cfaR. Infect Immun. 1995;63:4933–8. doi: 10.1128/iai.63.12.4933-4938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hinnebusch BJ, Trunkle T, Bosio CM, Suo Z, Tighe M, Harmsen A, Becker T, Crist K, Walters N, Avci R, Pascual DW. Oral vaccination with Salmonella simultaneously expressing Yersinia pestis F1 and V antigens protects against bubonic and pneumonic plague. J Immunol. 2007;178:1059–67. doi: 10.4049/jimmunol.178.2.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, et al. Expression of Escherichia coli virulence usher protein attenuates wild-type. Salmonella Virulence. 2012;3:29–41. doi: 10.4161/viru.3.1.18447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Thornburg T, Holderness K, Suo Z, Cao L, Lim T, Avci R, Pascual DW. Serum antibodies protect against intraperitoneal challenge with enterotoxigenic Escherichia coli. J Biomed Biotechnol. 2011;2011:632396. doi: 10.1155/2011/632396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolghadr B, Klingl A, Koerdt A, Driessen AJ, Rachel R, Albers SV. Appendage-mediated surface adherence of Sulfolobus solfataricus. J Bacteriol. 2010;192:104–10. doi: 10.1128/JB.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.